Introduction
Lung cancer is one of the most common cancer types
in terms of incidence and mortality at present globally (1), and among all the different types,
non-small cell lung cancer (NSCLC) accounts for 80–85% (2). NSCLC may be characterized by the driver
gene mutation, particularly by epidermal growth factor receptor
(EGFR) and kirsten rat sarcoma viral oncogene homolog (KRAS)
mutations. It has previously been reported that EGFR and KRAS
mutation occurs in 59.4 and 7.4%, respectively, of all Asian lung
adenocarcinoma cases (3). A number of
EGFR tyrosine kinase inhibitors (TKIs) have become the first line
therapy for lung adenocarcinoma with sensitive EGFR mutations
(4,5).
Even though there has been progress in molecular target therapy,
the 5-year survival rate remains <15% (6), which is largely due to TKI-resistance
and metastasis (7,8). For the purpose of improving
understanding of the mechanisms of resistance to TKIs, the
molecular categorization of patients with EGFR/KRAS mutations in
NSCLC is required.
Vascular endothelial growth factor (VEGF) signaling
serves a pivotal function in tumor angiogenesis and is associated
with an increased tumor recurrence and metastasis (9–12). As a
major regulator of angiogenesis, VEGFA binds to the VEGF receptor
(VEGFR)1 and VEGFR2, which are important members of the family of
receptor tyrosine kinases (RTKs). They stimulate multiple pathways,
including mitogen-activated protein kinases, phosphoinositide
3-kinases (PI3Ks), and protein kinase B (Akt) (13–15), to
promote recurrence and metastases. VEGFR1 signaling regulates
endothelial cell survival and VEGFR2 signaling regulates the
differentiation of endothelial cells into capillary tubes (16). By inhibiting the VEGFA-VEGFR signaling
pathway, a number of strategies for the therapy of different types
of cancer have been established, including bevacizumab, an antibody
that targets VEGFA, and diverse inhibitors of RTKs. However,
recurrence due to resistance to the therapy remains inevitable in a
number of tumors (17,18). In order to overcome resistance and
combine more rationally the two types of targeted therapy, it is
crucial to firstly reveal the association among different driver
genes associated with targeted efficacy and drug-resistance.
In the present study, the aim was to establish the
association between VEGFA, VEGFR1 and VEGFR2 expression and
EGFR/KRAS mutations. Furthermore, this study aimed to determine the
potential benefit of TKIs and anti-angiogenesis therapy, and
elucidate potential ‘cross resistance’ occurrences to the
aforementioned therapies due to associated driver gene mutations
and protein expression in lung adenocarcinoma. It may provide a
more improved understanding of how to treat various patients with
KRAS or a subtype of the EGFR mutation with anti-angiogenesis
therapy.
Materials and methods
Sample collection
A total of 204 patients with lung adenocarcinoma who
underwent surgery at Cancer Hospital of Tianjin Medical University
(Tianjin, China) between January 2013 and December 2015 were
selected for the study. There were 99 females (48.5%) and 105 males
(51.5%); 93 patients (45.6%) with age >60 years and 111 patients
(54.4%) with age ≤60 years (median age, 58 years; age range 30–76
years). Collection and use of tumor tissue samples for research
received written informed consent from all patients prior to the
study and was ethically approved by Ethics and Scientific Committee
of Tianjin Medical University Cancer Hospital. Each specimen was
confirmed as lung adenocarcinoma by pathological diagnosis.
Clinicopathological features of each patient comprised sex, age,
smoking status, lymph node metastasis and clinical stage. Tumor
clinical stage was identified according to the International
Association for the Study of Lung Cancer 2009 TNM tumor staging
system (19). Smoking history was
marked as either yes or no (patients were defined as non-smokers if
they had never smoked in their lifetime).
Immunohistochemistry (IHC)
IHC was performed on formalin-fixed
paraffin-embedded (FFPE) specimens (4 µm) by using antibodies for
VEGFA (mouse monoclonal IgG; 1:200; cat no. ab1316), VEGFR1 (rabbit
monoclonal IgG; 1:50; cat no. ab32152) (both from Abcam, Cambridge,
UK) and VEGFR2 (rabbit monoclonal IgG; 1:200; cat no. 55B11; Cell
Signaling Technologies, Inc., Danvers, MA, USA). Following baking
in a 65°C oven (Fuzhou Maixin Biotech Co., Ltd., Fuzhou, China) for
1 h, the FFPE specimens were deparaffinized in xylene, rehydrated
in graded alcohol and then washed with phosphate buffer saline
(PBS) three times for 5 min. For the purpose of antigen retrieval,
the sections were boiled for 3 min at 100°C in citric acid-based
buffer at pH 6.0 for VEGFA antigen and EDTA-based buffer at pH 9.0
for VEGFR1 and VEGFR2 antigens. Then, the slides were cooled to
room temperature and rinsed with PBS 3 times for 5 min.
Subsequently, the activity of endogenous peroxidase was blocked by
3% hydrogen peroxide for 20 min at room temperature, and the slides
were incubated with primary antibodies at 4°C overnight (for >12
h). Subsequently, slides were rinsed using PBS three times for 5
min at room temperature and incubated in horseradish
peroxidase-conjugated secondary antibodies (Polymer detection kit
for mouse and rabbit; used as supplied); PV-6000; OriGene
Technologies, Inc. (Beijing, China) for 1 h at 37°C. Each section
was washed as before and visualized using the chromogen
diaminobenzidine. Finally, prior to being dehydrated and mounted,
the slides were counterstained with hematoxylin for two min at room
temperature and then were observed at −200 magnification using a
light microscope.
DNA extraction and
amplification-refractory mutation system (ARMS) assay
Each case was analyzed for the presence of EGFR and
KRAS mutations. DNA extraction was applied to the FFPE sections,
which was performed using a QIAamp DNA FFPE Tissue kit (Qiagen
GmbH, Hilden, Germany) according to the manufacturer's protocol.
The presence of EGFR and KRAS mutations was detected using the
Human EGFR Gene Mutation Detection kit and the Human KRAS Gene
Mutation Detection kit (Fluorescent polymerase chain reaction; both
from Beijing ACCB Biotech Ltd., Beijing, China), which was approved
by the State Food and Drug Administration for clinical application
in China. Polymorphisms of the EGFR gene in exon 19 (E19del), 20
(T790M S768I and E20ins) and 21 (L861Q and L858R), and KRAS gene in
exon 2 were detected. Analysis of the presence of these mutations
was performed using a LightCycler480 (Roche Diagnostic, Basel,
Switzerland).
Interpretation of protein
expression
Two independent well-experienced pathologists
without knowledge of the clinicopathological information of each
patient assessed the VEGFA, VEGFR1 and VEGFR2 expression. For the
expression of VEGFA and VEGFR1, each slide was evaluated according
to the staining intensity and percentage of positive tumor cells.
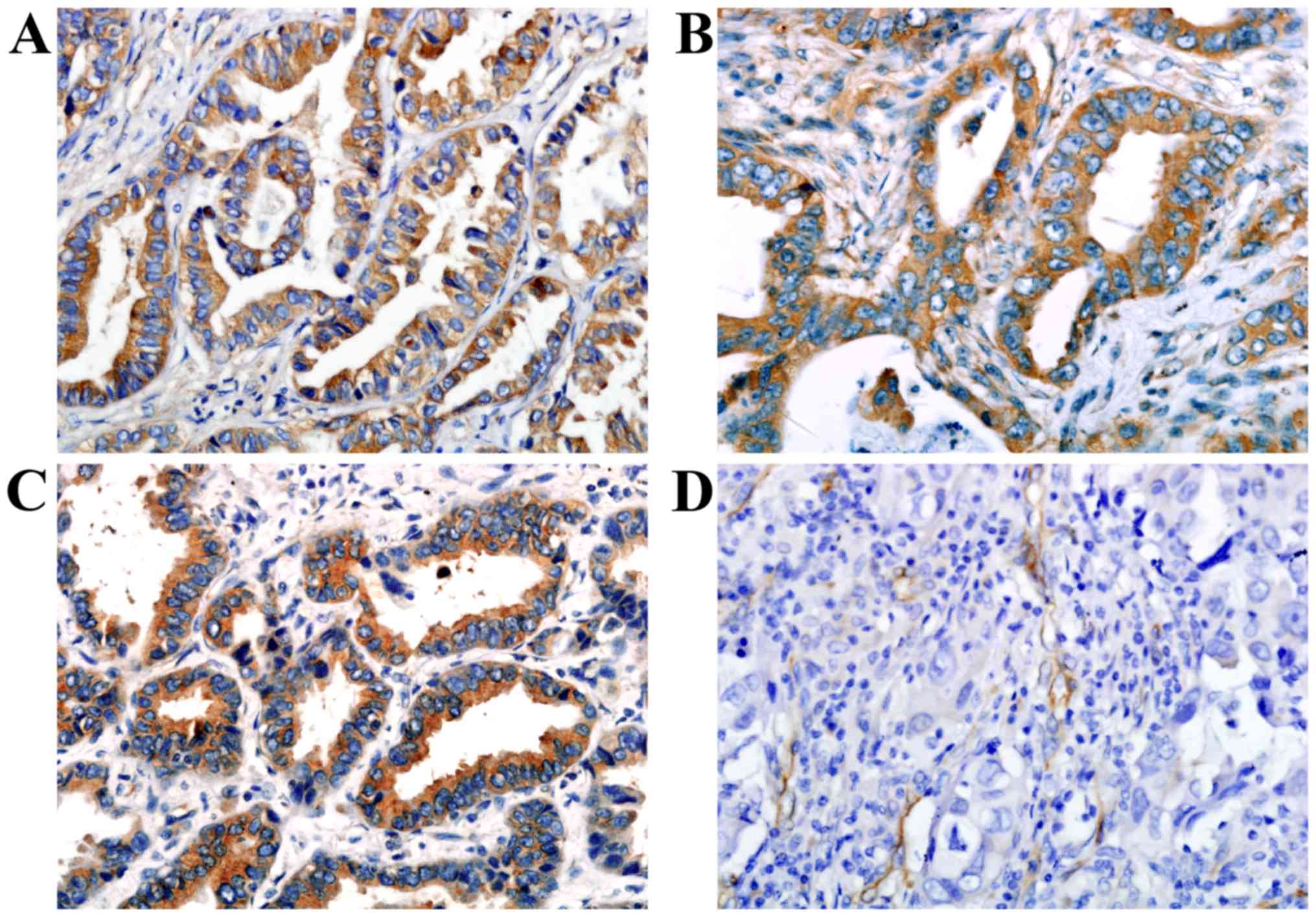
Scores for the staining intensity were classified as 0 (negative),
1 (weak), 2 (moderate) and 3 (strong) (Fig. 1). Scores for the percentage of tumor
cells for 0–10, 11–25, 26–50, 51–75 and >75% were classified as
0, 1, 2, 3 and 4, respectively. The scores of the staining
intensity was multiplied by the scores of the percentage of stained
cells (0–100%). The finally weighted scores of 0–1, 2–3, 4–6 and
7–12 were classified as-, +, ++ and +++, respectively (20). The samples that had weighted scores of
0–1 were classified as negative expression and the remaining
samples which had weighted scores >2 were classified as positive
expression.
Samples were defined as positive for VEGFR2
cytoplasmic staining when ≥5% of the tumor cells presented weak,
moderate or strong expression. Samples were defined as positive for
VEGFR2 vascular staining when the number of positive vessels was
>2 (21). Positive expression of
VEGFR2 was defined as either positivity in the tumor cells or in
the tumor stromal vasculature.
Statistical analysis
SPSS statistical software (version 17; SPSS, Inc.,
Chicago, IL, USA) was used for statistical analysis. P<0.05 was
considered to indicate a statistically significant difference.
Associations between clinicopathological variables (sex, age,
smoking status, lymph node metastasis and clinical stage) and
EGFR/KRAS mutant status or VEGFA/VEGFR1/VEGFR2 expression were
analyzed using Pearson's χ2 tests, which was also used
to evaluate the association between VEGFA/VEGFR1/VEGFR2 expression
and mutations in EGFR and KRAS. The correlation between VEGFA,
VEGFR1 and VEGFR2 expression was analyzed using Spearman's rank
correlation coefficient.
Results
VEGFA, VEGFR1 and VEGFR2 expression,
and their association with the clinicopathological characteristics
of patients with lung adenocarcinoma
VEGFA and VEGFR1 staining were localized primarily
to the tumor cell cytoplasm, while VEGFR2 was localized to the
tumor cell cytoplasm and tumor stromal vasculature (Fig. 1). Of the 204 adenocarcinoma samples,
140/204 (68.6%), 141/204 (69.1%) and 98/204 (48.0%) were identified
as for positive VEGFA, VEGFR1 and VEGFR2 expression, respectively.
No significant associations were revealed between the expression of
each and age, sex, smoking history, lymph node metastasis or
clinical stage (Table I). Of all
VEGFR1 positive cases, 77/141 (54.6%) exhibited VEGFR2 positive
expression. Of all VEGFR1 negative cases, 42/63 (66.7%) exhibited
VEGFR2 negative expression. VEGFR1 expression was significantly
correlated with VEGFR2 expression (r=0.247; P<0.001; Table II). However, no associations between
VEGFA expression and its receptors were revealed.
 | Table I.Association between
clinicopathological characteristics and VEGFA, VEGFR1 or VEGFR2
expression in patients with lung adenocarcinoma. |
Table I.
Association between
clinicopathological characteristics and VEGFA, VEGFR1 or VEGFR2
expression in patients with lung adenocarcinoma.
|
|
| VEGFA | VEGFR1 | VEGFR2 |
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
characteristics | No. | Positive No.
(%) | P-value | Positive No.
(%) | P-value | Positive No.
(%) | P-value |
|---|
| Sex |
|
|
|
|
|
|
|
|
Male | 105 | 67 (63.8) | 0.127 | 71 (67.6) | 0.633 | 49 (46.7) | 0.686 |
|
Female | 99 | 73 (73.7) |
| 70 (70.7) |
| 49 (49.5) |
|
| Age (years) |
|
|
|
|
|
|
|
|
≤60 | 111 | 80 (72.1) | 0.247 | 78 (70.3) | 0.697 | 50 (45.0) | 0.350 |
|
>60 | 93 | 60 (72.1) |
| 63 (67.7) |
| 48 (51.6) |
|
| Smoking
history |
|
|
|
|
|
|
|
|
Non-smokers | 108 | 80 (72.1) | 0.075 | 74 (68.5) | 0.844 | 53 (49.1) | 0.754 |
|
Smokers | 96 | 60 (62.5) |
| 67 (69.8) |
| 45 (46.9) |
|
| Lymph node
metastasis |
|
|
|
|
|
|
|
|
Absent | 135 | 91 (67.4) | 0.599 | 97 (71.9) | 0.237 | 68 (50.4) | 0.351 |
|
Present | 69 | 49 (70.1) |
| 44 (63.8) |
| 30 (43.5) |
|
| TNM stage |
|
|
|
|
|
|
|
|
I+II | 141 | 95 (67.4) | 0.727 | 102 (72.3) | 0.136 | 68 (48.2) | 0.936 |
|
III | 63 | 44 (69.8) |
| 39 (61.9) |
| 30 (47.6) |
|
| I | 120 | 80 (66.7) | 0.471 | 87 (72.5) | 0.211 | 59 (49.2) | 0.700 |
|
II+III | 84 | 60 (71.4) |
| 54 (64.3) |
| 39 (46.4) |
|
 | Table II.Correlation between VEGFR1 and VEGFR2
expression in lung adenocarcinoma. |
Table II.
Correlation between VEGFR1 and VEGFR2
expression in lung adenocarcinoma.
|
| VEGFR2 |
|
|
|
|---|
|
|
|
|
|
|
|---|
| VEGFR1 | Negative | Positive | Total | rs | P-value |
|---|
| – | 42 | 21 | 63 | 0.247 | P<0.001 |
| + | 17 | 9 | 26 |
|
|
| ++ | 19 | 23 | 42 |
|
|
| +++ | 28 | 45 | 73 |
|
|
| Total | 106 | 98 | 204 |
|
|
EGFR/KRAS mutations and the
association with the clinicopathological characteristics of
patients with lung adenocarcinoma
Among the total 204 cases, 104 (51.0%) mutated EGFR
and 19 (9.3%) mutated KRAS (exon 2) were identified. Of the 104
cases, there were 44 exon-19 mutations, 13 exon-20 mutations and 47
exon-21 mutations.
In female patients, EGFR mutation frequency was
62.6% (62/99) which was significantly higher compared with male
patients (40.0%; 42/105; P=0.001), and in non-smokers, the
frequency of EGFR mutations was 63.0% (68/108) which was
significantly higher compared with smokers (37.5%; 36/96;
P<0.001). It was concluded that there was a significant
association between EGFR mutation status and sex and smoking
history, but there was no notable association between EGFR
mutations and age, lymph node metastasis, or clinical stage
involvement (Table II). In male
patients, KRAS mutation frequency was 13.3% (14/105) which was
significantly higher compared with female patients (5.1%; 5/99;
P=0.042) and in smokers, the frequency of KRAS mutations was 13.5%
(13/96) which was significantly higher compared with non-smokers
(5.6%, 6/108; P=0.050). It was concluded that there were
significant associations between KRAS mutation status and sex and
smoking history. However, there was no significant association
identified between KRAS mutation status and age, lymph node
metastasis or clinical stage involvement (Table III).
 | Table III.Association between
clinicopathological characteristics and EGFR and KRAS mutations in
patients with lung adenocarcinoma. |
Table III.
Association between
clinicopathological characteristics and EGFR and KRAS mutations in
patients with lung adenocarcinoma.
|
|
| Mutant EGFR | Mutant KRAS |
|---|
|
|
|
|
|
|---|
| Clinicopathological
characteristics | No. | No. (%) | P-value | No. (%) | P-value |
|---|
| Sex |
|
|
|
|
|
|
Male | 105 | 42 (40.0) | 0.001 | 14 (13.3) | 0.042 |
|
Female | 99 | 62 (62.6) |
| 5 (5.1) |
|
| Age (years) |
|
|
|
|
|
|
≤60 | 111 | 58 (52.3) | 0.691 | 5 (4.5) | 0.100 |
|
>60 | 93 | 46 (49.5) |
| 14 (15.1) |
|
| Smoking
history |
|
|
|
|
|
|
Non-smokers | 108 | 68 (63.0) | <0.001 | 6 (5.6) | 0.050 |
|
Smokers | 96 | 36 (37.5) |
| 13 (13.5) |
|
| Lymph node
metastasis |
|
|
|
|
|
|
Absent | 135 | 71 (52.6) | 0.519 | 11 (8.1) | 0.423 |
|
Present | 69 | 33 (47.8) |
| 8 (11.6) |
|
| TNM stage |
|
|
|
|
|
|
I+II | 141 | 73 (51.8) | 0.735 | 14 (9.9) | 0.651 |
|
III | 63 | 31 (49.2) |
| 5 (7.9) |
|
| I | 120 | 65 (54.2) | 0.277 | 10 (8.3) | 0.565 |
|
II+III | 84 | 39 (46.4) |
| 9 (10.7) |
|
Correlation between each subtype of
EGFR mutation and the expression of VEGFA, VEGFR1 and VEGFR2
EGFR 20 and 21 exon mutation frequency in
VEGFA-positive samples was 8.6% (12/140) and 29.3% (41/140),
respectively. This was significantly higher compared with in the
VEGFA-negative samples (1.6%, 1/64; P=0.033) and (9.4%, 6/64;
P=0.002). EGFR 19 exon mutation frequency in VEGFA-positive samples
was 20.0% (28/140), which was insignificantly lower compared with
VEGFA-negative samples (25.0%; 16/64; P=0.420). Additionally, the
EGFR 19 exon mutation frequency in VEGFR1-positive samples was
15.6% (22/141), significantly lower compared with VEGFR1-negative
samples (31.7%; 20/63; P=0.008). A high level of VEGFA and VEGFR1
co-expression was significantly correlated with EGFR 21 exon
mutation (P<0.001). However, there was no significant
associations between VEGFR2 expression or the co-expression of
VEGFA/VEGFR1/VEGFR2 and each subtype of EGFR mutation status
(Table IV).
 | Table IV.Association between subtype of
EGFR/KRAS mutation and expression of VEGFA, VEGFR1 and VEGFR2 in
lung adenocarcinoma. |
Table IV.
Association between subtype of
EGFR/KRAS mutation and expression of VEGFA, VEGFR1 and VEGFR2 in
lung adenocarcinoma.
|
|
| Mutant EGFR | Mutant KRAS |
|---|
|
|
|
|
|
|---|
|
|
| EGFR 19 exon | EGFR 20 exon | EGFR 21 exon | KRAS 2 exon |
|---|
|
|
|
|
|
|
|
|---|
| Protein | No. | No. (%) | P-value | No. (%) | P-value | No. (%) | P-value | No. (%) | P-value |
|---|
| VEGFA |
|
|
|
|
|
|
|
|
|
|
Positive | 140 | 28 (20.0) | 0.420 | 12 (8.6) | 0.033 | 41 (29.3) | 0.002 | 14 (10.0) | 0.618 |
|
Negative | 64 | 16 (25.0) |
| 1 (1.6) |
| 6 (9.4) |
| 5 (7.8) |
|
| VEGFR1 |
|
|
|
|
|
|
|
|
|
|
Positive | 141 | 22 (15.6) | 0.008 | 7 (5.0) | 0.233 | 36 (25.5) | 0.206 | 16 (11.3) | 0.114 |
|
Negative | 63 | 20 (31.7) |
| 6 (9.5) |
| 11 (17.5) |
| 3 (4.8) |
|
| VEGFR2 |
|
|
|
|
|
|
|
|
|
|
Positive | 98 | 23 (23.5) | 0.526 | 7 (7.1) | 0.665 | 23 (23.5) | 0.776 | 12 (12.2) | 0.166 |
|
Negative | 106 | 21 (19.8) |
| 6 (5.7) |
| 24 (22.6) |
| 7 (6.6) |
|
|
VEGFA/VEGFR1a |
|
|
|
|
|
|
|
|
|
|
Positive | 87 | 14 (16.1) | 0.101 | 8 (9.2) | 0.792 | 30 (34.5) | <0.001 | 12 (13.8) | 0.058 |
|
Negative | 117 | 30 (25.6) |
| 7 (6.0) |
| 17 (14.5) |
| 7 (6.0) |
|
|
VEGFA/VEGFR1/VEGFR2b |
|
|
|
|
|
|
|
|
|
|
Positive | 53 | 9 (17.0) | 0.345 | 4 (7.5) | 0.689 | 15 (28.3) | 0.290 | 9 (16.7) | 0.035 |
|
Negative | 151 | 35 (23.2) |
| 9 (6.0) |
| 32 (21.2) |
| 10 (6.6) |
|
Association between KRAS mutation, and
the expression of VEGFA, VEGFR1 and VEGFR2
KRAS mutation frequency in the VEGFA-positive
samples was 10.0% (14/140), which was higher compared with the
negative samples (7.8%; 5/64), but with no statistical significance
between them (P=0.618). KRAS mutation frequency in the
VEGFR1-positive samples was 11.3% (16/141), which was
insignificantly higher compared with the VEGFR1-negative samples
(4.8%; 3/63) (P=0.114). KRAS mutation frequency in the
VEGFR2-positive samples was 12.2% (12/98), being insignificantly
higher compared with the VEGFR2 samples (6.6%; 7/106) (P=0.166).
However, it was revealed that a high level of co-expression of
VEGFA, VEGFR1 and VEGFR2 was significantly associated with KRAS
mutation (P=0.035; Table IV).
Discussion
According to characterizations by the driver gene
mutation, patients with NSCLC have different features and may
benefit from targeted therapies. There has been great improvements
in the targeted therapeutic outcome for selected patient groups
based on driver gene mutations (22).
However, the application of TKIs remains with numerous limitations
at present. A majority of patients with KRAS or EGFR (exon 20)
mutations may not benefit from EGFR inhibition (23–25).
Meanwhile, due to the complex network that drives KRAS tumors, a
combinatorial multi-target/multi-pathway inhibitory approach may be
necessary to modulate cell growth in patients with KRAS mutant
NSCLC (26–30). The present study focused on EGFR/KRAS
mutations in patients with lung adenocarcinoma and the expression
of a number of angiogenic proteins, and analyzed the
clinicopathological features of these patients in order to better
define their characteristics. It may provide further evidence for
the use of certain molecular markers for targeted therapy, namely
EGFR/KRAS, VEGFA, VEGFR1 and VEGFR2.
In all 204 cases of patients with lung
adenocarcinoma included in the present study, high expression rates
of VEGFA (68.6%), VEGFR1 (69.1%) and VEGFR2 (48%) were identified.
The mutation rates of EGFR exons (19–21) and
KRAS exon 2 were 51.0 and 9.3%, respectively. The proportion of
exon 19, 20 and 21 mutations were 21.6, 6.4 and 23.0%,
respectively. Consistent with previous studies (3,31), EGFR
mutations occur more frequently in non-smokers and female patients,
and KRAS mutations exist more commonly in smokers and male
patients. It was revealed that the expression of VEGFR1 was
significantly correlated with that of VEGFR2. However, no
associations were revealed between the expression of VEGFA and
receptors.
Reinmuth et al (32) observed that mutant EGFR tumors,
without exposing mutant subtypes, represented a higher level of
VEGFA expression. Clarke et al (33) demonstrated that mutant EGFR enhanced
the induction of VEGF by hypoxia and insulin-like growth factor-1
via a PI3 kinase-dependent pathway. However, the association
between different subtypes of EGFR mutation status and VEGFA or RTK
expression have seldom been revealed. In the present study, it was
revealed that all patients with lung adenocarcinoma harboring
either EGFR 20 or 21 exon mutations had a high level of VEGFA
expression. However, there was no association between EGFR 19
mutation and VEGFA expression. It may provide the suggestion that
patients harboring either EGFR 20 or 21 exon mutations ought to
have the priority of anti-VEGFA targeted therapy.
In the present study, it was observed that the high
level of co-expression of VEGFA and VEGFR1 were significantly
associated with the EGFR 21 mutation. Zhang et al (16) revealed that VEGF-induced accumulation
of VEGFR1 occurs through Akt and ERK signaling. Owing to the high
level of co-expression of VEGFA and VEGFR1, it may provide the
potential for EGFR 21 mutant patients to receive inhibitors of the
Akt and ERK signaling pathway to downregulate VEGFR1, which further
reduces the combination of VEGFA and VEGFR1. Notably, it was
identified that the EGFR 19 exon mutation frequency in the
VEGFA-positive cases was only 20% (28/140), lower compared with
that in the negative samples (25%, 16/64), despite the lack of
statistical significance between them (P=0.42). In addition, lower
VEGFR1 expression was significantly associated with the EGFR 19
exon mutation. Liu et al (34)
demonstrated that compared with patients with EGFR exon 21
mutations, patients with EGFR exon 19 mutations exhibit an
increased objective response rate, progression-free survival time
and overall survival time following EGFR-TKI therapy. Further
studies may be required to explore whether there is a potential
association between the two phenomena.
Additionally, there was no significant association
identified between VEGFR2 expression and each subtype of EGFR or
KRAS mutation. To the best of our knowledge, only a few previous
studies have reported the potential association between KRAS gene
status and RTK (VEGFR1 and VEGFR2) expression (35,36), and
there were no associated reports on the correlation of EGFR gene
status and RTK (VEGFR1 and VEGFR2) expression. Schimanski et
al (35) found that KRAS mutation
can increase the expression of VEGFR1 and VEGFR2 in colorectal
cancer, but the mechanisms remain unknown. Further validation of
the associations identified between RTK expression, EGFR and KRAS
mutant status in a larger cohort of patients with lung
adenocarcinoma, in addition to further studies on the mechanism are
warranted.
A number of studies have reported that VEGFA
expression may be upregulated by oncogene activation of KRAS in
different tumor types (37) and KRAS
mutation upregulates VEGF through PI3K-dependent pathways in colon
cancer cells (38). However, in the
present study, no significant association between KRAS mutation
status and individual expression of VEGFA, VEGFR1 or VEGFR2 was
identified. Notably, the co-expression of VEGFA, VEGFR1 and VEGFR2
presented in 26% of the cases, and demonstrated a statistically
significant association with the presence of KRAS mutations (16.7%
with KRAS mutations vs. 6.6% with KRAS wild type). The different
results from previous studies may be ascribed to a number of
reasons. First, the pathological features varied among different
studies, and retrospectively collected data resulted in a potential
bias, such as selection bias. Second, the heterogeneity of tumor
tissue among studies resulted in different conclusions. Finally, it
suggests that only in a number of ‘more active’ neoplasms with KRAS
stimulation may evident associations be identified between KRAS
gene and numerous proteins, including VEGFA and RTKs.
Conclusively, the upregulation of VEGFA may be
associated with different types of EGFR mutation. Low level
expression of VEGFR1 is more likely to be associated with EGFR 19
exon mutations. High level co-expression of VEGFA and VEGFR1 is
associated with EGFR 21 exon mutations, and the high level of
co-expression of VEGFA, VEGFR1 and VEGFR2 is associated with KRAS
mutations. It remains requisite to evaluate the exact benefit of
anti-angiogenesis therapy in patients with different RTK
expression. However, confirmation of the different subtypes of EGFR
and KRAS mutation status may provide the reference to predict
anti-angiogenesis therapeutic effects, and the resistance by
neoplasm.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Tianjin
Municipal Science and Technology Commission Project (grant nos.
11JCYBJC11300 and 12ZCDZSY15600), the CSCO Project (grant no.
Y-S2014-011) and the Tianjin Municipal Key Technology R&D
Program of the Ministry of Science and Technology (grant no.
13ZCZCSY20300).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KL and XY contributed to the conception and design
of the study and wrote the manuscript. XY, JY, XZ and TQ performed
the experiments. XY, KL and XW analyzed clinical data, and KL
performed quality control. All authors reviewed the manuscript and
approved the final authorship.
Ethics approval and consent to
participate
This study was approved by the Ethics and Scientific
Committee of Tianjin Medical University Cancer Hospital. According
to the rules set by the Declaration of Helsinki, all patients knew
the purpose of the study. Collection and use of tumor tissue
samples for research received written informed consent from all
patients prior to the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li S, Choi YL, Gong Z, Liu X, Lira M, Kan
Z, Oh E, Wang J, Ting JC, Ye X, et al: Comprehensive
characterization of oncogenic drivers in asian lung adenocarcinoma.
J Thorac Oncol. 11:2129–2140. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maemondo M, Inoue A, Kobayashi K, Sugawara
S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I,
et al: Gefitinib or chemotherapy for non-small-cell lung cancer
with mutated EGFR. N Engl J Med. 362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rosell R, Carcereny E, Gervais R,
Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R,
Pallares C, Sanchez JM, et al: Erlotinib versus standard
chemotherapy as first-line treatment for European patients with
advanced EGFR mutation-positive non-small-cell lung cancer
(EURTAC): A multicentre, open-label, randomised phase 3 trial.
Lancet Oncol. 13:239–246. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun S, Schiller JH, Spinola M and Minna
JD: New molecularly targeted therapies for lung cancer. J Clin
Invest. 117:2740–2750. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mantovani A, Romero P, Palucka AK and
Marincola FM: Tumour immunity: Effector response to tumour and role
of the microenvironment. Lancet. 371:771–783. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Solan MJ and Werner-Wasik M: Prognostic
factors in non-small cell lung cancer. Semin Surg Oncol. 21:64–73.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ferrara N: The role of VEGF in the
regulation of physiological and pathological angiogenesis. Exs. pp.
209–231. 2005
|
|
10
|
Mattern J, Koomagi R and Volm M:
Association of vascular endothelial growth factor expression with
intratumoral microvessel density and tumour cell proliferation in
human epidermoid lung carcinoma. Br J Cancer. 73:931–934. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Seto T, Higashiyama M, Funai H, Imamura F,
Uematsu K, Seki N, Eguchi K, Yamanaka T and Ichinose Y: Prognostic
value of expression of vascular endothelial growth factor and its
flt-1 and KDR receptors in stage I non-small-cell lung cancer. Lung
Cancer. 53:91–96. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hicklin DJ and Ellis LM: Role of the
vascular endothelial growth factor pathway in tumor growth and
angiogenesis. J Clin Oncol. 23:1011–1027. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lohela M, Bry M, Tammela T and Alitalo K:
VEGFs and receptors involved in angiogenesis versus
lymphangiogenesis. Curr Opin Cell Biol. 21:154–165. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Coultas L, Chawengsaksophak K and Rossant
J: Endothelial cells and VEGF in vascular development. Nature.
438:937–945. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shibuya M: Differential roles of vascular
endothelial growth factor receptor-1 and receptor-2 in
angiogenesis. J Biochem Mol Biol. 39:469–478. 2006.PubMed/NCBI
|
|
16
|
Zhang Z, Neiva KG, Lingen MW, Ellis LM and
Nor JE: VEGF-dependent tumor angiogenesis requires inverse and
reciprocal regulation of VEGFR1 and VEGFR2. Cell Death Differ.
17:499–512. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kowanetz M and Ferrara N: Vascular
endothelial growth factor signaling pathways: Therapeutic
perspective. Clin Cancer Res. 12:5018–5022. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ellis LM and Hicklin DJ: VEGF-targeted
therapy: Mechanisms of anti-tumour activity. Nat Rev Cancer.
8:579–591. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chansky K, Sculier JP, Crowley JJ, Giroux
D, van Meerbeeck J and Goldstraw P; International Staging Committee
and Participating Institutions: The International Association for
the study of lung cancer staging project. Prognostic factors and
pathologic TNM stage in surgically managed non-small cell lung
cancer. Zhongguo Fei Ai Za Zhi. 4:792–801. 2009.(In Chinese).
|
|
20
|
Chen P, Zhu J, Liu DY, Li HY, Xu N and Hou
M: Over-expression of survivin and VEGF in small-cell lung cancer
may predict the poorer prognosis. Med Oncol. 31:7752014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Holzer TR, Fulford AD, Reising LO,
Nedderman DM, Zhang X, Benjamin LE, Schade AE and Nasir A:
Profiling of vascular endothelial growth factor receptor
heterogeneity identifies protein expression-defined subclasses of
human non-small cell lung carcinoma. Anticancer Res. 36:3277–3288.
2016.PubMed/NCBI
|
|
22
|
de Mello RA, Madureira P, Carvalho LS,
Araújo A, O Brien M and Popat S: EGFR and KRAS mutation, and ALK
fusioin: current developments and personalized therapies for
patients with advanced non-small-cell ling cancer.
Pharmacogenomics. 14:1765–1777. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Paez JG, Jänne PA, Lee JC, Tracy S,
Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et
al: EGFR mutations in lung cancer: Correlation with clinical
response to gefitinib therapy. Science. 304:1497–1500. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, et al: Activating mutations in the
epidermal growth factor receptor underlying responsiveness of
non-small-cell lung cancer to gefitinib. N Engl J Med.
350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mitsudomi T, Kosaka T, Endoh H, Horio Y,
Hida T, Mori S, Hatooka S, Shinoda M, Takahashi T and Yatabe Y:
Mutations of the epidermal growth factor receptor gene predict
prolonged survival after gefitinib treatment in patients with
non-small-cell lung cancer with postoperative recurrence. J Clin
Oncol. 23:2513–2520. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
E J, Xing J, Gong H, He J and Zhang W:
Combine MEK inhibition with PI3K/mTOR inhibition exert inhibitory
tumor growth effect on KRAS and PIK3CA mutation CRC xenografts due
to reduced expression of VEGF and matrix metallopeptidase-9. Tumour
Biol. 36:1091–1097. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hirai H, Sootome H, Nakatsuru Y, Miyama K,
Taguchi S, Tsujioka K, Ueno Y, Hatch H, Majumder PK, Pan BS and
Kotani H: MK-2206, an allosteric Akt inhibitor, enhances antitumor
efficacy by standard chemotherapeutic agents or molecular targeted
drugs in vitro and in vivo. Mol Cancer Ther. 9:1956–1967. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Engelman JA, Chen L, Tan X, Crosby K,
Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y,
et al: Effective use of PI3K and MEK inhibitors to treat mutant
Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med.
14:1351–1356. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Meng J, Dai B, Fang B, Bekele BN, Bornmann
WG, Sun D, Peng Z, Herbst RS, Papadimitrakopoulou V, Minna JD, et
al: Combination treatment with MEK and AKT inhibitors is more
effective than each drug alone in human non-small cell lung cancer
in vitro and in vivo. PLoS One. 5:e141242010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Corcoran RB, Cheng KA, Hata AN, Faber AC,
Ebi H, Coffee EM, Greninger P, Brown RD, Godfrey JT, Cohoon TJ, et
al: Synthetic lethal interaction of combined BCL-XL and MEK
inhibition promotes tumor regressions in KRAS mutant cancer models.
Cancer Cell. 23:121–128. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
de Mello RA, Marques DS, Medeiros R and
Araujo AM: Epidermal growth factor receptor and K-Ras in non-small
cell lung cancer-molecular pathways involved and targeted
therapies. World J Clin Oncol. 2:367–376. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Reinmuth N, Jauch A, Xu EC, Muley T,
Granzow M, Hoffmann H, Dienemann H, Herpel E, Schnabel PA, Herth
FJ, et al: Correlation of EGFR mutations with chromosomal
alterations and expression of EGFR, ErbB3 and VEGF in tumor samples
of lung adenocarcinoma patients. Lung Cancer. 62:193–201. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Clarke K, Smith K, Gullick WJ and Harris
AL: Mutant epidermal growth factor receptor enhances induction of
vascular endothelial growth factor by hypoxia and insulin-like
growth factor-1 via a PI3 kinase dependent pathway. Br J Cancer.
84:1322–1329. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu Y, Ren Z, Wang J and Zhang S:
Epidermal growth factor receptor-tyrosine kinase inhibitor therapy
is especially beneficial to patients with exon 19 deletion compared
with exon 21 L858R mutation in non-small-cell lung cancer:
Systematic review and meta analysis. Thorac Cancer. 7:406–414.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Schimanski CC, Zimmermann T, Schmidtmann
I, Gockel I, Lang H, Galle PR, Moehler M and Berger MR: K-ras
mutation status correlates with the expression of VEGFR1, VEGFR2,
and PDGFRalpha in colorectal cancer. Int J Colorectal Dis.
25:181–186. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mancikova V, Inglada-Pérez L,
Curras-Freixes M, de Cubas AA, Gómez Á, Letón R, Kersten I,
Leandro-García LJ, Comino-Méndez I, Apellaniz-Ruiz M, et al: VEGF,
VEGFR3, and PDGFRB protein expression is influenced by RAS
mutations in medullary thyroid carcinoma. Thyroid. 24:1251–1255.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Krajnović M, Marković B, Knežević-Ušaj S,
Nikolić I, Stanojević M, Nikolić V, Šiljić M, Jovanović Ćupić S and
Dimitrijević B: Locally advanced rectal cancers with simultaneous
occurrence of KRAS mutation and high VEGF expression show invasive
characteristics. Pathol Res Pract. 212:598–603. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kang YH, Kim KS, Yu YK, Lim SC, Kim YC and
Park KO: The relationship between microvessel count and the
expression of vascular endothelial growth factor, p53, and K-ras in
non-small cell lung cancer. J Korean Med Sci. 16:417–423. 2001.
View Article : Google Scholar : PubMed/NCBI
|