|
1
|
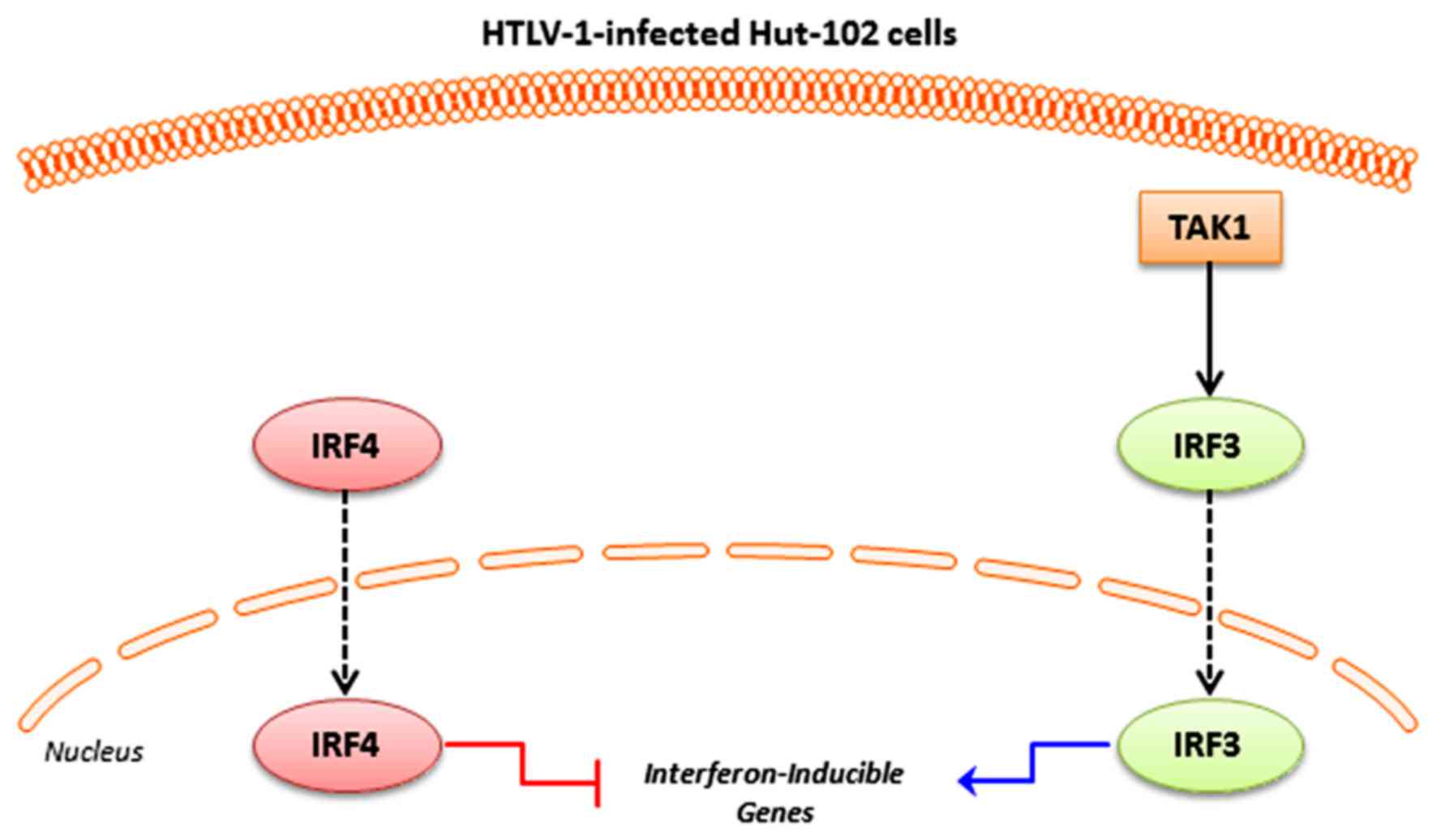
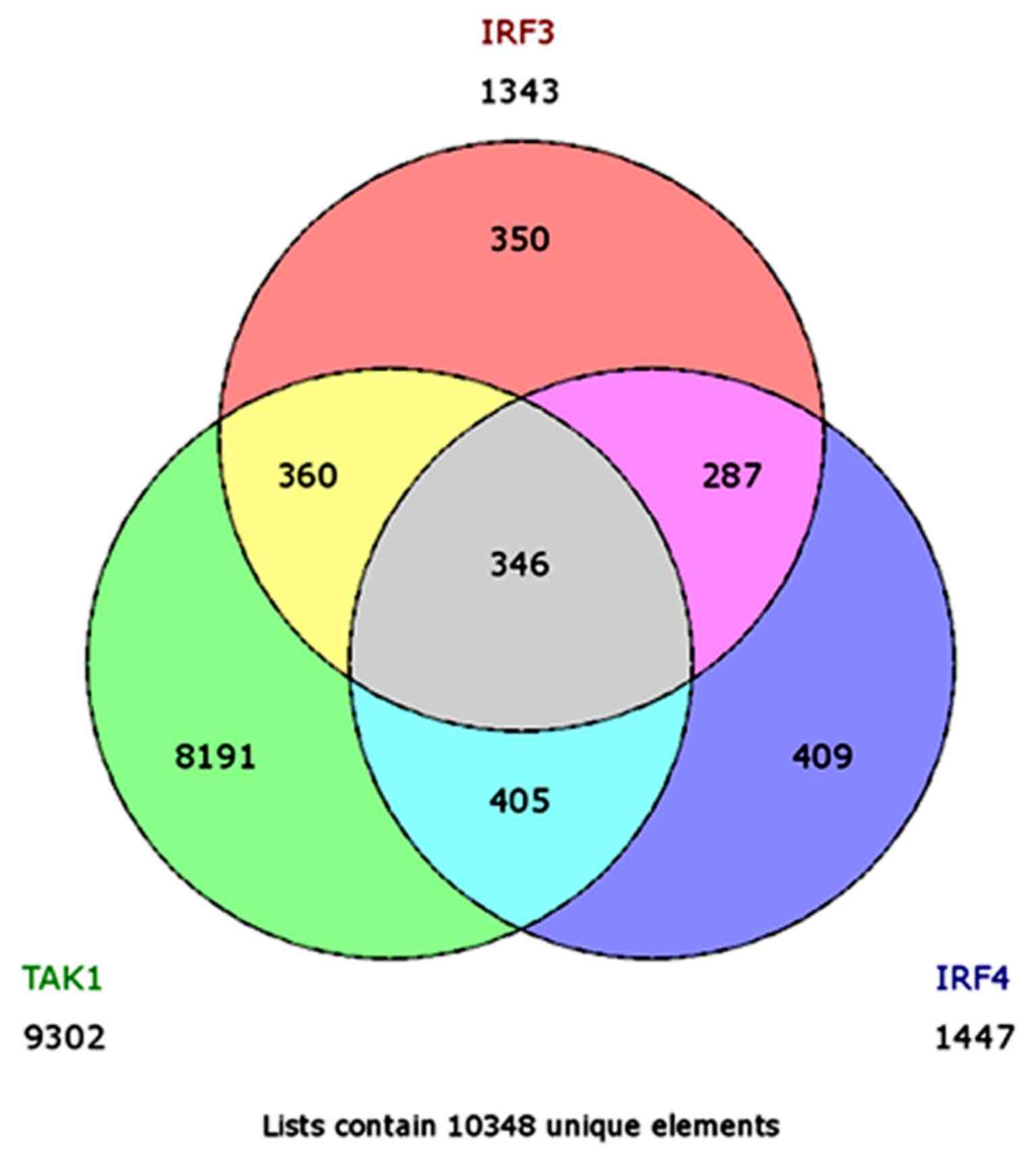
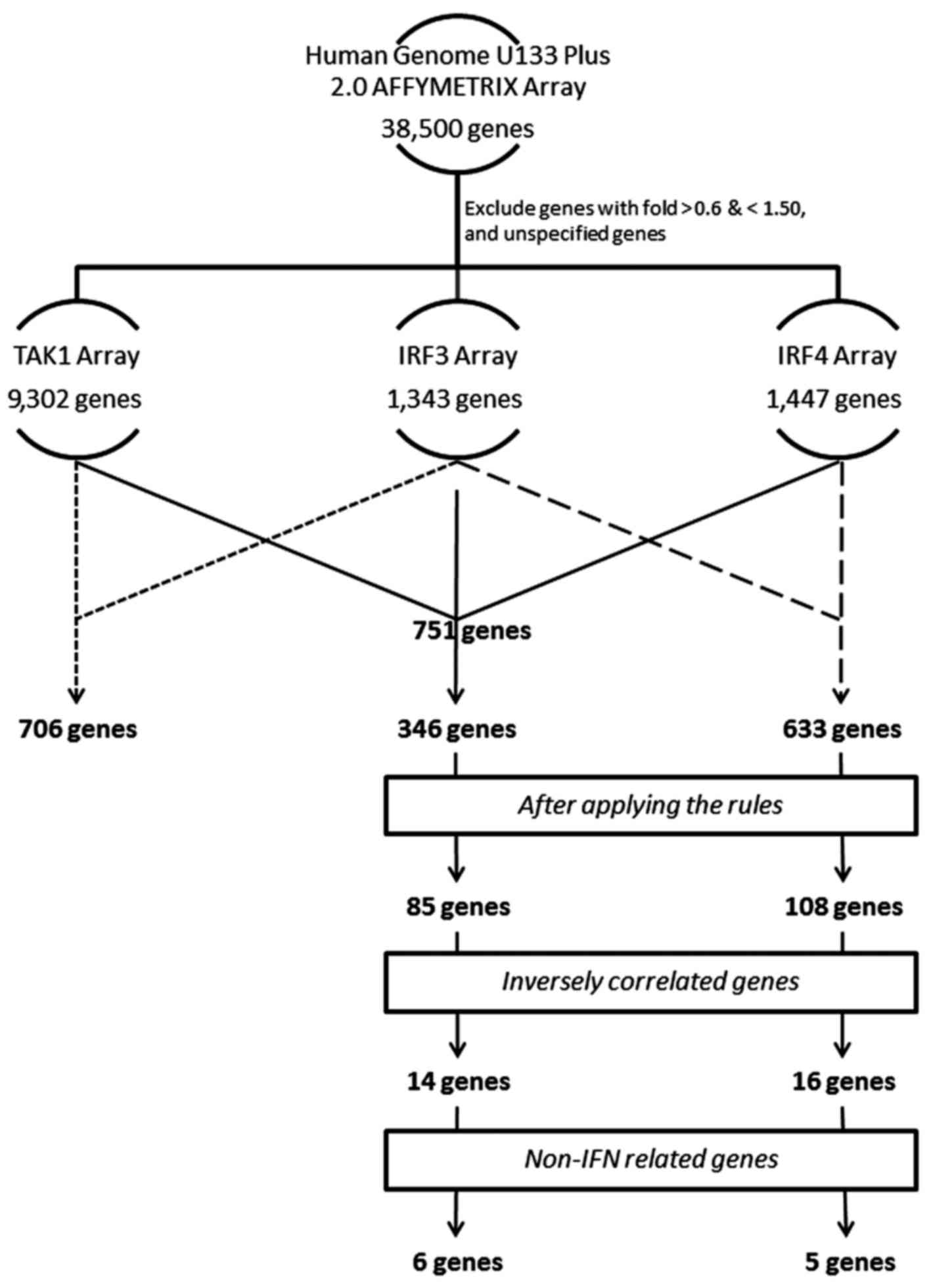
Coffin JM: The discovery of HTLV-1, the
first pathogenic human retrovirus. Proc Natl Acad Sci USA.
112:15525–15529. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn
PA, Minna JD and Gallo RC: Detection and isolation of type C
retrovirus particles from fresh and cultured lymphocytes of a
patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA.
77:7415–7419. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Refaat A, Zhou Y, Suzuki S, Takasaki I,
Koizumi K, Yamaoka S, Tabuchi Y, Saiki I and Sakurai H: Distinct
roles of transforming growth factor-beta-activated kinase 1
(TAK1)-c-Rel and interferon regulatory factor 4 (IRF4) pathways in
human T cell lymphotropic virus 1-transformed T helper 17 cells
producing interleukin-9. J Biol Chem. 286:21092–21099. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Suzuki S, Zhou Y, Refaat A, Takasaki I,
Koizumi K, Yamaoka S, Tabuchi Y, Saiki I and Sakurai H: Human T
cell lymphotropic virus 1 manipulates interferon regulatory signals
by controlling the TAK1-IRF3 and IRF4 pathways. J Biol Chem.
285:4441–4446. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee JH and Paull TT: Activation and
regulation of ATM kinase activity in response to DNA double-strand
breaks. Oncogene. 26:7741–7748. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang X, Halicka HD and Darzynkiewicz Z:
Detection of histone H2AX phosphorylation on Ser-139 as an
indicator of DNA damage (DNA double-strand breaks). Curr Protoc
Cytom Chapter. 7:Unit 7.27. 2004. View Article : Google Scholar
|
|
7
|
Canman CE, Lim DS, Cimprich KA, Taya Y,
Tamai K, Sakaguchi K, Appella E, Kastan MB and Siliciano JD:
Activation of the ATM kinase by ionizing radiation and
phosphorylation of p53. Science. 281:1677–1679. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ahmed M and Rahman N: ATM and breast
cancer susceptibility. Oncogene. 25:5906–5911. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gadsby DC, Vergani P and Csanády L: The
ABC protein turned chloride channel whose failure causes cystic
fibrosis. Nature. 440:477–483. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jones AM and Helm JM: Emerging treatments
in cystic fibrosis. Drugs. 69:1903–1910. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Than BLN, Linnekamp JF, Starr TK,
Largaespada DA, Rod A, Zhang Y, Bruner V, Abrahante J, Schumann A,
Luczak T, et al: CFTR is a tumor suppressor gene in murine and
human intestinal cancer. Oncogene. 36:35042017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xie C, Jiang XH, Zhang JT, Sun TT, Dong
JD, Sanders AJ, Diao RY, Wang Y, Fok KL, Tsang LL, et al: CFTR
suppresses tumor progression through miR-193b targeting urokinase
plasminogen activator (uPA) in prostate cancer. Oncogene.
32:2282–2291, 2291.e1-7. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu J, Yong M, Li J, Dong X, Yu T, Fu X and
Hu L: High level of CFTR expression is associated with tumor
aggression and knockdown of CFTR suppresses proliferation of
ovarian cancer in vitro and in vivo. Oncol Rep. 33:2227–2234. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang JT, Jiang XH, Xie C, Cheng H, Da
Dong J, Wang Y, Fok KL, Zhang XH, Sun TT, Tsang LL, et al:
Downregulation of CFTR promotes epithelial-to-mesenchymal
transition and is associated with poor prognosis of breast cancer.
Biochim Biophys Acta. 1833:2961–2969. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jhala N, Jhala D, Vickers SM, Eltoum I,
Batra SK, Manne U, Eloubeidi M, Jones JJ and Grizzle WE: Biomarkers
in Diagnosis of pancreatic carcinoma in fine-needle aspirates. Am J
Clin Pathol. 126:572–579. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Srivastava SK, Bhardwaj A, Singh S, Arora
S, Wang B, Grizzle WE and Singh AP: MicroRNA-150 directly targets
MUC4 and suppresses growth and malignant behavior of pancreatic
cancer cells. Carcinogenesis. 32:1832–1839. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mehrotra P, Riley JP, Patel R, Li F, Voss
L and Goenka S: PARP-14 functions as a transcriptional switch for
Stat6-dependent gene activation. J Biol Chem. 286:1767–1776. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Iansante V, Choy PM, Fung SW, Liu Y, Chai
JG, Dyson J, Del Rio A, D'Santos C, Williams R, Chokshi S, et al:
PARP14 promotes the Warburg effect in hepatocellular carcinoma by
inhibiting JNK1-dependent PKM2 phosphorylation and activation. Nat
Commun. 6:78822015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Aberg K, Saetre P, Jareborg N and Jazin E:
Human QKI, a potential regulator of mRNA expression of human
oligodendrocyte-related genes involved in schizophrenia. Proc Natl
Acad Sci USA. 103:7482–7487. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zong FY, Fu X, Wei WJ, Luo YG, Heiner M,
Cao LJ, Fang Z, Fang R, Lu D, Ji H and Hui J: The RNA-binding
protein QKI suppresses cancer-associated aberrant splicing. PLoS
Genet. 10:e10042892014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen AJ, Paik JH, Zhang H, Shukla SA,
Mortensen R, Hu J, Ying H, Hu B, Hurt J, Farny N, et al: STAR
RNA-binding protein Quaking suppresses cancer via stabilization of
specific miRNA. Genes Dev. 26:1459–1472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kume K, Iizumi Y, Shimada M, Ito Y, Kishi
T, Yamaguchi Y and Handa H: Role of N-end rule ubiquitin ligases
UBR1 and UBR2 in regulating the leucine-mTOR signaling pathway.
Genes Cells. 15:339–349. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Taylor PR, Brown GD, Reid DM, Willment JA,
Martinez-Pomares L, Gordon S and Wong SY: The beta-glucan receptor,
dectin-1, is predominantly expressed on the surface of cells of the
monocyte/macrophage and neutrophil lineages. J Immunol.
169:3876–3882. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huysamen C and Brown GD: The fungal
pattern recognition receptor, Dectin-1, and the associated cluster
of C-type lectin-like receptors. FEMS Microbiol Lett. 290:121–128.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Saijo S and Iwakura Y: Dectin-1 and
Dectin-2 in innate immunity against fungi. Int Immunol. 23:467–472.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Activation of Dectin-1 on Macrophages
Promotes Pancreatic Cancer. Cancer Discov. 7:5492017.
|
|
27
|
Li J, Bench AJ, Vassiliou GS, Fourouclas
N, Ferguson-Smith AC and Green AR: Imprinting of the human L3MBTL
gene, a polycomb family member located in a region of chromosome 20
deleted in human myeloid malignancies. Proc Natl Acad Sci USA.
101:7341–7346. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pagano A, Letourneur F, Garcia-Estefania
D, Carpentier JL, Orci L and Paccaud JP: Sec24 proteins and sorting
at the endoplasmic reticulum. J Biol Chem. 274:7833–7840. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li B, Huang MZ, Wang XQ, Tao BB, Zhong J,
Wang XH, Zhang WC and Li ST: TMEM140 is associated with the
prognosis of glioma by promoting cell viability and invasion. J
Hematol Oncol. 8:892015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Camicia R, Winkler HC and Hassa PO: Novel
drug targets for personalized precision medicine in
relapsed/refractory diffuse large B-cell lymphoma: A comprehensive
review. Mol Cancer. 14:2072015. View Article : Google Scholar : PubMed/NCBI
|