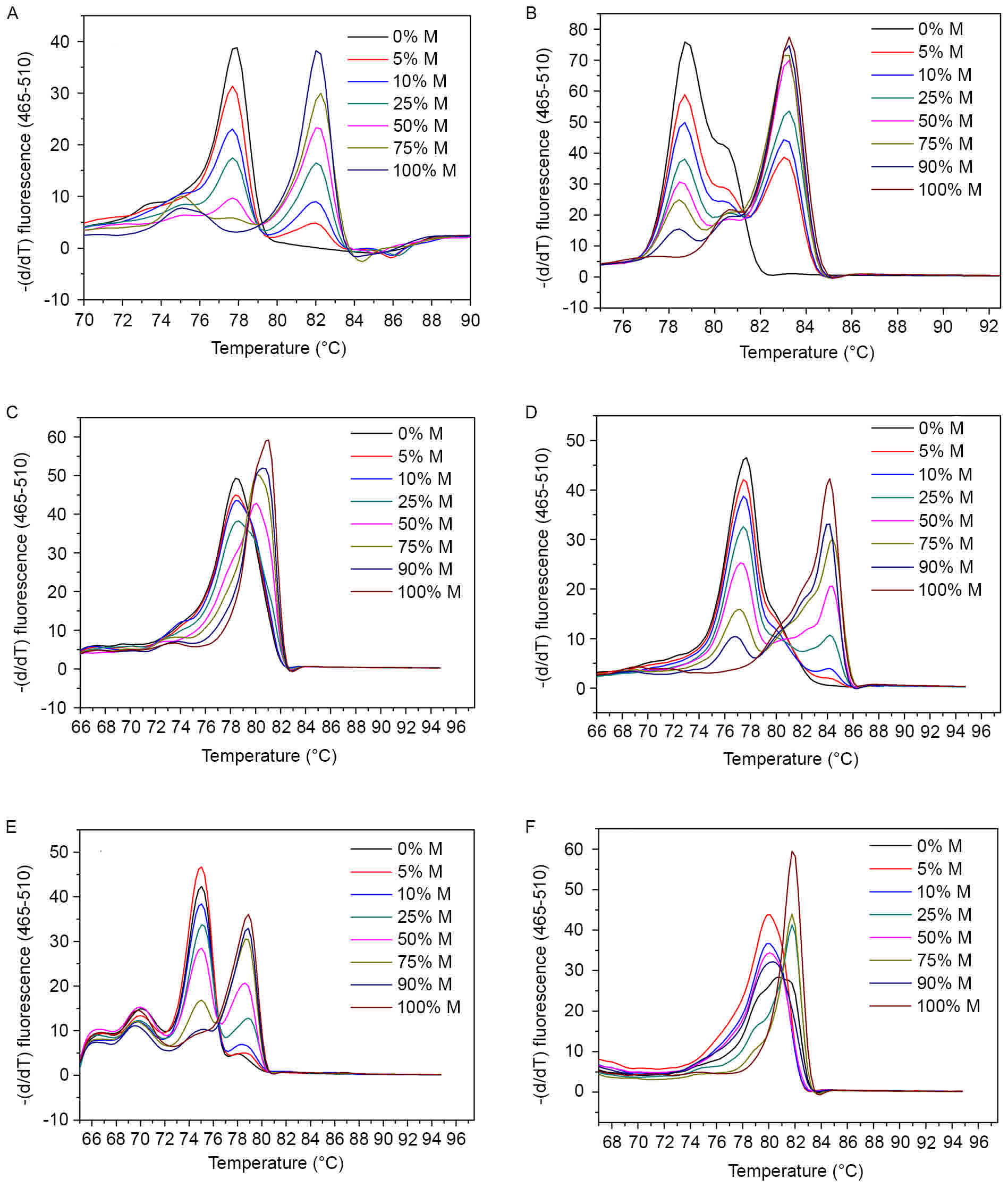
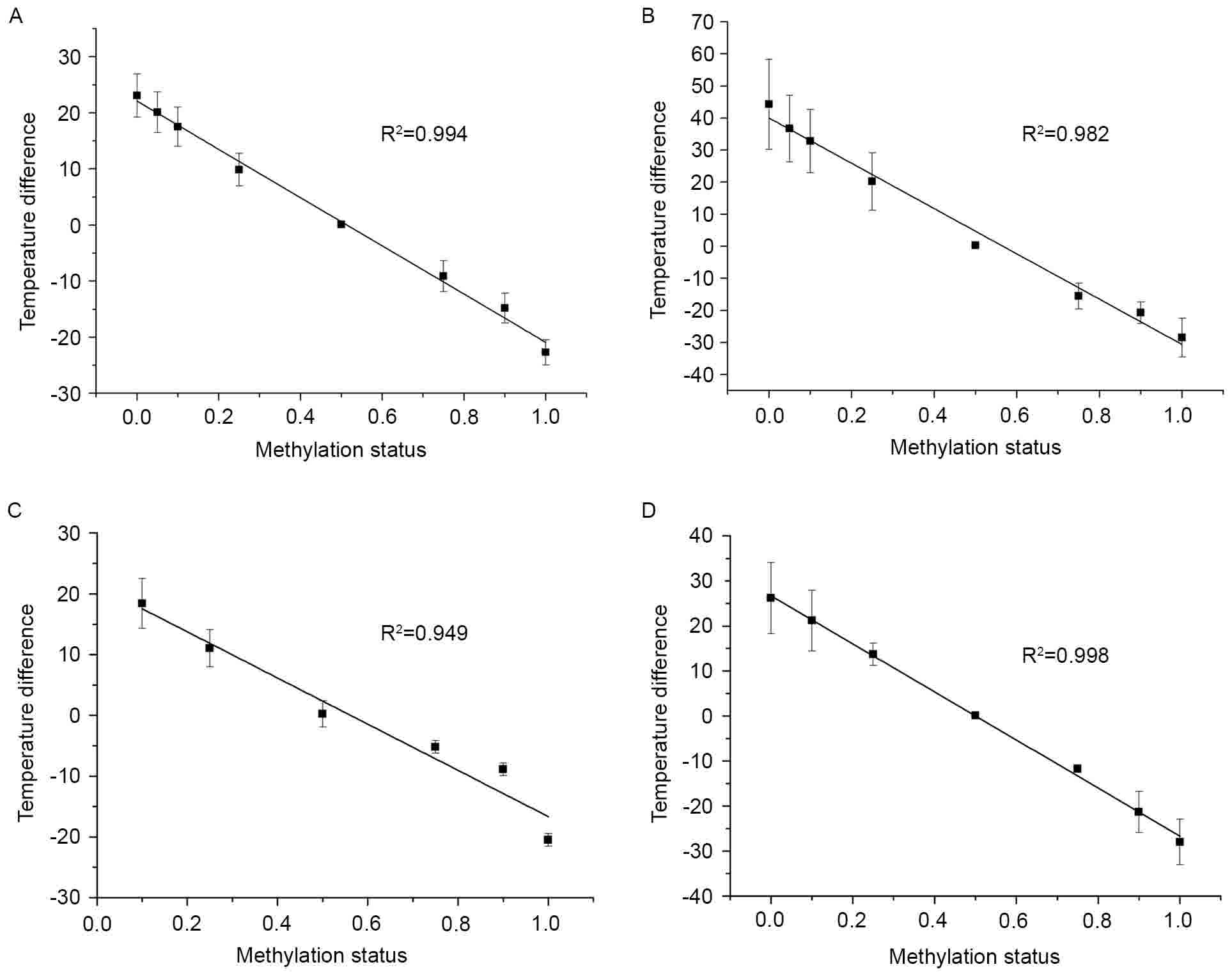
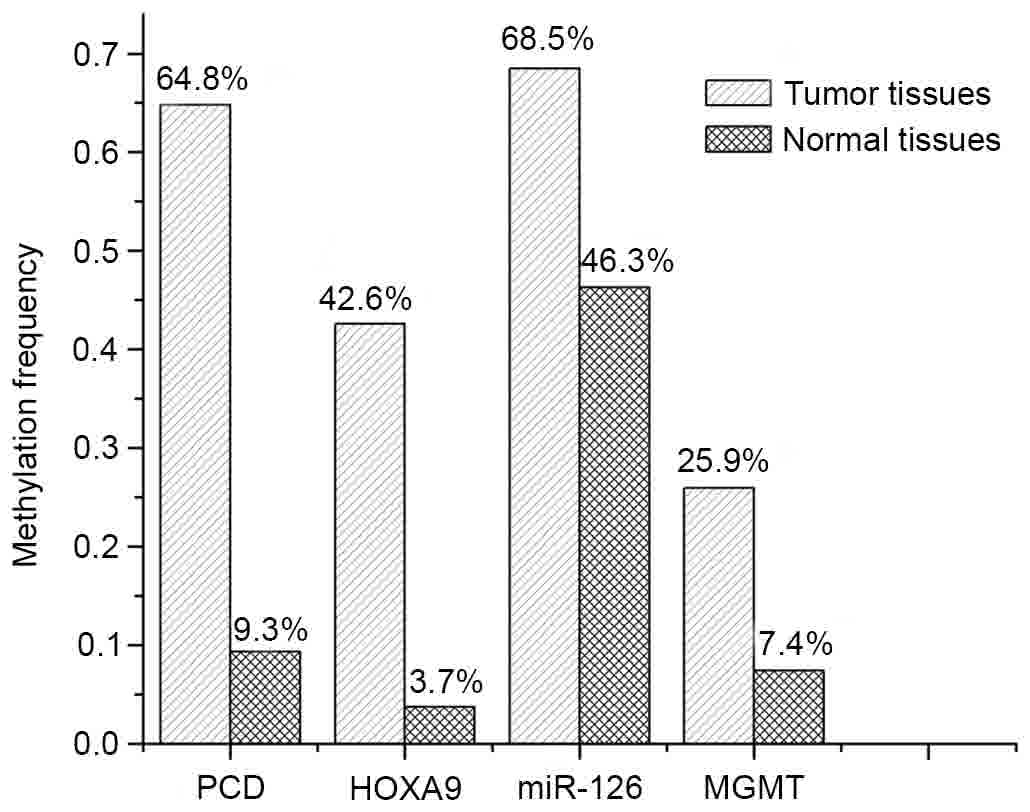
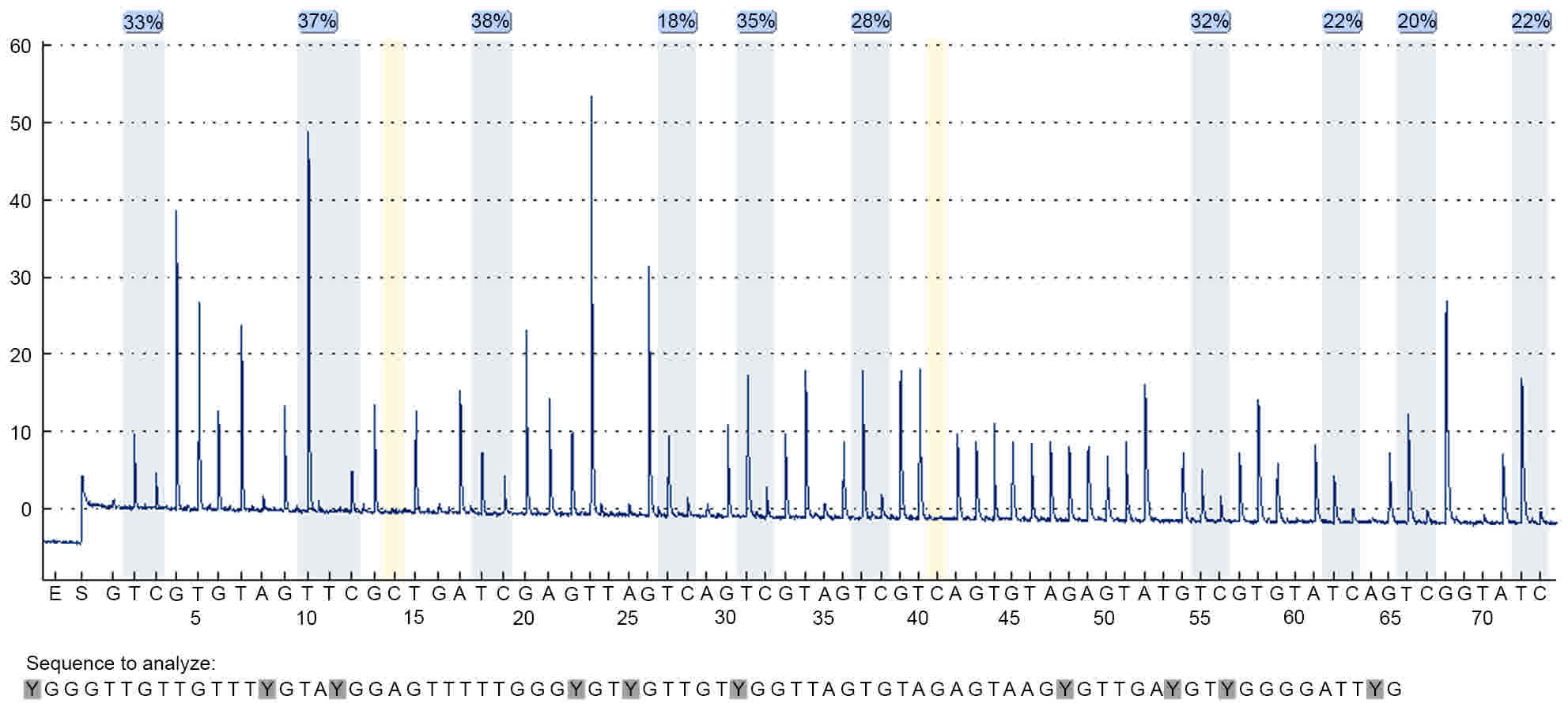
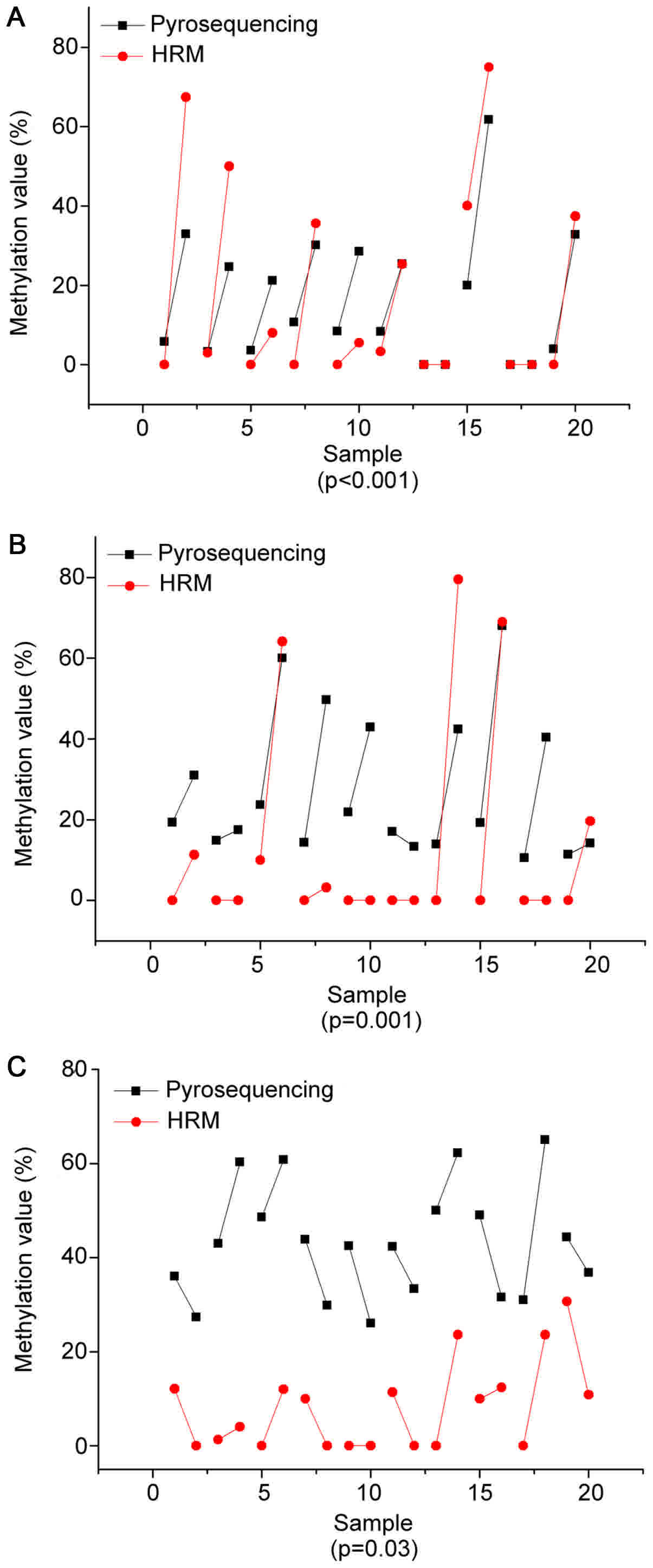
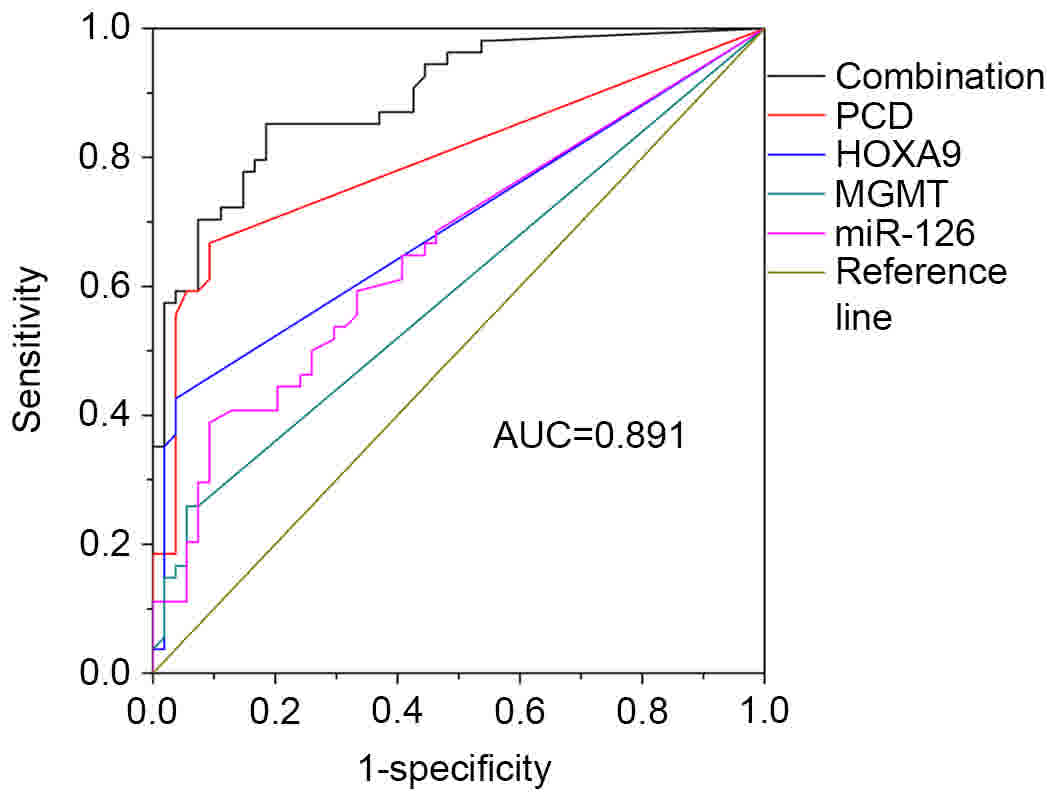
|
1
|
Hubaux R, Thu KL, Vucic EA, Pikor LA, Kung
SH, Martinez VD, Mosslemi M, Becker-Santos DD, Gazdar AF, Lam S and
Lam WL: Microtubule affinity-regulating kinase 2 is associated with
DNA damage response and cisplatin resistance in non-small cell lung
cancer. Int J Cancer. 137:2072–2082. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Peters S, Adjei AA, Gridelli C, Reck M,
Kerr K and Felip E: ESMO Guidelines Working Group: Metastatic
non-small-cell lung cancer (NSCLC): ESMO Clinical Practice
Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 23
Suppl 7:vii56–64. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dillman RO, Herndon J, Seagren SL, Eaton
WL Jr and Green MR: Improved survival in stage III non-small-cell
lung cancer: Seven-year follow-up of cancer and leukemia group B
(CALGB) 8433 trial. J Natl Cancer Inst. 88:1210–1215. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sandoval J, Mendez-Gonzalez J, Nadal E,
Chen G, Carmona FJ, Sayols S, Moran S, Heyn H, Vizoso M, Gomez A,
et al: A prognostic DNA methylation signature for stage I
non-small-cell lung cancer. J Clin Oncol. 31:4140–4147. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lister R, Pelizzola M, Dowen RH, Hawkins
RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al:
Human DNA methylomes at base resolution show widespread epigenomic
differences. Nature. 462:315–322. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Herman JG, Graff JR, Myöhänen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: Anovel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Eads CA, Danenberg KD, Kawakami K, Saltz
LB, Blake C, Shibata D, Danenberg PV and Laird PW: Methy Light: A
high-throughput assay to measure DNA methylation. Nucleic Acids
Res. 28:e322000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Du Y, Zhou Y and Wu Q: MS-HRM to detect
serum DNA methylation of intrauterine growth retardation children.
Engineering. 4:106–109. 2012. View Article : Google Scholar
|
|
9
|
Doerks T, Copley RR, Schultz J, Ponting CP
and Bork P: Systematic identification of novel protein domain
families associated with nuclear functions. Genome Res. 12:47–56.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gitan RS, Shi H, Chen CM, Yan PS and Huang
TH: Methylation-specific oligonucleotide microarray: A new
potential for high-throughput methylation analysis. Genome Res.
12:158–164. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wojdacz TK: Methylation-sensitive
high-resolution melting in the context of legislative requirements
for validation of analytical procedures for diagnostic
applications. Expert Rev Mol Diagn. 12:92012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wojdacz TK and Dobrovic A:
Methylation-sensitive high resolution melting (MS-HRM): A new
approach for sensitive and high-throughput assessment of
methylation. Nucleic Acids Res. 35:e412007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wolf P, Hu YC, Doffek K, Sidransky D and
Ahrendt SA: O6-Methylguanine-DNA methyltransferase promoter
hypermethylation shifts the p53 mutational spectrum in non-small
cell lung cancer. Cancer Res. 61:8113–8117. 2001.PubMed/NCBI
|
|
14
|
Watanabe K, Emoto N, Hamano E, Sunohara M,
Kawakami M, Kage H, Kitano K, Nakajima J, Goto A, Fukayama M, et
al: Genome structure-based screening identified epigenetically
silenced microRNA associated with invasiveness in non-small-cell
lung cancer. Int J Cancer. 130:2580–2590. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Boosani CS and Agrawal DK: Methylation and
microRNA-mediated epigenetic regulation of SOCS3. Mol Biol Rep.
42:853–872. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Irimia M, Fraga MF, Sanchez-Cespedes M and
Esteller M: CpG island promoter hypermethylation of the
Ras-effector gene NORE1A occurs in the context of a wild-type K-ras
in lung cancer. Oncogene. 23:8695–8699. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang KH, Lin CJ, Liu CJ, Liu DW, Huang RL,
Ding DC, Weng CF and Chu TY: Global methylation silencing of
clustered proto-cadherin genes in cervical cancer: Serving as
diagnostic markers comparable to HPV. Cancer Med. 4:43–55. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Haller F, Zhang JD, Moskalev EA, Braun A,
Otto C, Geddert H, Riazalhosseini Y, Ward A, Balwierz A, Schaefer
IM, et al: Combined DNA methylation and gene expression profiling
in gastrointestinal stromal tumors reveals hypomethylation of SPP1
as an independent prognostic factor. Int J Cancer. 136:1013–1023.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Son JW, Jeong KJ, Jean WS, Park SY, Jheon
S, Cho HM, Park CG, Lee HY and Kang J: Genome-wide combination
profiling of DNA copy number and methylation for deciphering
biomarkers in non-small cell lung cancer patients. Cancer Lett.
311:29–37. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Saito Y, Friedman JM, Chihara Y, Egger G,
Chuang JC and Liang G: Epigenetic therapy upregulates the tumor
suppressor microRNA-126 and its host gene EGFL7 in human cancer
cells. Biochem Biophys Res Commun. 379:726–731. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang P, Yang D, Zhang H, Wei X, Ma T,
Cheng Z, Hong Q, Hu J, Zhuo H, Song Y, et al: Early detection of
lung cancer in serum by a panel of MicroRNA biomarkers. Clin Lung
Cancer. 16:313–319.e1. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Crawford M, Brawner E, Batte K, Yu L,
Hunter MG, Otterson GA, Nuovo G, Marsh CB and Nana-Sinkam SP:
MicroRNA-126 inhibits invasion in non-small cell lung carcinoma
cell lines. Biochem Biophys Res Commun. 373:607–612. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Esteller M, Garcia-Foncillas J, Andion E,
Goodman SN, Hidalgo OF, Vanaclocha VV, Baylin SB and Herman JG:
Inactivation of the DNA-repair gene MGMT and the clinical response
of gliomas to alkylating agents. New Engl J Med. 343:1350–1354.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu JY, Wang J, Lai JC, Cheng YW, Yeh KT,
Wu TC, Chen CY and Lee H: Association of O6-methylguanine-DNA
methyltransferase (MGMT) promoter methylation with p53 mutation
occurrence in non-small cell lung cancer with different histology,
gender and smoking status. Ann Surg Oncol. 15:3272–3277. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Barclay JL, Anderson ST, Waters MJ and
Curlewis JD: SOCS3 as a tumor suppressor in breast cancer cells and
its regulation by PRL. Int J Cancer. 124:1756–1766. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rigby RJ, Simmons JG, Greenhalgh CJ,
Alexander WS and Lund PK: Suppressor of cytokine signaling 3
(SOCS3) limits damage-induced crypt hyper-proliferation and
inflammation-associated tumorigenesis in the colon. Oncogene.
26:4833–4841. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Moshnikova A, Frye J, Shay JW, Minna JD
and Khokhlatchev AV: The growth and tumor suppressor NORE1A is a
cytoskeletal protein that suppresses growth by inhibition of the
ERK pathway. J Biol Chem. 281:8143–8152. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hesson L, Dallol A, Minna JD, Maher ER and
Latif F: NORE1A, a homologue of RASSF1A tumour suppressor gene is
inactivated in human cancers. Oncogene. 22:947–954. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin TC, Jiang SS, Chou WC, Hou HA, Lin YM,
Chang CL, Hsu CA, Tien HF and Lin LI: Rapid assessment of the
heterogeneous methylation status of CEBPA in patients with acute
myeloid leukemia by using high-resolution melting profile. J Mol
Diagn. 13:514–519. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Newman M, Blyth BJ, Hussey DJ, Jardine D,
Sykes PJ and Ormsby RJ: Sensitive quantitative analysis of murine
LINE1 DNA methylation using high resolution melt analysis.
Epigenetics. 7:92–105. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang X, Dai W, Kwong DL, Szeto CY, Wong
EH, Ng WT, Lee AW, Ngan RK, Yau CC, Tung SY and Lung ML: Epigenetic
markers for noninvasive early detection of nasopharyngeal carcinoma
by methylation-sensitive high resolution melting. Int J Cancer.
136:E127–E135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wojdacz TK, Møller TH, Thestrup BB,
Kristensen LS and Hansen LL: Limitations and advantages of MS-HRM
and bisulfite sequencing for single locus methylation studies.
Expert Rev Mol Diagn. 10:575–580. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhu J and Yao X: Use of DNA methylation
for cancer detection: Promises and challenges. Int J Biochem Cell
Biol. 41:147–154. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma Y, Bai Y, Mao H, Hong Q, Yang D, Zhang
H, Liu F, Wu Z, Jin Q, Zhou H, et al: A panel of promoter
methylation markers for invasive and noninvasive early detection of
NSCLC using a quantum dots-based FRET approach. Biosens
Bioelectron. 85:641–648. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hwang JA, Lee BB, Kim Y, Hong SH, Kim YH,
Han J, Shim YM, Yoon CY, Lee YS and Kim DH: HOXA9 inhibits
migration of lung cancer cells and its hypermethylation is
associated with recurrence in non-small cell lung cancer. Mol
Carcinog. 54 Suppl 1:E72–E80. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang T, Chen X, Hong Q, Deng Z, Ma H, Xin
Y, Fang Y, Ye H, Wang R, Zhang C, et al: Meta-analyses of gene
methylation and smoking behavior in non-small cell lung cancer
patients. Sci Rep. 5:88972015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Colella S, Shen L, Baggerly KA, Issa JP
and Krahe R: Sensitive and quantitative universal Pyrosequencing
methylation analysis of CpG sites. Biotechniques. 35:146–150.
2003.PubMed/NCBI
|
|
38
|
Candiloro IL, Mikeska T and Dobrovic A:
Assessing combined methylation-sensitive high resolution melting
and pyrosequencing for the analysis of heterogeneous DNA
methylation. Epigenetics. 6:500–507. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Quillien V, Lavenu A, Karayan-Tapon L,
Carpentier C, Labussiere M, Lesimple T, Chinot O, Wager M, Honnorat
J, Saikali S, et al: Comparative assessment of 5 methods
(methylation-specific polymerase chain reaction, MethyLight,
pyrosequencing, methylation-sensitive high-resolution melting and
immunohistochemistry) to analyze
O6-methylguanine-DNA-methyltranferase in a series of 100
glioblastoma patients. Cancer. 118:4201–4211. 2012. View Article : Google Scholar : PubMed/NCBI
|