Introduction
Gastric cancer is the fourth most common human
cancer and the second most prevalent cause of cancer mortality
worldwide (1). In total, ~1,000,000
cases of gastric cancer are newly diagnosed and ~740,000
mortalities occur every year (2).
Multiple risk factors have been identified to be involved in the
initiation and progression of gastric cancer, including old age,
smoking, alcohol consumption, obesity, unhealthy diet, low economic
status, pernicious anemia, other chronic gastric diseases and
Helicobacter pylori infection (3,4). The
current comprehensive care strategy for gastric cancer is a
combination of surgery, chemotherapy and radiotherapy (5). However, >50% of patients with
advanced-stage gastric cancer succumb to recurrence or metastasis,
despite undergoing curative gastrectomy (6). Gastric cancer typically has a 5-year
overall survival rate of ~15% and the median overall survival time
for advanced-stage gastric cancer specifically is <1 year
(7). Therefore, fully understanding
the molecular mechanisms behind the progression of gastric cancer
is imperative for the development of novel therapeutic
strategies.
microRNAs (miRNAs/miRs) are endogenous, non-coding
RNA molecules 21–23 nucleotides in length that can degrade mRNAs
and suppress protein expression (8).
miRNAs directly bind to the 3′-untranslated region (3′UTR) of their
target mRNAs in a base pairing manner to induce mRNA degradation or
suppress translation of the target proteins (9). miRNAs are involved in a variety of
physiological and pathological processes, including cell
proliferation, differentiation, apoptosis, invasion, migration and
survival (10,11). Differential expression of miRNAs
between normal and tumor tissues has been reported in numerous
human cancer types, suggesting a possible link between aberrant
miRNA expression and the initiation and development of cancer
(12). Furthermore, it has been
demonstrated that miRNAs can function as oncogenes or tumor
suppressors in gastric cancer and have significant regulatory
functions in the disease. In a previous study, restored expression
of the tumor suppressor miR-let-7a significantly inhibited gastric
cancer cell growth, migration and invasion through the negative
regulation of pyruvate kinase M2 (13). Furthermore, oncogenic miR-543 was
demonstrated to promote the proliferation of gastric cancer via
direct targeting of sirtuin 1 (14).
Therefore, investigating the function of miRNAs in the tumor
biology of gastric cancer may provide novel diagnostic and
therapeutic targets for the treatment of this disease. In the
present study, miR-139 expression in gastric cancer tissues and
cell lines was detected and the effect of miR-139 overexpression on
gastric cancer cell proliferation, migration and invasion was
investigated.
Materials and methods
Tissue specimens
The present study was approved by The Research
Ethics Committee of The People's Hospital of Xuyi (Huai'an, China).
A total of 30 patients (age range, 49–72 years; 19 males, 11
females) diagnosed with primary gastric cancer at The People's
Hospital of Xuyi were included in the study between June 2014 and
February 2016. Written informed consent was obtained from all
patients. No participants had received chemotherapy or radiotherapy
prior to surgery. Gastric cancer tissue and adjacent normal gastric
tissue samples were collected, rapidly frozen in liquid nitrogen
and kept at −80°C until use.
Cell culture and oligonucleotide
transfection
Human gastric cell lines (BGC-823, MGC-803 and
SGC-7901) and the human immortalized gastric epithelial cell line
(GES-1) were all purchased from American Type Culture Collection
(Manassas, VA, USA). Cells were routinely cultured in RPMI 1640
medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific,
Inc.) in a humidified chamber with 5% CO2 at 37°C.
miR-139 mimic, miR-139 mimic negative control (NC),
ROCK1 expression vector (pcDNA3.1-ROCK1) and control expression
vector (pcDNA3.1-Control) were chemically synthesized by Shanghai
GenePharma Co., Ltd. (Shanghai, China). The miR-139 mimics sequence
was 5′-UCUACAGUGCACGUGUCUCCAGU-3′ and the NC sequence was
5′-UUCUCCGAACGUGUCACGUTT-3′.
Gastric cancer cells were transfected with miRNA
mimics (100 pmol) or vector (2 µg) with Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Following
transfection (48 h), RT-qPCR was performed to determine miR-139 or
ROCK1 mRNA expression. Cell proliferation, migration and invasion
assays were conducted at 24 and 48 h post-transfection,
respectively. Western blot analysis was carried out at 72 h
following transfection.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissues or cell lines
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturers instructions. The relative
expression of miR-139 was determined by RT-qPCR using the TaqMan
Universal Master Mix (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The thermocycling conditions were as follows: 50°C for 2
min, 95°C for 10 min; 40 cycles of denaturation at 95°C for 15 sec;
annealing/extension at 60°C for 60 sec. RNU6b was used to normalize
miR-139 expression. For ROCK1 mRNA expression, reverse
transcription was performed using the RevertAid™ H Minus First
Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.),
followed by PCR using SYBR Green Premix Ex Taq (Takara
Biotechnology Co., Ltd., Dalian, China). The amplification was
performed with cycling conditions as follows: 5 min at 95°C,
followed by 40 cycles of 95°C for 30 sec and 65°C for 45 sec.
β-actin was used as an endogenous control for ROCK1 mRNA
expression. The primers were designed as follows: miR-139 forward,
5′-CAGCGGTCTACAGTGCACGT-3′ and reverse, 5′-CAGTGCAGGGTCCGAGGT-3′ U6
forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′ ROCK1 forward,
5′-AGGAAGGCGGACATATTGATCCCT-3′ and reverse,
5′-AGACGATAGTTGGGTCCCGGC-3′ and β-actin forward,
5′-CCTGGCACCCAGCACAAT-3′ and reverse, 5′-GCTGATCCACATCTGCTGGAA-3′.
The relative expression of miR-139 and ROCK1 mRNA was analyzed
using the 2−ΔΔCq method (15).
Cell proliferation assay
Cell proliferation was determined by MTT assay.
Transfected cells were collected and seeded into 96-well plates at
a density of 5×103 cells/well with 200 µl RPMI 1640
medium (Gibco; Thermo Fisher Scientific, Inc.). Following
incubation at 37°C for 24–96 h, cells were incubated for 4 h at
37°C with 20 µl MTT solution (5 mg/ml; Sigma-Aldrich; Merck KGaA).
Dimethyl sulfoxide (150 µl) was subsequently added to dissolve the
crystals for 20 min at room temperature. The absorbance was
measured at 490 nm with a microplate reader (Bio-Rad Laboratories,
Inc., Hercules, CA, USA).
Cell migration and invasion assay
For the cell invasion and migration assays,
Transwell plates (pore size, 8 µm) with and without a Matrigel
coating were respectively used (BD Biosciences, Franklin Lakes, NJ,
USA). A total of 1×105 transfected cells in 100 µl
FBS-free RPMI 1640 medium (Gibco; Thermo Fisher Scientific, Inc.)
were added into the upper chamber, and the lower chamber was filled
with 600 µl RMPI 1640 medium (Gibco; Thermo Fisher Scientific,
Inc.) containing 20% FBS (Gibco; Thermo Fisher Scientific, Inc.).
After incubation for 48 h, cells on the upper surface of Transwell
plates were removed with a cotton-tipped swab and cells migrating
to the bottom of the filter were fixed with 100% methanol at room
temperature for 15 min and stained with 0.1% crystal violet at room
temperature for 15 min. The migrated or invaded cells were counted
in five fields using a light microscope (magnification, ×100; BX51;
Olympus Corporation, Tokyo, Japan).
Bioinformatics analysis
TargetScan (http://www.targetscan.org/) and PicTar (http://www.pictar.org/) were used to predict the
target genes of miR-139.
Luciferase reporter assay
pGL3-ROCK1-3′UTR-wild-type (Wt) or
pGL3-ROCK1-3′UTR-Mutant (Mut) luciferase reporter vectors (Shanghai
GenePharma Co., Ltd.), along with miR-139 mimics or NC, were
transfected into gastric cancer cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). At 48 h post-transfection, the cells were
assayed for luciferase activity using the Dual-Luciferase Reporter
assay system (Promega Corporation, Madison, WI, USA) according to
the manufacturer's instructions. The firefly luciferase activity
was normalized to Renilla luciferase activity.
Western blot analysis
At 78 h post-transfection, total protein was
isolated from transfected cells using radioimmunoprecipitation
assay lysis buffer (1% 4-nonylphenyl poly (ethylene glycol), 0.1%
SDS, 100 µg/ml phenylmethylsulfonyl fluoride and 0.5% sodium
deoxycholate in PBS). Protein concentration was determined using a
bicinchoninic acid assay kit (Beyotime Institute of Biotechnology,
Haimen, China). Protein (20 µg) was separated using SDS-PAGE (10%
gel) and transferred to polyvinylidene fluoride membranes (Merck
KGaA). Membranes were blocked with 5% skimmed milk in TBS/0.1%
Tween for 1 h and subsequently incubated at 4°C overnight with
primary antibodies against ROCK1 (1:1,000 dilution; catalog no.
sc-17794; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) or GADPH
(1:1,000 dilution; catalog no. sc-47778; Santa Cruz Biotechnology,
Inc.). Membranes were then incubated with goat anti-mouse
horseradish peroxidase-conjugated secondary antibody (1:10,000
dilution; catalog no. sc-2005; Santa Cruz Biotechnology, Inc.).
Protein bands were visualized using an enhanced chemiluminescence
kit (Thermo Fisher Scientific, Inc.) and quantified with LabWorks
Image Acquisition and Analysis Software (version 3; UVP, LLC,
Phoenix, AZ, USA).
Statistical analysis
Statistical analysis was performed using SPSS 15.0
(SPSS Inc., Chicago, IL, USA). Data is presented as the mean ±
standard deviation. Statistical significance was determined using
Student's t-test or one-way analysis of variance followed by the
Student-Newman-Keuls test. P<0.05 was considered to indicate a
statistically significant difference.
Results
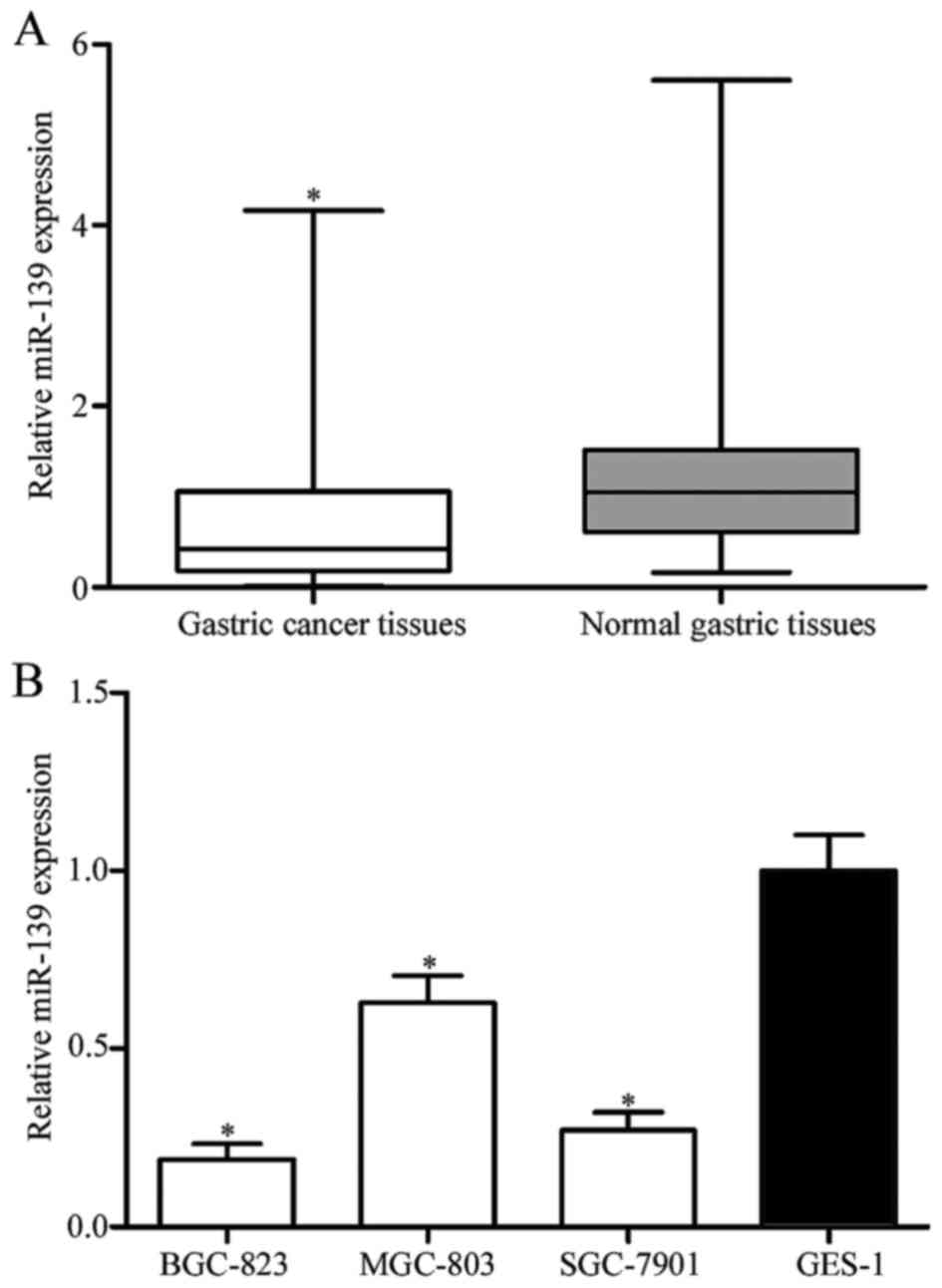
miR-139 expression is downregulated in
human gastric cancer cell lines and tissues
miR-139 expression levels in gastric cancer and
adjacent normal gastric tissues were detected by RT-qPCR. miR-139
expression was significantly downregulated in gastric cancer
tissues compared with their matched adjacent normal gastric tissues
(Fig. 1A; P<0.05). miR-139
expression levels were subsequently determined in BGC-823, MGC-803,
SGC-7901 and GES-1 cell lines. Significantly reduced expression
levels were observed in the gastric cancer cell lines compared with
that in the GES-1 cell line, indicating that miR-139 expression may
be downregulated in gastric cancer (Fig.
1B; P<0.05). BGC-823 and SGC-7901 cell lines exhibited
relatively lower miR-139 expression among these three cell lines,
and therefore were selected for subsequent experiments.
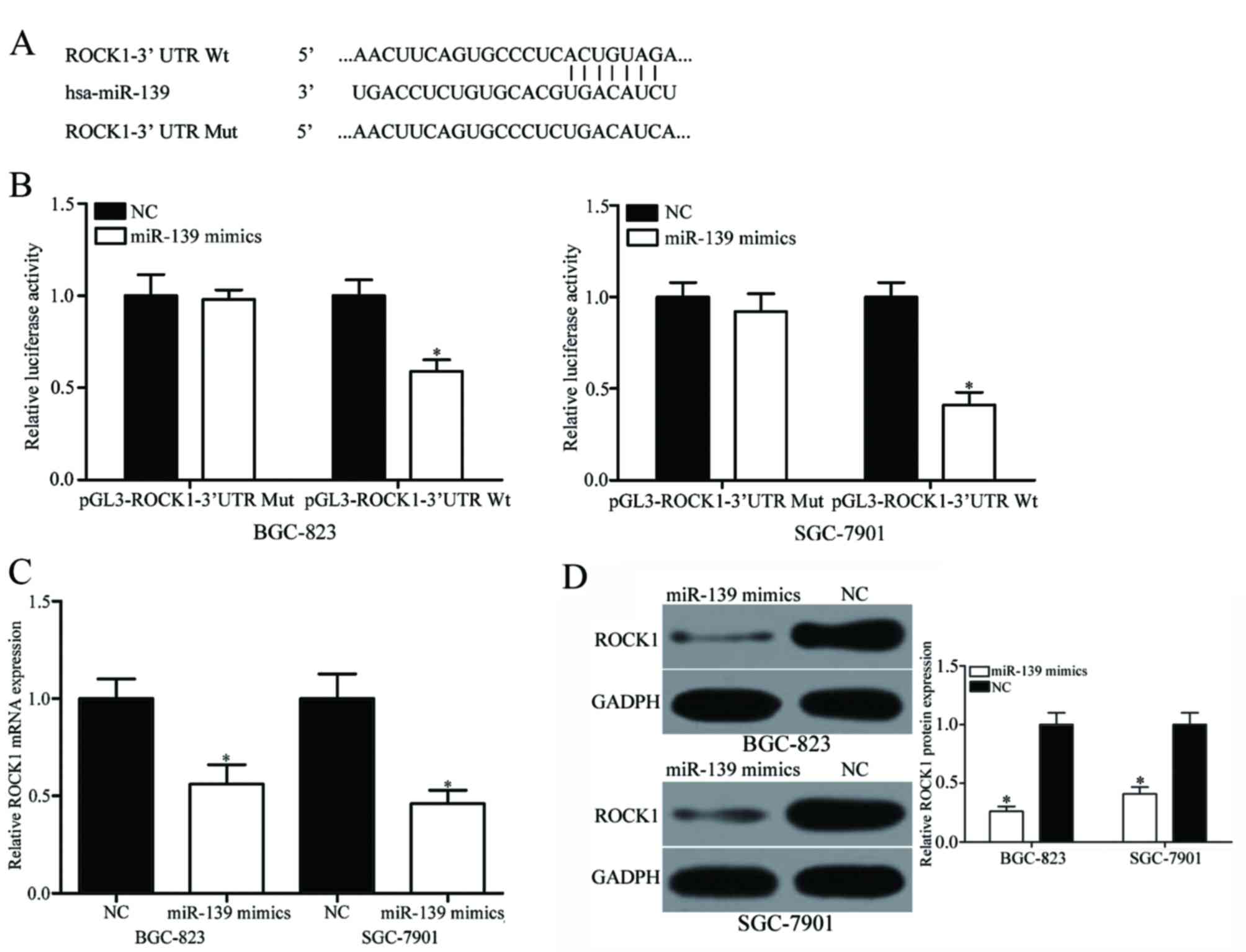
ROCK1 is a direct target of
miR-139
Bioinformatics analysis was performed to further
understand the mechanisms underlying the role of miR-139 in gastric
cancer. The 3′UTR of ROCK1 was confirmed to contain the binding
site of miR-139 at positions 538–545 (Fig. 2A). A luciferase reporter assay was
performed to validate whether ROCK1 was a direct target of miR-139.
Results revealed that miR-139 significantly reduced the luciferase
activity in the pGL3-ROCK1-3′UTR-Wt compared with the NC (Fig. 2B; P<0.05), whereas
pGL3-ROCK1-3′UTR-Mut activity was not significantly different.
RT-qPCR and western blot analysis revealed that restored miR-139
expression significantly decreased ROCK1 mRNA (Fig. 2C; P<0.05) and protein (Fig. 2D; P<0.05) expression in BGC-823 and
SGC-7901 cell lines. Only the BGC-823 and SGC-7901 cell lines were
used as these exhibited relatively lower miR-139 expression among
the three cell lines and were therefore selected for subsequent
experiments. These results indicated that miR-139 may regulate
ROCK1 expression in gastric cancer through interaction with the
3′UTR of ROCK1 mRNA.
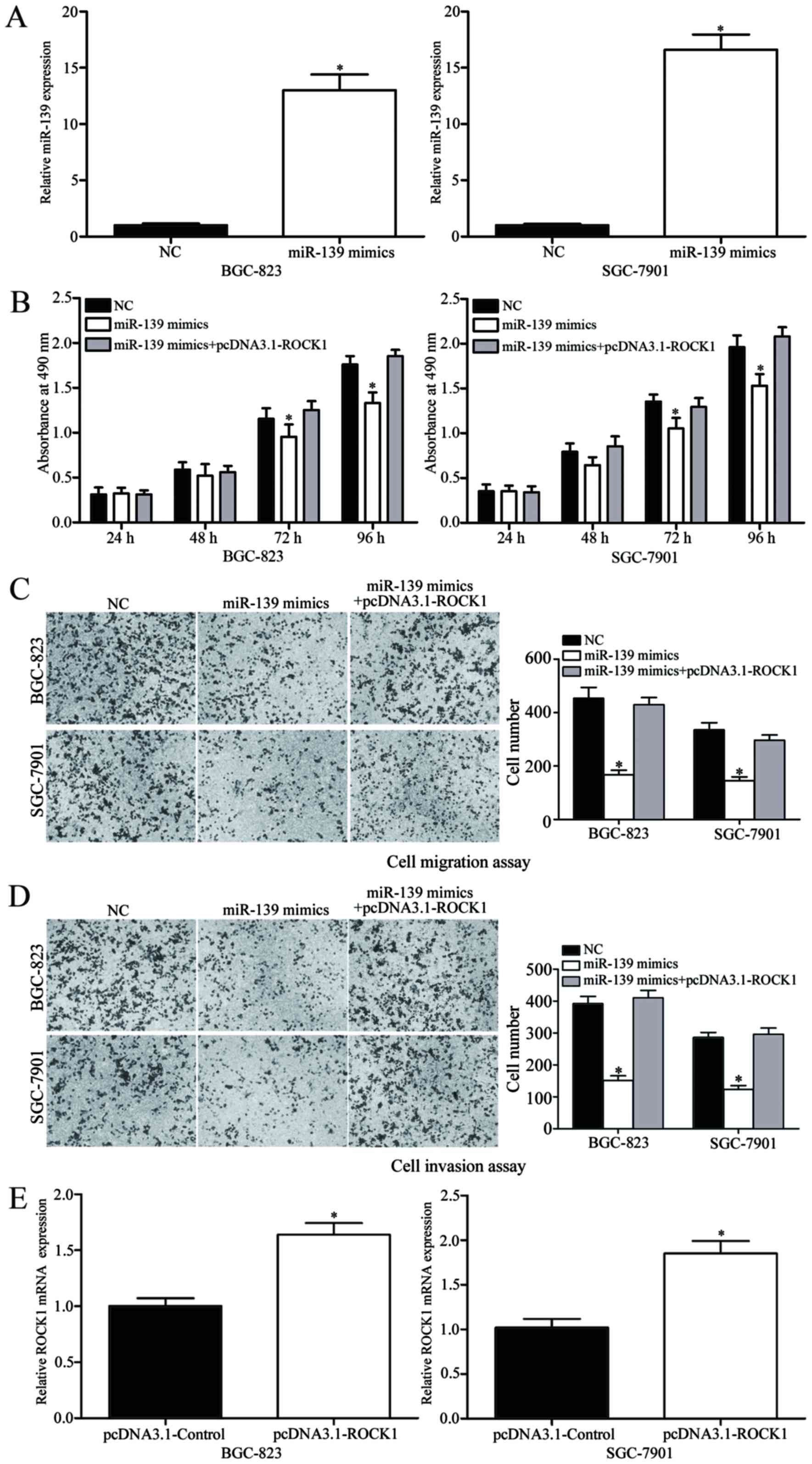
miR-139 inhibits gastric cancer cell
proliferation, migration and invasion by repressing ROCK1
expression
The downregulation of miR-139 observed in gastric
cancer cell lines and tissues suggested that it may physiologically
act as a tumor suppressor. Therefore, miR-139 mimics were
transfected into SGC-7901 and BGC-823 cell lines. Significantly
increased miR-139 expression compared with the NC was confirmed by
RT-qPCR after 24 h (Fig. 3A;
P<0.05). The effect of miR-139 expression on gastric cancer
proliferation was investigated by MTT assay. Results demonstrated
that BGC-823 and SGC-7901 cell lines transfected with miR-139
mimics had significantly reduced cell proliferation at 72 and 96 h
compared with the NC group (Fig. 3B;
P<0.05). Cell migration and invasion assays indicated that
restored miR-139 expression significantly decreased the migration
(Fig. 3C; P<0.05) and invasion
(Fig. 3D; P<0.05) abilities of the
BGC-823 and SGC-7901 cell lines. Taken together, these results
suggested that miR-139 acts as a tumor suppressor by inhibiting
cell proliferation, migration and invasion.
The present study also investigated whether
increased ROCK1 expression was able to attenuate the antitumor
activity of miR-139 in gastric cancer cell lines. ROCK1
overexpression was established by transfection and confirmed by
RT-qPCR (Fig. 3E; P<0.05).
Subsequent assays demonstrated that ROCK1 overexpression reversed
the suppressive effects induced by miR-139 on gastric cancer cell
proliferation (Fig. 3B; P<0.05),
migration (Fig. 3C; P<0.05) and
invasion (Fig. 3D; P<0.05). These
results indicated that miR-139 may inhibit gastric cancer
proliferation, migration and invasion partially through the
downregulation of ROCK1.
Discussion
To the best of our knowledge, the present study
provides novel evidence of the downregulation of miR-139 in gastric
cancer tissues and cell lines compared with adjacent normal gastric
tissues and the normal gastric epithelial GES-1 cell line,
respectively. The aim of the present study was to elucidate the
biological function of miR-139 in gastric cancer carcinogenesis and
progression. Restored miR-139 expression significantly inhibited
gastric cancer proliferation, migration and invasion. Therefore,
miR-139 may function as a tumor suppressor in gastric cancer.
Recent studies have revealed that low miR-139
expression occurs in various types of human cancer, including
glioma (16), tongue squamous cell
carcinoma (17), breast cancer
(18), non-small cell lung cancer
(19) and colon cancer (20). Furthermore, miR-139 expression has
been demonstrated to associate with clinicopathological factors.
Wong et al (21) reported that
low miR-139 expression in hepatocellular carcinoma was
significantly associated with poor patient prognosis and metastatic
tumor characteristics, including venous invasion, microsatellite
formation, an absence of tumor encapsulation and reduced
differentiation. Shen et al (22) demonstrated that miR-139 expression was
also downregulated in colorectal cancer tissues compared with that
in adjacent non-tumorous tissues, and decreased miR-139 expression
has also been associated with colorectal cancer progression and
metastasis. Furthermore, in a study by Liu et al (23), miR-139 expression was demonstrated to
be reduced in esophageal squamous cell carcinoma tissues, and this
reduction was associated with lymph node metastasis. These findings
indicate that increasing miR-139 expression may be useful in the
prevention of cancer progression.
Several lines of evidences have demonstrated the
role of miR-139 in a range of physiological and pathological
processes. Wang et al (15)
and Li et al (24) reported
that miR-139 may inhibit glioma cell proliferation and invasion
in vitro and in vivo. It has also been reported that
miR-139 can suppress glioblastoma cell proliferation, migration and
invasion, and induce apoptosis (25,26). In
non-small cell lung cancer, miR-139 decreases the proliferation and
invasion of cancer cells (18).
Additionally, miR-139 suppresses epithelial-mesenchymal transition,
migration and invasion in hepatocellular carcinoma, and the
incidence and severity of pulmonary metastasis in orthotopic liver
tumors in mice is also reduced (20,27).
Furthermore, Zhang et al (28)
revealed that colorectal cancer cell viability, colony formation,
nude mice tumor growth, cell migration and invasion are repressed
by miR-139, and Luo et al (29) demonstrated that proliferation,
invasion and metastasis of laryngeal squamous carcinoma cells are
also reduced. miR-139 functions as a tumor suppressor in esophageal
squamous cell carcinoma by cell proliferation, migration and
invasion inhibition, apoptosis induction and G0/G1 phase cell cycle
arrest (22). These results
demonstrated that miR-139 is a promising target for cancer
therapy.
To investigate the molecular mechanisms underlying
the cell proliferation, migration and invasion inhibition by
miR-139, the target genes of miR-139 in gastric cancer were
investigated. Bioinformatics analysis with TargetScan and PicTar
revealed that the conserved binding sites on the 3′UTR of ROCK1
mRNA can be recognized by miR-139, suggesting that ROCK1 expression
may be regulated by miR-139 by binding to the 3′UTR of ROCK.
Luciferase reporter assays were performed to validate this
hypothesis. Vectors containing the wild-type or the mutant 3′UTR
segments of ROCK1 and miR-139 mimic or NC were co-transfected into
gastric cancer cells. The luciferase activity of the
pGL3-ROCK1-3′UTR-Wt- and miR-139 mimic-transfected gastric cells
was lower than that of cells transfected with pGL3-ROCK1-3′UTR-Mut
and miR-139 mimic, confirming that miR-139 binds to the 3′UTR of
ROCK1 mRNA. The ability of miR-139 to modulate ROCK1 mRNA and
protein expression was subsequently investigated. RT-qPCR and
western blot analysis revealed that restored miR-139 expression
resulted in significantly decreased ROCK1 mRNA and protein
expression. Furthermore, ROCK1 overexpression attenuated the
effects of miR-139 in gastric cancer cells, indicating that ROCK1
is a functional target of miR-139 in gastric cancer. To the best
our knowledge, the present study is the first to investigate
miR-139/ROCK1 interactions in gastric cancer.
In conclusion, the downregulation of miR-139 in
gastric cancer tissues and cell lines was confirmed, and ectopic
miR-139 expression repressed the proliferation, migration and
invasion of gastric cancer cells. ROCK1 was validated as a novel
direct target gene of miR-139 in gastric cancer. Taken together,
the findings of the present study demonstrate that miR-139
downregulation is key in the initiation and progression of gastric
cancer, and that miR-139 has therapeutic potential for gastric
cancer.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar
|
|
2
|
Zhang YZ, Zhang LH, Gao Y, Li CH, Jia SQ,
Liu N, Cheng F, Niu DY, Cho WC, Ji JF and Zeng CQ: Discovery and
validation of prognostic markers in gastric cancer by genome-wide
expression profiling. World J Gastroenterol. 17:1710–1717. 2011.
View Article : Google Scholar
|
|
3
|
Tkachenko MA, Zhannat NZ, Erman LV,
Blashenkova EL, Isachenko SV, Isachenko OB, Graham DY and Malaty
HM: Dramatic changes in the prevalence of Helicobacter pylori
infection during childhood: A 10-year follow-up study in Russia. J
Pediatr Gastroenterol Nutr. 45:428–432. 2007. View Article : Google Scholar
|
|
4
|
Wang F, Meng W, Wang B and Qiao L:
Helicobacter pylori-induced gastric inflammation and gastric
cancer. Cancer Lett. 345:196–202. 2014. View Article : Google Scholar
|
|
5
|
Yan C, Yu J, Liu Y, Kang W, Ma Z and Zhou
L: MiR-32 promotes gastric carcinoma tumorigenesis by targeting
Kruppel-like factor 4. Biochem Biophys Res Commun. 467:913–920.
2015. View Article : Google Scholar
|
|
6
|
Kim SJ, Wang YG, Lee HW, Kang HG, La SH,
Choi IJ, Irimura T, Ro JY, Bresalier RS and Chun KH: Up-regulation
of neogenin-1 increases cell proliferation and motility in gastric
cancer. Oncotarget. 5:3386–3398. 2014. View Article : Google Scholar
|
|
7
|
Delaunoit T: Latest developments and
emerging treatment options in the management of stomach cancer.
Cancer Manag Res. 3:257–266. 2011. View Article : Google Scholar
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar
|
|
9
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tahara H, Kay MA, Yasui W and Tahara E:
MicroRNAs in Cancer: The 22nd hiroshima cancer seminar/the 4th
Japanese Association for RNA interference joint international
symposium, 30 August 2012, grand prince hotel hiroshima. Jpn J Clin
Oncol. 43:579–582. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yates LA, Norbury CJ and Gilbert RJ: The
long and short of microRNA. Cell. 153:516–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang R, Yang C, Ma X, Wang Y, Luo D, Huang
C, Xu Z, Liu P and Yang L: MiR-let-7a inhibits cell proliferation,
migration and invasion by down-regulating PKM2 in gastric cancer.
Oncotarget. 7:5972–5984. 2016.PubMed/NCBI
|
|
14
|
Li J, Dong G, Wang B, Gao W and Yang Q:
miR-543 promotes gastric cancer cell proliferation by targeting
SIRT1. Biochem Biophys Res Commun. 469:15–21. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang H, Yan X, Ji LY, Ji XT, Wang P, Guo
SW and Li SZ: miR-139 functions as an antioncomir to repress glioma
progression through targeting IGF-1 R, AMY-1 and PGC-1β. Technol
Cancer Res Treat. 16:497–511. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Duz MB, Karatas OF, Guzel E, Turgut NF,
Yilmaz M, Creighton CJ and Ozen M: Identification of miR-139-5p as
a saliva biomarker for tongue squamous cell carcinoma: A pilot
study. Cell Oncol (Dordr). 39:187–193. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang HD, Sun DW, Mao L, Zhang J, Jiang
LH, Li J, Wu Y, Ji H, Chen W, Wang J, et al: MiR-139-5p inhibits
the biological function of breast cancer cells by targeting Notch1
and mediates chemosensitivity to docetaxel. Biochem Biophys Res
Commun. 465:702–713. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu W, Hang M, Yuan CY, Wu FL, Chen SB and
Xue K: MicroRNA-139-5p inhibits cell proliferation and invasion by
targeting insulin-like growth factor 1 receptor in human non-small
cell lung cancer. Int J Clin Exp Pathol. 8:3864–3870.
2015.PubMed/NCBI
|
|
20
|
Liu X, Duan B, Dong Y, He C, Zhou H, Sheng
H, Gao H and Zhang X: MicroRNA-139-3p indicates a poor prognosis of
colon cancer. Int J Clin Exp Pathol. 7:8046–8052. 2014.PubMed/NCBI
|
|
21
|
Wong CC, Wong CM, Tung EK, Au SL, Lee JM,
Poon RT, Man K and Ng IO: The microRNA miR-139 suppresses
metastasis and progression of hepatocellular carcinoma by
down-regulating Rho-kinase 2. Gastroenterology. 140:322–331. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shen K, Liang Q, Xu K, Cui D, Jiang L, Yin
P, Lu Y, Li Q and Liu J.: MiR-139 inhibits invasion and metastasis
of colorectal cancer by targeting the type I insulin-like growth
factor receptor. Biochem Pharmacol. 84:320–330. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu R, Yang M, Meng Y, Liao J, Sheng J, Pu
Y, Yin L and Kim SJ: Tumor-suppressive function of miR-139-5p in
esophageal squamous cell carcinoma. PLoS One. 8:e770682013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li RY, Chen LC, Zhang HY, Du WZ, Feng Y,
Wang HB, Wen JQ, Liu X, Li XF, Sun Y, et al: MiR-139 inhibits Mcl-1
expression and potentiates TMZ-induced apoptosis in glioma. CNS
Neurosci Ther. 19:477–483. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dai S, Wang X, Li X and Cao Y:
MicroRNA-139-5p acts as a tumor suppressor by targeting ELTD1 and
regulating cell cycle in glioblastoma multiforme. Biochem Biophys
Res Commun. 467:204–210. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yue S, Wang L, Zhang H, Min Y, Lou Y, Sun
H, Jiang Y, Zhang W, Liang A, Guo Y, et al: miR-139-5p suppresses
cancer cell migration and invasion through targeting ZEB1 and ZEB2
in GBM. Tumour Biol. 36:6741–6749. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qiu G, Lin Y, Zhang H and Wu D: miR-139-5p
inhibits epithelial-mesenchymal transition, migration and invasion
of hepatocellular carcinoma cells by targeting ZEB1 and ZEB2.
Biochem Biophys Res Commun. 463:315–321. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang L, Dong Y, Zhu N, Tsoi H, Zhao Z, Wu
CW, Wang K, Zheng S, Ng SS, Chan FK, et al: microRNA-139-5p exerts
tumor suppressor function by targeting NOTCH1 in colorectal cancer.
Mol Cancer. 13:1242014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Luo HN, Wang ZH, Sheng Y, Zhang Q, Yan J,
Hou J, Zhu K, Cheng Y, Xu YL, Zhang XH, et al: MiR-139 targets
CXCR4 and inhibits the proliferation and metastasis of laryngeal
squamous carcinoma cells. Med Oncol. 31:7892014. View Article : Google Scholar : PubMed/NCBI
|