Introduction
Breast cancer, a heterogeneous tumor, may be divided
into different subtypes with various manifestations. Among them,
triple-negative breast cancer (TNBC) involves tumors negative for
estrogen receptor (ER), progesterone receptor and human epidermal
growth factor receptor 2. The primary characteristics of TNBC
include: High incidence in young females; poor histological
differentiation; early visceral metastasis; reduced disease-free
survival; and high mortality rates (1,2). Despite
the aggressive clinical behavior and poor overall prognosis of
TNBC, the treatment of patients with TNBC remains inferior due to a
lack of known target genes and hormone receptors.
Immunotherapy, which aims to strengthen the capacity
of the immune system rather than methods that attack cancer cells
directly, such as chemotherapy and radiotherapy, is expected to
prolong patient survival. It is considered that immunotherapy may
revolutionize the treatment of cancer.
The provision of antigen-presenting cells (APCs) and
co-stimulation signals are essential in the activation of T
lymphocytes. Dendritic cells (DCs) are one of the most powerful
APCs and serve an important role in activating resting T
lymphocytes (3). The major
histocompatibility complex (MHC) combines with the surface proteins
of tumor antigens to form peptide-MHC, and DCs assist T cells in
recognizing the antigen, thus stimulating a cluster of
differentiation (CD)8+ cytotoxic T cell (CTL) response
and a CD4+ helper T cell response (4). DCs have the ability to activate T cells
by highly expressing MHC I/II molecules and by expressing a variety
of co-stimulating molecules; however, in patients with cancer, the
tumor microenvironment competes with their immune system to
maintain the growth and reproduction of cancer cells (5). The normal immune function becomes
damaged and exhibits the following features: Absence or dysfunction
of the MHC molecule; decline in the number of DCs and their
presentation capacity; and inhibition of T lymphocytes (5). All of these features contribute to an
imbalance of the immune system, and DC-based immunotherapy assists
in the restoration of this balance, providing a novel cancer
treatment option (6,7).
Different surface antigens were determined between
tumor cells and healthy cells in the 1960s, and these
tumor-associated antigens became the basis of cancer treatment
(8). Currently, DC vaccines for
breast cancer primary include a DC-fusion vaccine and a specific
antigen-loaded DC vaccine (9–12). Fusion vaccines present all of the
antigens on the surface of breast cancer cells to T cells, in order
to induce an immune response, whilst antigen-loaded DC vaccines
stably present antigens and continuously stimulate DCs to provide a
stronger antitumor effect (13,14).
Overall, identifying a specific antigen marker may result in a
novel breakthrough in TNBC treatment.
Runt-associated transcription factor 2 (Runx2) is a
type of oncogene that is unusually increased in prostate cancer and
breast cancer, and is associated with cancer progression (15–17). Runx2
is expressed in various mammary epithelial cell lines indicating a
potential role in the development of mammary glands; however,
downregulation is required in pregnancy for full differentiation
(18). Abnormal Runx2 expression
drives epithelial to mesenchymal transition in normal mammary
epithelial cells (19), disturbing
normal mammary gland structures, such that the normal breast tissue
exhibits malignant features and promoting the development of breast
cancer by enhancing tumor cell proliferation (20,21). Runx2
has the ability to also directly influence downstream effectors,
including matrix metalloproteinase (MMP) 9, MMP13 (22–24),
vascular endothelial growth factor (VEGF), parathyroid
hormone-associated protein levels (25), bone sialoprotein and osteopontin
(22), which results in a more
aggressive breast cancer (26,27). In
addition, the pro-angiogenic effect of Runx2 serves an important
role in promoting migration and invasion in tumors (28,29). Runx2
is also associated with hormones in breast cancer (26,30). The
loss of ERα may lead to enhanced invasiveness in breast cancer
cells (31); therefore, ERα is
considered to be a key protective factor for breast cancer cells,
whilst Runx2 and ERα antagonize each other through the ERαDNA
binding domain and partially through SNAI2 (26,27,30). In a
study on 123 patients with breast cancer, Runx2 was more active in
patients with c-erbB2 (HER2/c-neu)-negative breast cancer than the
positive ones (39 vs. 17%) (32).
These data may indicate that Runx2 may be a novel biomarker for
patients with TNBC.
In the present study, Runx2 served as a target to
induce immunotherapy against TNBC by preparing a Runx2-DC vaccine.
The aims were to evaluate the efficacy of the vaccine in activating
autologous lymphocytes in vitro and to observe the specific
anti-TNBC effects of the vaccine with the goal of providing a novel
therapeutic strategy for patients with TNBC.
Materials and methods
The use of human subjects was specifically approved
by the Clinical Research Ethics Committee of the Third Affiliated
Hospital of Sun Yat-sen University (Guangzhou, China). Guangzhou
Blood Center (Guangzhou, China) supplied the blood and recorded the
informed consent. Prior to donating blood, the volunteers were
informed and provided written informed consent for the scientific
research use of blood samples.
Cell cultures
MDA-MB-231 cells exhibited greater Runx2 expression
than non-TNBC cell lines in previous studies (32–34), thus
MDA-MB-231 was selected as the focus of the present study. Human
breast epithelial cell line MCF10A was purchased from the National
Infrastructure of Cell Line Resource (China; http://www.cellresource.cn/index.aspx). The cell line
was cultured in D/F 12 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 5% horse serum (Gibco;
Thermo Fisher Scientific, Inc.), insulin (10 µg/ml), hydrocortisone
(0.5 µg/ml) and epidermal growth factor (20 ng/ml) (PeproTech
China, Suzhou, China). The TNBC cell line MDA-MB-231 and the MCF7
cell line were purchased from the American Type Culture Collection
(ATCC) and cultured in Dulbecco's modified Eagle's medium (DMEM)
(Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine
serum (FBS) (Gibco; Thermo Fisher Scientific, Inc.). 293FT cells
(purchased from the ATCC) were also cultured in DMEM containing 10%
FBS. All the cell lines were negative for mycoplasma and were
maintained in a humidified environment at 37°C with 5%
CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from all three cell lines (MDA-MB-231,
MCF7 and MCF10A) were extracted by trizol (Invitrogen; Thermo
Fisher Scientific, Inc.) and transcribed into cDNA according to the
reverse transcription kit (PrimeScript RT Master Mix Perfect Real
Time; Takara Biotechnology Co., Ltd., Dalian, China) protocols. The
resultant cDNA mixed with the SYBR® Green PCR mix
(Takara Biotechnology Co., Ltd.) and primers of the target genes
were amplified and analyzed with the Applied Biosystems 7500 FAST
system (Thermo Fisher Scientific, Inc., Waltham MA, USA) according
to the manufacturer's protocol. The reaction conditions were as
follows: 95°C for 30 sec; followed by 95°C for 3 sec and 60°C for
30 sec for 40 cycles. GAPDH was used as an internal control. The
relative quantification 2−∆∆Cq method was used to
analyze the PCR data (35). The
primers were the following: Runx2 forward, 5′-CGGCCCTCCCTGAACTCT-3′
and reverse, 5′-TGCCTGCCTGGGGTCTGTA-3′; GAPDH forward,
5′-ATGTTCGTCATGGGTGTGAA-3′ and reverse, 5′-TGTGGTCATGAGTCCTTCCA-3′.
All experiments were repeated in triplicate.
Western blot analysis
Runx2 protein expression was evaluated in all three
cell lines. Cells were lysed in lysis buffer (Sigma-Aldrich; Merck
KGaA, Damstadt, Germany) and quantified via a bicinchoninic acid
assay (Thermo Fisher Scientific, Inc.). A total of 20 µg of extract
was loaded and resolved on a 10% SDS-PAGE gel and transferred to a
polyvinylidene fluoride (PVDF) membrane using the Bio-Rad protein
transferring apparatus (Bio-Rad Laboratories, Inc., Hercules, CA,
USA). The PVDF membrane was removed and blocked with 5% bovine
serum albumin (Gibco; Thermo Fisher Scientific, Inc.) for 1–2 h at
room temperature (RT). The membrane was incubated with a monoclonal
antibody against Runx2 (1:1,000; cat. no. ab769560; Abcam,
Cambridge, UK) overnight at 4°C, followed by secondary antibody
incubation (Rabbit Anti-mouse horseradish peroxidase conjugated;
1:2,000; cat. no. bs-0296R-HRP; Beijing Boaosen Biotechnology Co.,
Ltd., Beijing, China) at RT for 1 h and SuperSignal West Pico Trial
Kit for subsequent detection (Thermo Fisher Scientific, Inc.). The
internal reference was GAPDH. The protein expression were detected
by ChemiDoc MP V3 Western Workflow for Midi Gels (Bio-Rad
Laboratories, Inc.) and analyzed by GraphPad Prism v.5.0 (GraphPad
Software, USA).
Immunocytochemistry
A total of 2×104 cells/ml in each cell
line group were seeded on disinfected coverslips in 6-well plates,
followed by fixation with 4% poly-stained formaldehyde for 15 min
at RT until the cells were grown to 60–70% confluence. Following
this, the catalase was eliminated from the cells by incubating with
0.5% Trion X-100 at RT for 20 min, followed by 3%
H2O2 at 37°C fo between 10 and 15 min. The
cells were blocked with 5% goat serum (Beijing Boaosen
Biotechnology Co., Ltd.) for 5–10 min at RT. Anti-Runx2 mouse mAb
(cat. no. ab76956; Abcam, Cambridge, UK) at a 1:1,000 dilution was
used as the primary antibody for the treatment group while antibody
diluent (Beijing Boaosen Biotechnology Co., Ltd.) was added in
negative group, and incubated at 37°C for 1 h. Next, the sections
were incubated with a secondary antibody (ChemMate EnVision
Detection kit, horseradish peroxidase/DAB conjugated, rabbit/mouse,
cat. no. GK500705/10; Dako; Agilent Technologies, Inc., Santa
Clara, Clara, CA, USA) for 30 min at 37°C, processed with the
3,3′-diaminobenzidine provided in the manufacturer's kit and
counterstained using hematoxylin for 1–2 mins at RT. Electron
microscopy (Olympus Corporation, Tokyo, Japan) was used to observe
the cells at a magnification of ×100.
Generation and culture of DCs
Human peripheral venous blood was obtained from the
Guangzhou Blood Center and heparinized under aseptic conditions.
Ficoll-Hypaque gradient centrifugation at 450 × g, 22°C for 25 min
was used to isolate peripheral blood mononuclear cells (PBMCs) from
peripheral blood. The isolated cells were replenished in sterile
6-well plates of 1×105 cells/well with serum-free
RPMI-1640 medium (Gibico, USA) at 37°C in a 5% CO2
incubator for 2 h. Non-adherent cells were collected for culture
reserves. Adherent cells were cultured in RPMI-1640 medium
containing recombinant human granulocyte macrophage-colony
stimulating factor (100 ng/ml) (Peprotech China) and recombinant
human interleukin-4 (rhIL-4; 50 ng/ml) (Peprotech China). The
medium was changed every other day. On day five, the
pro-inflammatory mediator tumor necrosis factor (TNF-α; 20 ng/ml)
(Peprotech China) was added, and the cells were incubated for
another two days. Finally, the suspension of cells were collected
as mature DCs.
Flow cytometry analysis
All of the day-7 DCs were collected and washed with
PBS, followed by blocking with 5% bovine serum albumin (Gibco;
Thermo Fisher Scientific, Inc.) for 20 min on ice, and then
incubating with 1:100 dilutions of antibodies against
CD11c-phycoerythrin (PE; cat. no. 555392; BD Biosciences, Franklin
Lakes, NJ, USA), CD83-fluorescein isothiocyanate (FITC; cat. no.
556910; BD Biosciences), CD86-FITC (cat. no. 557343; BD
Biosciences) and human leukocyte antigen-antigen D related
(HLA-DR)-PE (cat. no. 555561; BD Biosciences) and their isotype
controls IgG-1k-PE (cat. no. 562538; BD Biosciences) and
IgG-1k-FITC (cat. no. 555786; BD Biosciences) for 30 min at 4°C.
Cells were analyzed by a FACScan flow cytometer with CellQuest Pro
software (version 5.1; BD Biosciences) following a wash with
PBS.
DC transfection and detection
Runx2 lentivirus vector was purchased from Novo
Biological Company (Shanghai, China), and amplified according to
the plasmid extraction kit (Endo-free Plasmid Mini Kit II; Omega
Bio-Tek, Guangzhou, China). Plenti6 plasmids (150 ng/ul;
Invitrogen; Thermo Fisher Scientific, Inc.) were used to pack Runx2
lentivirus according to the manufacturer's specification. After
48–72 h, the supernatant was collected following centrifuging at
500 × g for 15 min at 4°C and passing through a 0.45 um filter. The
293FT cells were cultured in sterile 96-well plates of
1×105/ml in a humidified environment at 37°C with 5%
CO2. According to the expected titer of the virus, 8
sterile EP tubes were prepared and 90 µl serum-free DMEM was added
to each tube. In the first tube, 10 µl virus was added. A total of
10 µl was taken from the first tune and mixed with the second tube,
and this was repeated in the same manner until the last tube. Next,
90 µl diluted virus from each tube was joined with the 293FT cells
at day 2. Subsequent to 4 days, the results were observed under a
fluorescence microscope, then the last two wells in which
fluorescence could be observed were selected and the number of
fluorescent cells were counted. With Y being the number of
fluorescent cells counted in the last well as Y and X being the
number of fluorescent cells in the second-to-last well, the titer
(TU/ml)=(X+Y*10)*1,000/2/the virus fluid content (µl) of X. The
prepared day-2 DCs were divided into three groups as follows: Runx2
lentivirus was added to one group (transfected group); 293 cell
lysate (Abgent, USA) was added to another group (control group);
and no treatment was added to the last group (blank control group).
The cells were placed in RPMI-1640 medium 24 h following infection,
and DCs were cultured as aforementioned. After 48–72 h, green
fluorescent protein was observed by a fluorescence microscope at
×40 magnification. Transfection efficiency was assessed by RT-qPCR
and western blot analysis using the methods aforementioned.
Autologous T-lymphocyte culture
Nylon wool was used to purify the non-adherent PBMCs
as previously described, and B and NK cells were then removed,
leaving autologous T lymphocytes. T cells were cultured in
RPMI-1640 medium containing 10% FBS and rhIL-2 (20 ng/ml) at 37°C
in a 5% CO2 atmosphere.
Cytokine detection by ELISA
The cells were divided into TNBC groups
(DC-transfection group+MDA-MB-231; control group+MDA-MB-231; and
blank control group+MDA-MB-231), luminal breast cancer groups
(DC-transfection group+MCF7; control group+MCF7; and blank control
group+MCF7), normal breast cell groups (DC-transfection
group+MCF10A; control group+MCF10A; and blank control
group+MCF10A), effector cell groups (DC-transfection group; control
group; and blank control group) and target cell groups (MDA-MB-231;
MCF7; and MCF10A). The cells in each group were co-cultured with T
lymphocytes extracted previously at a ratio of 1:20 for 72 h in
24-well plates. The supernatant was collected to detect the
secretion of IL-12 and interferon (IFN)-γ using an ELISA kit (cat.
no. SEA111Hu for IL-12, SEA049Hu for IFN-γ; Wuhan USCN Business
Co., Ltd., Wuhan, China) in triplicate.
Cytotoxicity assay
The tumor-specific CTLs were stimulated following
the co-culture of each group of cells with T lymphocytes, and nylon
wool was used to harvest the CTLs again. MDA-MB-231 was selected as
the research focus, and the CTLs of the TNBC groups were incubated
with MDA-MB-231 at a ratio of 20:1 in a 96-well plate for 72 h as
the reactive cells. In order to understand what proportion of the
effective target ratio is more appropriate, the ratio was also set
to 5:1 and 10:1. At the time, the sole target cell group, sole
effector cell group were being cultured. PBS was used to dissolve
the purple formazan 2–3 times. Following the addition of MTT in
each group for 4 h at 37°C, the supernatant was removed. Absorbance
optical density (OD) values were detected using an enzyme-labeled
instrument at 490 nm. Detection in the MCF7 and MCF10A groups was
performed in a similar manner. The cytotoxicity rate was calculated
by the following formula: [1-(OD value of reactive cell well-OD
value of effector cell well)/(OD value of target cell well)]
×100%.
Statistical analysis
SPSS 19.0 (IBM Corp., Armonk, NY, USA) software was
used to analyze all the measured data. The data of western blot
were analyzed by GraphPad Prism v.5.0 (GraphPad Software, USA). The
other statistical differences were calculated by a two-way analysis
of variance and LSD post hoc test between groups. The data is
presented as the mean ± standard error of the mean, and three
repeats were conducted. P<0.05 was considered to indicate a
statistically significant difference.
Results
Runx2 expression in breast cells
Runx2 is considered to be associated with a
particular subtype of breast cancer cells (36); however, whether it can be regarded as
a biomarker of TNBC has not yet been concluded. Therefore, RT-qPCR
was used to detect the expression of Runx2 in various breast cancer
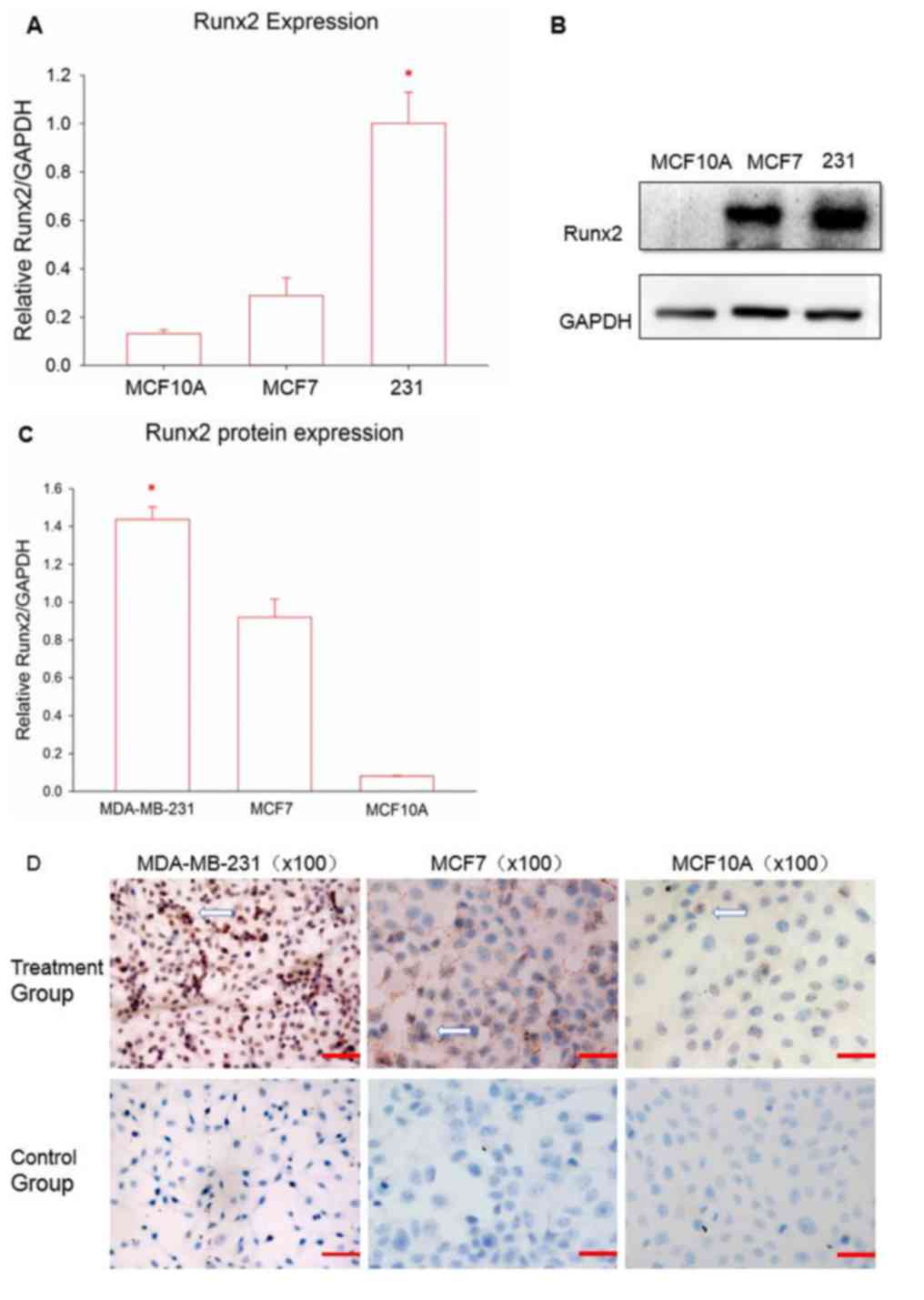
cell lines. Fig. 1A demonstrates the
results of Runx2 mRNA expression. Runx2 expression in the TNBC cell
line MDA-MB-231 was significantly higher compared with the other
cell lines, followed by the luminal breast cell line MCF7 and then
the normal breast cell line MCF10A (P<0.05). Following this,
western blot analysis was performed to examine whether Runx2
protein expression followed the same patterns. Runx2 protein levels
remained significantly higher in the TNBC cells, compared with the
other two cell lines (P<0.05; Fig. 1B
and C); Runx2 expression was also greater in MCF7 cells,
compared with MCF10A cells. Next, immunocytochemistry was used to
elucidate the distribution of the Runx2 protein (Fig. 1D). All of the cell lines demonstrated
positive staining for Runx2; however, the levels and distribution
were different. Normal breast cells indicated reduced Runx2
expression, compared with the other breast cancer cell lines, and
Runx2 was situated in the cell membrane, similar to MCF7. Runx2 in
the MDA-MB-231 cell line was almost completely distributed
throughout the nucleus with a high positive rate.
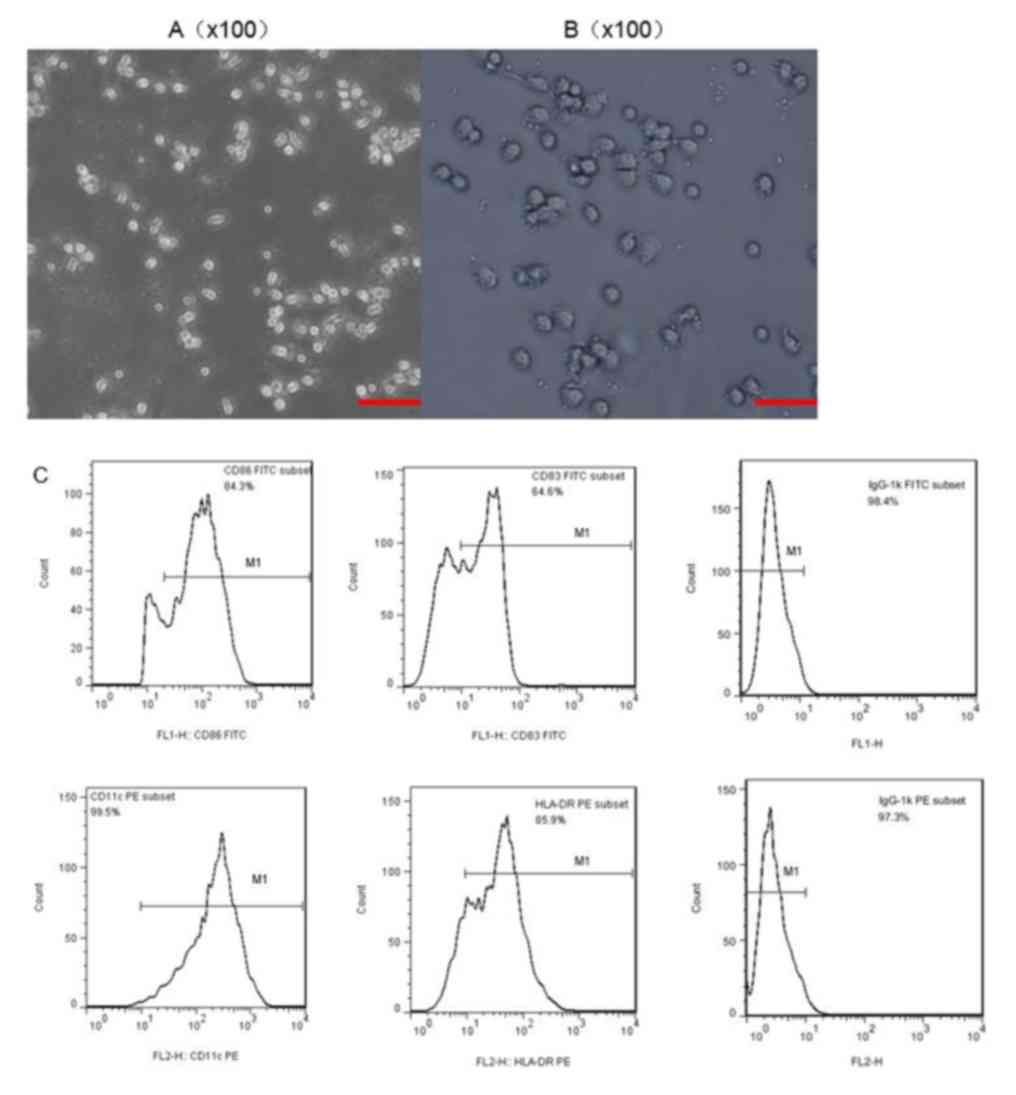
Morphology and phenotype of DCs
The DCs were all extracted from the PBMCs of healthy
volunteers, and were supplied and isolated at the Guangzhou Blood
Center. In the culture process, pro-inflammatory cytokines
stimulated DC maturation. When observed under a microscope,
immature DCs exhibited adherent growth, and clusters consisted of
small, round cells with relatively little cytoplasm (Fig. 2A). DCs gradually transformed from an
adherent to a semi-suspended state when cultured for 3 days, and
they began to have a small amount of fine outgrowths on their
surface (Fig. 2B). The typical
morphology of mature DCs acquired following 7 days of incubation
revealed a dispersed suspension state with irregular cell
morphology and a coarse dendritic surface, with a significantly
larger volume and more abundant cytoplasm, compared with immature
DCs. Flow cytometry was used to analyze the phenotype of the DCs.
The monoclonal antibodies CD83, CD86, HLA-DR and CD11c were
selected to stain the DCs, and all of them were present at high
levels (Fig. 2C); thus indicating
that mature DCs were successfully obtained.
DC transfection and identification of
transfection efficiency
To prepare the Runx2-DC vaccine, a Runx2 plasmid was
used to package the lentivirus and infect the DCs. Eventually, a
lentivirus was added to day-2 immature DCs, due to there being no
influence on the state of maturity, the ability to stimulate an
immune response and also due to the highest transfection efficiency
achieved in day-2 DCs (37).
Following transfection for 48 h, significant expression of green
fluorescent protein was observed using a fluorescence microscope
(Fig. 3A and B). The expression was
more notable following 72 h, indicating successful transduction.
Following 7 days of incubation, the transfected DCs demonstrated
typical mature cell morphology with suspended growth and clear
dendritic protrusions on the cell surface (Fig. 3C). Whether Runx2 was stably expressed
in DCs has not been previously confirmed; therefore, RT-qPCR and
western blot (Fig. 3D and E) analysis
were used to assess the Runx2 expression of transfected groups at
the mRNA and protein levels. It was concluded that Runx2 was
successfully integrated into the DCs at the nucleic acid level due
to the transfected group expressing a significant amount of Runx2
and due to the control groups expressing almost no Runx2
(P<0.05). In addition, the Runx2 protein was notably expressed
in the transfected group, but almost no significant expression was
observed in the other two control groups. The Runx2-DC vaccine was
then established.
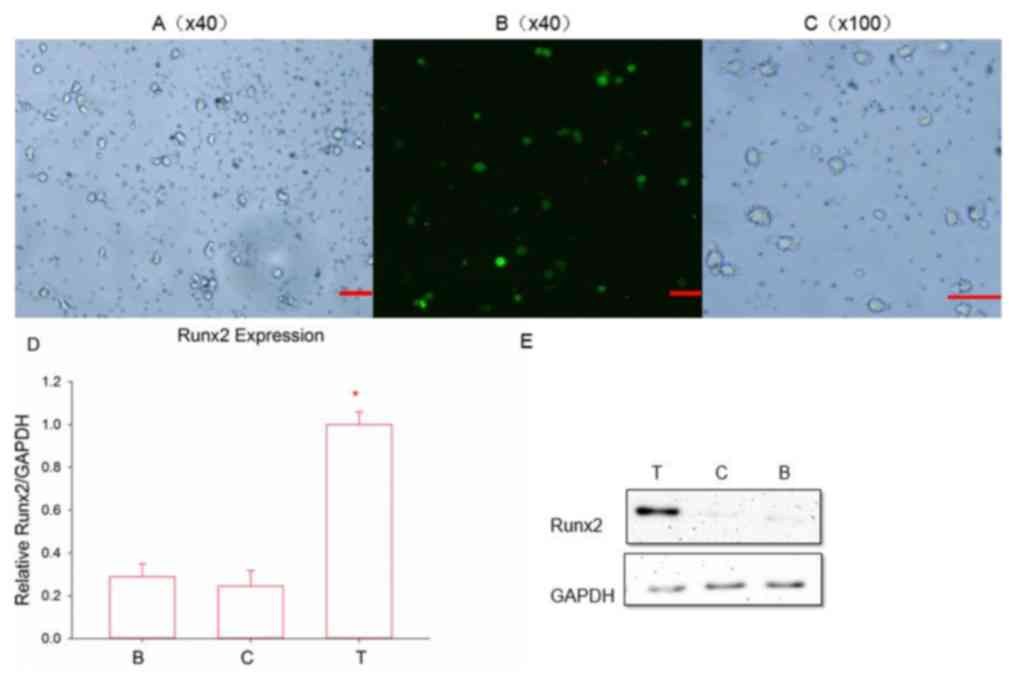 | Figure 3.(A) Fluorescence signals and Runx2
emitted by infected cells. Magnification, ×40; scale bar, 100 µm.
(B) Following 48 h of transfection. Magnification, ×40; scale bar,
100 µm. (C) Following 7 days of culture, transfected DCs became
mature. Magnification, ×100; scale bar, 100 µm. Reverse
transcription-quantitative polymerase chain reaction and western
blot analysis, respectively, were used to detect the (D) mRNA and
(E) protein expression of Runx2. *P<0.05 vs. the two control
groups according to analysis of variance and an LSD-post hoc test.
T, transfected group; C, negative control group; B, blank control
group; Runx2, runt-associated transcription factor 2. |
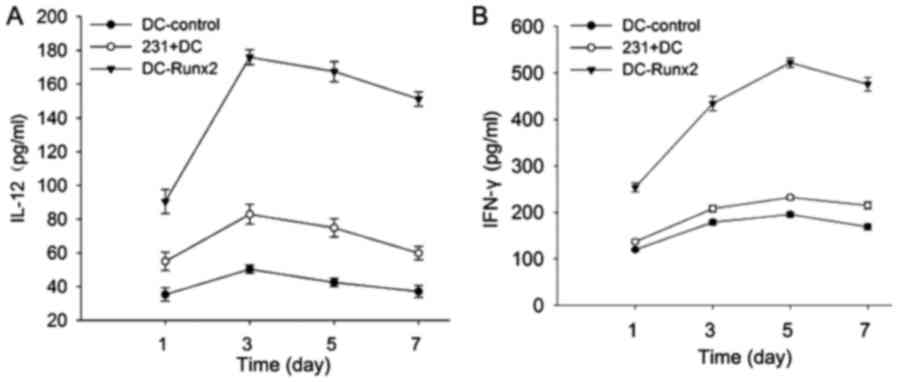
Secretion of IL-12 and IFN-γ in T
cells stimulated with the Runx2-DC vaccine
To detect T cell activation, the secretion of IL-12
and IFN-γ was measured in each group with an ELISA kit (Fig. 4). In the previous experiments, it was
determined that cytokine secretions reach a peak when co-cultured
with T lymphocytes and the vaccine or other control groups for 5
days. The supernatants were collected for detection using an ELISA
kit following 5 days of co-culture. The expression of IL-12 (pg/ml)
was as follows in each group: 170.33±5.03 (DC-Runx2); 42.43±2.48
(DC-control); and 75.00±3.00 (231-DC) (P<0.05). The secretion of
IFN-γ in each of the three groups was as follows: 517.15±5.88
(DC-Runx2); 195.63±5.51 (DC-control); and 232.57±4.73 (231-DC)
(P<0.05). Thus, the Runx2-DC vaccine may stimulate T lymphocytes
to secrete more cytokines (P<0.05).
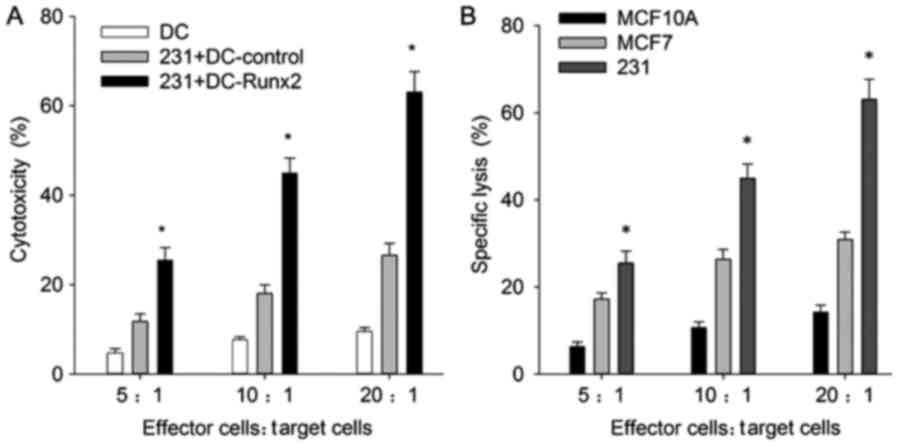
Cytotoxicity against TNBC provided by
the Runx2-DC vaccine
An objective of the present study was to determine
whether the Runx2-DC vaccine would be efficacious for cytotoxicity
against TNBC; therefore, the cytotoxicity rates for different cell
lines were analyzed. For MDA-MB-231, the cytotoxicity rates were
positively associated with the effective target ratio, and at the
ratio of 20:1 the cytotoxicity rates of the transfected, control
and blank group were 60.02±3.51, 25.57±3.64 and 26.45±1.30%,
respectively (Fig. 5A). The
transfected group indicated a significant increase in cytotoxicity
efficiency in MDA-MB-231 cells, compared with the non-transfected
groups (P<0.01); thus, the Runx2-DC vaccine may induce more
powerful cytotoxicity of CTLs.
To ensure that the anti-TNBC cytotoxicity was
specific, the transfected CTLs were co-cultured with MCF7 and
MCF10A, and the following cytotoxicity rates at the effective
target ratio of 20:1 were calculated: 30.91±1.70 and 14.27±1.60,
respectively (Fig. 5B). The
cytotoxicity rates showed the same correlation with effective
target ratio. These results revealed that the vaccine had
significantly stronger specific cytotoxicity effects against TNBC,
compared with the non-TNBC and normal breast cell groups
(P<0.05). It is notable that the vaccine has strong cytotoxicity
against breast cell lines, but also affects normal breast
cells.
Discussion
Based on microarray and proteomic studies, Runx2 was
determined to be one of the most upregulated genes in invasive
breast cancers (38,39). Runx2 has the ability to promote cell
proliferation and inhibit apoptosis caused by P53 in tumor cells,
thus advancing the progression of breast cancer (21). Runx2 has also been reported to mediate
the invasion of the human breast cancer cell lines MDA-MB-231 and
MCF7 (36). Following Runx2
inhibition or knockout in breast cancer, the tumor phenotypes move
gradually toward normal phenotypes, with suppression of tumor cell
proliferation and growth (20,21). It
was previously determined that the expression of Runx2 was lower in
the normal breast cells and significantly increased in breast
cancer cells (20,21); therefore, abnormal expression of Runx2
in tumors may be associated with tumor progression. McDonald et
al (36) described abundant
Runx2-specific expression in TNBC using microarray analysis and
determined that it was associated with a lower survival rate;
however, other research has indicated that Runx2 expression is more
notable in ER-positive cells (32).
Based on these characteristics, Pratap et al (20) demonstrated that Runx2 had a higher
expression in breast cancer cells MDA-MB-231 compared with that in
MCF10A cells (20). Comparing between
MDA-MB-231 and MCF7 cells, higher expression of Runx2 was present
in the highly invasive MDA-MB-231 cells (32,33). With
a higher Runx2 expression level in MDA-MB-231 cells, compared with
non-TNBC cell lines demonstrated in previous studies (32–34),
MDA-MB-231 was selected as the research focus. Similar to these
previous data, higher Runx2 expressions was demonstrated in the
breast cancer cell lines compared with the normal breast cancer
cell line, and a stronger expression of Runx2 in TNBC was
determined, compared with the MCF7 and MCF10A cell lines,
indicating that Runx2 may be used as a novel target for TNBC
immunotherapy.
In the present study, Runx2 indicated a wide variety
of distribution in different breast cancer cells. Positive
expression of Runx2 in the cytoplasm and nucleus was observed in
breast cancer cells. Runx2 primarily existed in the nucleus in TNBC
and in the cytoplasm in luminal breast cancer cells, whilst Runx2
was typically present in the cytoplasm in normal breast cell lines.
Different distributions may be associated with the function of
Runx2. As a transcription factor, Runx2 was enriched in terminal
end buds, which were essential for the branching of the developing
gland in mammary epithelial and breast cancer cell lines (40). It was considered that Runx2 expressed
in the cytoplasm of normal breast cells promoted differentiation;
therefore, Runx2 may exert more functions, including tumor cell
proliferation, migration and invasion, in the nucleus.
As they are concentrated in the periphery of
carcinomas, DCs may come into contact and identify tumor
cell-specific antigens with dendrites on their surface (41). Their major functions include: Intake,
processing and presentation of antigens; participation in
T-lymphocyte differentiation; promotion of B-lymphocyte
proliferation directly or indirectly; and regulation of innate
immunity (42). However, patients
with cancer lack DC and co-stimulatory signals, genetic instability
and heterogeneity of tumor cell antigens, which all contribute to a
valid T cell activation and an effective immune response (42). Transfusion or vaccination of patients
with cancer with antigen-sensitized DCs in vitro may assist
in breaking down these barriers.
The method of transducing Runx2 into DCs is
challenging. As non-dividing cells, DCs are difficult to transduce
with viral vectors, and their phenotype and function may change
following transfection (43–45). A previous study indicated that
lentivirus may lead to efficient transfection and preservation of
cell maturation and function with day-2 immature DCs (46). Unlike the adenoviral vector,
lentiviruses may also avoid vector-mediated immune stimulation
(47). The present data confirmed
that Runx2 may be loaded into DCs without affecting cell morphology
and maturity, and that lentivirus vector transfection provided
stable expression of Runx2.
A valid CD8+ T cell response is associated with
positive prognosis for tumor control and clearance (48). CD8+ T cells do not have the ability to
produce the cytokines IFN-γ, IL-2 and TNF, which means that they
gradually deplete in the tumor microenvironment, thus losing the
ability to eliminate tumor cells (49). Following effective identification of
tumor antigen presented by DCs, CD4+ T cells have the ability to
activate the CTL response (50). CD4+
T cells differentiate into Th1 and Th2 cells, Th1 cells effectively
stimulate and mediate cellular immunity, which is primarily induced
by IL-12. Under normal circumstances, Th1 and Th2 cells are in
equilibrium. Changes to the equilibrium help the tumor cells to
escape the immune response, thus leading to tumor growth (51,52). DCs
drive naive T cells to differentiate into various phenotypes,
including Th1, Th2, Th17, Treg or CTLs (53). The unique ability to regulate specific
types of cellular immune responses gives priority to DCs in cancer
immunotherapy, where Th1-induced tumor-specific CTLs are
particularly desirable (54,55).
Integrated DCs and T cells secrete IL-12 and IL-18
to promote T cell proliferation and aggregation, and release
anti-angiogenic factors, including IL-12 and IFN-γ, to inhibit
tumor angiogenesis (56). The
cytokine IFN-γ strengthens cellular immunity by inducing increased
MHC expression and promoting T-lymphocyte differentiation (57). For example, CD8+ T cell
differentiation is partially dependent on IFN-γ (48,58). It is
also important for innate immunity due to its actions in enhancing
monocyte-macrophage activation, the efficiency of natural killer
(NK) cells and stimulating VEGF secretion. IL-12 serves an
essential role in promoting cell division of T lymphocytes,
inducing Th1 cell differentiation, and stimulating NK cell and B
cell activity (57,59). This may reflect a change in the
quantity and function of T cells, reaching a peak value following
24–72 h in vitro according to a previous study (60).
In the present study, the transfection group
secreted more IL-12, activating and polarizing Th1 cells and
inducing NK cells, compared with the non-transfection group.
Subsequently, Th1 cells have the ability to effectively activate
CTLs, providing a large amount of IFN-γ, perforin and granzyme,
enhancing the cytotoxic effect on tumor cells. This result was
confirmed in subsequent experiments, with the sustained expression
of the Runx2 antigen significantly enhancing the antitumor effect
on TNBC by inducing the CTL and Th1 response. Different immune
responses in anti-MDA-MB-231 and anti-MCF7 cells were induced by
the Runx2-DC vaccine, and anti-MDA-MB-231 cells demonstrated the
strongest effect among the different cell lines. The vaccine may
induce specific T cell-mediated immune responses against TNBC
following targeting and recognizing TNBC. To further confirm the
data, additional cell lines should be used. A more troublesome
problem is the damage to normal breast tissue caused by the vaccine
in vitro. The cytotoxicity against normal tissue requires
further study in vivo, and the use of regulatory T cells and
suppressor cells may potentially prevent this phenomenon.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Guangdong
Natural Science Foundation (grant no. 2014A030313193).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MT, YL and YH designed the study; QCZ, PZ and JKW
analyzed the data; and JNW and YR drafted the manuscript and helped
to analyzed the data.
Ethics approval and consent to
participate
The use of human subjects was specifically approved
by the Clinical Research Ethics Committee of the Third Affiliated
Hospital of Sun Yat-sen University (Guangzhou, China). Guangzhou
Blood Center (Guangzhou, China) supplied the blood and recorded the
informed consent. Prior to donating blood, the volunteers were
informed and provided written informed consent for the scientific
research use of blood samples.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
TNBC
|
triple-negative breast cancer
|
|
ER
|
estrogen receptor
|
|
DCs
|
dendritic cells
|
|
APC
|
antigen-presenting cells
|
|
MHC
|
major histocompatibility complex
|
|
CTL
|
cytotoxic T cell
|
|
MMP
|
matrix metalloproteinase
|
|
VEGF
|
vascular endothelial growth factor
|
|
FBS
|
fetal bovine serum
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
RT
|
room temperature
|
References
|
1
|
Liedtke C, Mazouni C, Hess KR, André F,
Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B,
Green M, et al: Response to neoadjuvant therapy and long-term
survival in patients with triple-negative breast cancer. J Clin
Oncol. 26:1275–1281. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bonasio R and von Andrian UH: Generation,
migration and function of circulating dendritic cells. Curr Opin
Immunol. 18:503–511. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Celluzzi CM, Mayordomo JI, Storkus WJ,
Lotze MT and Falo LD Jr: Peptide-pulsed dendritic cells induce
antigen-specific CTL-mediated protective tumor immunity. J Exp Med.
183:283–287. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kichler-Lakomy C, Budinsky AC, Wolfram R,
Hellan M, Wiltschke C, Brodowicz T, Viernstein H and Zielinski CC:
Deficiences in phenotype expression and function of dendritic cells
from patients with early breast cancer. Eur J Med Res. 11:7–12.
2006.PubMed/NCBI
|
|
6
|
Bennaceur K, Chapman J, Brikci-Nigassa L,
Sanhadji K, Touraine JL and Portoukalian J: Dendritic cells
dysfunction in tumour environment. Cancer Lett. 272:186–196. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bosco MC, Puppo M, Blengio F, Fraone T,
Cappello P, Giovarelli M and Varesio L: Monocytes and dendritic
cells in a hypoxic environment: Spotlights on chemotaxis and
migration. Immunobiology. 213:733–749. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rifkind RA, Hsu KC, Morgan C, Seegal Bc,
Knox Aw and Rose Hm: Use of ferritin-conjugated antibody to
localize antigen by electron microscopy. Nature. 187:1094–1095.
1960. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Koido S, Nikrui N, Ohana M, Xia J, Tanaka
Y, Liu C, Durfee JK, Lerner A and Gong J: Assessment of fusion
cells from patient-derived ovarian carcinoma cells and dendritic
cells as a vaccine for clinical use. Gynecol Oncol. 99:462–471.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Homma S, Kikuchi T, Ishiji N, Ochiai K,
Takeyama H, Saotome H, Sagawa Y, Hara E, Kufe D, Ryan JL, et al:
Cancer immunotherapy by fusions of dendritic and tumour cells and
rh-IL-12. Eur J Clin Invest. 35:279–286. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Berzofsky JA, Terabe M, Oh S, Belyakov IM,
Ahlers JD, Janik JE and Morris JC: Progress on new vaccine
strategies for the immunotherapy and prevention of cancer. J Clin
Invest. 113:1515–1525. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Figdor CG, de Vries IJ, Lesterhuis WJ and
Melief CJ: Dendritic cell immunotherapy: Mapping the way. Nat Med.
10:475–480. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Iwamoto M, Shinohara H, Miyamoto A,
Okuzawa M, Mabuchi H, Nohara T, Gon G, Toyoda M and Tanigawa N:
Prognostic value of tumor-infiltrating dendritic cells expressing
CD83 in human breast carcinomas. Int J Cancer J. 104:92–97. 2003.
View Article : Google Scholar
|
|
14
|
Zou W: Immunosuppressive networks in the
tumour environment and their therapeutic relevance. Nat Rev Cancer.
5:263–274. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Blyth K, Vaillant F, Jenkins A, McDonald
L, Pringle MA, Huser C, Stein T, Neil J and Cameron ER: Runx2 in
normal tissues and cancer cells: A developing story. Blood Cells
Mol Dis. 45:117–123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pratap J, Lian JB and Stein GS: Metastatic
bone disease: Role of transcription factors and future targets.
Bone. 48:30–36. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shore P: A role for Runx2 in normal
mammary gland and breast cancer bone metastasis. J Cell Biochem.
96:484–489. 2010. View Article : Google Scholar
|
|
18
|
Otto F, Thornell AP, Crompton T, Denzel A,
Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen
BR, et al: Cbfa1, a candidate gene for cleidocranial dysplasia
syndrome, is essential for osteoblast differentiation and bone
development. Cell. 89:765–771. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Owens TW, Rogers RL, Best S, Ledger A,
Mooney AM, Ferguson A, Shore P, Swarbrick A, Ormandy CJ, Simpson
PT, et al: Runx2 is a novel regulator of mammary epithelial cell
fate in development and breast cancer. Cancer Res. 74:5277–5286.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pratap J, Imbalzano KM, Underwood JM,
Cohet N, Gokul K, Akech J, van Wijnen AJ, Stein JL, Imbalzano AN,
Nickerson JA, et al: Ectopic runx2 expression in mammary epithelial
cells disrupts formation of normal acini structure: Implications
for breast cancer progression. Cancer Res. 69:6807–6814. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ozaki T, Wu D, Sugimoto H, Nagase H and
Nakagawara A: Runt-related transcription factor 2 (RUNX2) inhibits
p53-dependent apoptosis through the collaboration with HDAC6 in
response to DNA damage. Cell Death Disease. 4:e6102013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mendoza-Villanueva D, Deng W,
Lopez-Camacho C and Shore P: The Runx transcriptional co-activator,
CBFbeta, is essential for invasion of breast cancer cells. Mol
Cancer. 9:1712010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pratap J, Javed A, Languino LR, van Wijnen
AJ, Stein JL, Stein GS and Lian JB: The Runx2 osteogenic
transcription factor regulates matrix metalloproteinase 9 in bone
metastatic cancer cells and controls cell invasion. Mol Cell Biol.
25:8581–8591. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Selvamurugan N, Kwok S and Partridge NC:
Smad3 interacts with JunB and Cbfa1/Runx2 for transforming growth
factor-beta1-stimulated collagenase-3 expression in human breast
cancer cells. J Biol Chem. 279:27764–27773. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pratap J, Wixted JJ, Gaur T, Zaidi SK,
Dobson J, Gokul KD, Hussain S, van Wijnen AJ, Stein JL, Stein GS
and Lian JB: Runx2 transcriptional activation of Indian Hedgehog
and a downstream bone metastatic pathway in breast cancer cells.
Cancer Res. 68:7795–7802. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Khalid O, Baniwal SK, Purcell DJ, Leclerc
N, Gabet Y, Stallcup MR, Coetzee GA and Frenkel B: Modulation of
Runx2 activity by estrogen receptor-alpha: Implications for
osteoporosis and breast cancer. Endocrinology. 149:5984–5995. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chimge NO, Baniwal SK, Luo J, Coetzee S,
Khalid O, Berman BP, Tripathy D, Ellis MJ and Frenkel B: Opposing
effects of Runx2 and estradiol on breast cancer cell proliferation:
In vitro identification of reciprocally regulated gene signature
related to clinical letrozole responsiveness. Clin Cancer Res.
18:901–911. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun L, Vitolo M and Passaniti A:
Runt-related gene 2 in endothelial cells: Inducible expression and
specific regulation of cell migration and invasion. Cancer Res.
61:4994–5001. 2001.PubMed/NCBI
|
|
29
|
Pierce AD, Anglin IE, Vitolo MI, Mochin
MT, Underwood KF, Goldblum SE, Kommineni S and Passaniti A:
Glucose-activated RUNX2 phosphorylation promotes endothelial cell
proliferation and an angiogenic phenotype. J Cell Biochem.
113:282–292. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chimge NO, Baniwal SK, Little GH, Chen Y,
Kahn M, Tripathy D, Borok Z and Frenkel B: Regulation of breast
cancer metastasis by Runx2 and estrogen signaling: The role of
SNAI2. Breast Cancer Res. 13:R1272011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Herynk MH and Fuqua SA: Estrogen receptor
mutations in human disease. Endocr Rev. 25:869–898. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Das K, Leong DT, Gupta A, Shen L, Putti T,
Stein GS, van Wijnen AJ and Salto-Tellez M: Positive association
between nuclear Runx2 and oestrogen-progesterone receptor gene
expression characterises a biological subtype of breast cancer. Eur
J Cancer. 45:2239–2248. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lau QC, Raja E, Salto-Tellez M, Liu Q, Ito
K, Inoue M, Putti TC, Loh M, Ko TK, Huang C, et al: Runx3 is
frequently inactivated by dual mechanisms of protein
mislocalization and promoter hypermethylation in breast cancer.
Cancer Res. 66:6512–6520. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tandon M, Chen Z and Pratap J: Runx2
activates PI3K/Akt signaling via mTORC2 regulation in invasive
breast cancer cells. Breast Cancer Res. 16:R162014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
McDonald L, Ferrari N, Terry A, Bell M,
Mohammed ZM, Orange C, Jenkins A, Muller WJ, Gusterson BA, Neil JC,
et al: RUNX2 correlates with subtype-specific breast cancer in a
human tissue microarray, and ectopic expression of Runx2 perturbs
differentiation in the mouse mammary gland. Dis Model Mech.
7:525–534. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
He Y, Zhang J, Mi Z, Robbins P and Falo LD
Jr: Immunization with lentiviral vector-transduced dendritic cells
induces strong and long-lasting T cell responses and therapeutic
immunity. J Immunol. 174:3808–3817. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nagaraja GM, Othman M, Fox BP, Alsaber R,
Pellegrino CM, Zeng Y, Khanna R, Tamburini P, Swaroop A and Kandpal
RP: Gene expression signatures and biomarkers of noninvasive and
invasive breast cancer cells: Comprehensive profiles by
representational difference analysis, microarrays and proteomics.
Oncogene. 25:2328–2338. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Barnes GL, Javed A, Waller SM, Kamal MH,
Hebert KE, Hassan MQ, Bellahcene A, Van Wijnen AJ, Young MF, Lian
JB, et al: Osteoblast-related transcription factors Runx2
(Cbfa1/AML3) and MSX2 mediate the expression of bone sialoprotein
in human metastatic breast cancer cells. Cancer Res. 63:2631–2637.
2003.PubMed/NCBI
|
|
40
|
Kouros-Mehr H and Werb Z: Candidate
regulators of mammary branching morphogenesis identified by
genome-wide transcript analysis. Dev Dyn. 235:3404–3412. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hillenbrand EE, Neville AM and Coventry
BJ: Immunohistochemical localization of CD1a-positive putative
dendritic cells in human breast tumours. Br J Cancer. 79:940–944.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Banchereau J and Palucka AK: Dendritic
cells as therapeutic vaccines against cancer. Nat Rev Immunol.
5:296–306. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hegde NR, Chevalier MS and Johnson DC:
Viral inhibition of MHC class II antigen presentation. Trends
Immunol. 24:278–285. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Alcami A, Ghazal P and Yewdell JW: Viruses
in control of the immune system. Workshop on molecular mechanisms
of immune modulation: Lessons from viruses. EMBO Rep. 3:927–932.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yewdell JW and Hill AB: Viral interference
with antigen presentation. Nature Immunol. 3:1019–1025. 2002.
View Article : Google Scholar
|
|
46
|
Follenzi A and Naldini L: Generation of
HIV-1 derived lentiviral vectors. Methods Enzymol. 346:454–465.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Morelli AE, Larregina AT, Ganster RW,
Zahorchak AF, Plowey JM, Takayama T, Logar AJ, Robbins PD, Falo LD
and Thomson AW: Recombinant adenovirus induces maturation of
dendritic cells via an NF-kappaB-dependent pathway. J Virol.
74:9617–9628. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hwang ML, Lukens JR and Bullock TN:
Cognate memory CD4+ T cells generated with dendritic cell priming
influence the expansion, trafficking, and differentiation of
secondary CD8+ T cells and enhance tumor control. J Immunol.
179:5829–5838. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jiang Y, Li Y and Zhu B: T-cell exhaustion
in the tumor microenvironment. Cell Death Dis. 6:e17922015.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Guerder S and Matzinger P: A fail-safe
mechanism for maintaining self-tolerance. J Exp Med. 176:553–564.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Romagnani S: Human TH1 and TH2 subsets:
Doubt no more. Immunol Today. 12:256–257. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sheu BC, Lin RH, Lien HC, Ho HN, Hsu SM
and Huang SC: Predominant Th2/Tc2 Polarity of tumor-infiltrating
lymphocytes in human cervical cancer. J Immunol. 167:2972–2978.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Palucka K and Banchereau J: Cancer
immunotherapy via dendritic cells. Nat Rev Cancer. 12:265–277.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Huang H, Hao S, Li F, Ye Z, Yang J and
Xiang J: CD4+ Th1 cells promote CD8+ Tc1 cell survival, memory
response, tumor localizationand therapy by targeted delivery of
interleukin 2 via acquired pMHC I complexes. Immunology.
120:148–159. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Knutson KL and Disis ML: Tumor
antigen-specific T helper cells in cancer immunity and
immunotherapy. Cancer Immunol Immunother. 54:721–728. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Savai R, Schermuly RT, Pullamsetti SS,
Schneider M, Greschus S, Ghofrani HA, Traupe H, Grimminger F and
Banat GA: A combination hybrid-based vaccination/adoptive cellular
therapy to prevent tumor growth by involvement of T cells. Cancer
Res. 67:5443–5453. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Trinchieri G: Interleukin-12 and the
regulation of innate resistance and adaptive immunity. Nat Rev
Immunol. 3:133–146. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Janssen EM, Lemmens EE, Wolfe T, Christen
U, von Herrath MG and Schoenberger SP: CD4+ T cells are required
for secondary expansion and memory in CD8+ T lymphocytes. Nature.
421:852–856. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zobywalski A, Javorovic M, Frankenberger
B, Pohla H, Kremmer E, Bigalke I and Schendel DJ: Generation of
clinical grade dendritic cells with capacity to produce
biologically active IL-12p70. J Transl Med. 5:182007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhang P, Yi S, Li X, Liu R, Jiang H, Huang
Z, Liu Y, Wu J and Huang Y: Preparation of triple-negative breast
cancer vaccine through electrofusion with day-3 dendritic cells.
PLoS One. 9:e1021972014. View Article : Google Scholar : PubMed/NCBI
|