Introduction
Primary liver cancer is a malignant tumor, incidence
rate was the fifth and the mortality ranked third in 2007, and
approximately 85-90% was hepatocellular carcinoma (HCC).
Sub-Saharan Africa and Eastern Asia are the two most common places
for HCC, of which the incidence in China was more than half of the
whole world (1). Metastasis and
recurrence ofter occur in HCC due to factors, such as geographical
location, race, sex, environment, and molecular factors. Therefore,
identification of tumor biomarkers for early diagnosis is essential
for HCC patients.
MicroRNAs (miRNAs), a class of small regulatory RNA
molecules with ~22-28 nucleotides, inhibit target gene expression
in post-transcriptional regulation by targeting the 3′UTR of mRNAs
or cleaving their mRNA directly in order to inhibit protein
expression (2–5). miR-185 was reported to play important
roles in several human cancers, covering prostate carcinoma, lung
cancer, ovarian, pediatric renal, breast, cervical and colon
cancers (6–10). In prostate carcinoma cells, miR-185
downregulated androgen receptor (AR) to inhibit cell proliferation
and induced apoptosis by directly targeting it (6). miR-185 also inhibited colorectal cell
proliferation by targeting RhoA and CDC42 (11).
Cell division cycle 42 (CDC42) is a member of Rho
GTPase family, which belong to subfamily of 20-30 kDa GTP-binding
proteins of Ras superfamily (11–13). It
was reported that CDC42 is overexpressed in various carcinomas,
such as breast, colon, esophageal, bladder and liver cancer
(14–17). CDC42 impacts cell cycle during cell
division, leading to generation of multinucleated cells in primary
mouse embryonic fibroblasts (18).
Studies have found that the level in HCC tissues was higher than
paracancerous tissues, and was connected with metastasis in HCC
cells (19,20).
In our study, the relationship between miR-185 and
CDC42 and correlation in HCC was investigated. We analyzed the
miR-185 expression level in HCC tissues and liver cancer cells with
non-carcinomatous tissues and normal liver cells as control. The
effects of overexpression or knockdown of miR-185 on migration and
invasion of liver cancer cells HuH-7 was explored. Moreover, we
examined the influence of miR-185 expression on CDC42 and the
prognosis of patients. Finally, through recovery tests, we
determined that miR-185 inhibits the cell processes by regulating
CDC42 cell processes.
Materials and methods
Tissue samples and cell lines
Paired tumors and non-carcinomatous tissue were
available from 63 patients with surgery from Central Hospital of
Zibo. All the specimens were obtained with informed consent of the
patients and the study was approved by the Ethics Committee of
Central Hospital of Zibo (Zibo, China).
HuH-7 human hepatocellular carcinoma cell and the
human normal hepatocyte L-02 cell line were purchased from American
Type Culture Collection (ATCC; Manassas, VA, USA). Cells were
incubated at constant temperature of 37°C with 5%
CO2.
Western blotting
For western blotting, the first cells were lysed
using RIPA lysis buffer containing PMSF (both from Beyotime,
Shanghai, China) on ice. Following centrifugation with 12,000 × g
at 4°C, the concentration of protein was measured by BCA reagent
kit (Solarbio, Beijing, China) and the absorbance was measured by a
microplate reader. SDS-PAGE was applied for separation and transfer
onto a PVDF membrane, and it was incubated at 4°C overnight with
mouse anti-CDC42 monoclonal antibody (cat. no. ab41429; 1:1,000;
Abcam, Cambridge, UK). After washing with TBST buffer
(Tris-buffered saline with Tween-20, pH 8.0), the blots were
incubated with anti-mouse IgG (1:3,000; cat. no. no. SAB4600004
Novus Biologicals, Littleton, CO, USA) at room temperature for 2 h.
The protein signal was detected with chemiluminescence using the
Bio-Rad Gel Doc XR instrument (Bio-Rad Laboratories, Inc.,
Berkeley, CA, USA).
Transwell assay
Transwell assay was used to test the ability of
migration and invasion. The Transwell chambers (Costar, Corning,
NY, USA), were 8 µm in size, with or without Matrigel (Clontech,
Mountain View, CA, USA). The chambers within 200 µl cell suspension
was put into 24-well plate containing 500 µl medium. Culturing at
37°C with 5% CO2 for about 48 h before the cells moved
under the chambers. After fixed with methanol for 20 min, the cells
were stained using crystal violet and observed by microscope (BX51
Olympus, Shenzhen, China).
RT-qPCR
Total RNAs were extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Carlsbad, CA, USA) and
miRNAs using miRcute Extraction and Separation of miRNAs kit
(Tiangen, Beijing, China). Reverse transcription of purified RNAs
was used PrimeScript™ II 1st Strand cDNA Synthesis kit (Takara
Biotechnology Co., Ltd., Dalian, China) conducted by two steps at
37°C for 5 min and 42°C for 25 min in 20 µl reaction system. STEP
ONE RT-qPCR Apparatus (Applied Biosystems, Foster City, CA, USA)
and miRNA SYBR Green RT-qPCR kit (ABM, Inc., USA) were employed to
perform the quantitative real time PCR. The following thermocycling
conditions were used for PCR: 5 min at 95°C, followed by 40 cycles
of 95°C for 30 sec and 65°C for 45 sec. GAPDH and U6 were used as
normalization for CDC42 and miR-185, respectively. All the primers
were purchased from Genechem (Shanghai, China), which were CDC42
forward: 5′-GCTCTAGAGCCCTTAAGGGGAGGAG-3′ and reverse:
5′-GCTCTAGAAAAAATCCCTATTAACAC-3′; U6 forward:
5′-CTCGCTTCGGCAGCACA-3′ and reverse: 5′-AACGCTTCACGAATTTGCGT-3′;
miR-185 forward: 5′-CAATGGAGAGAAAGGCAGTTCC-3′ and reverse:
5′-AATCCATGAGAGATCCCTACCG-3′; GAPDH forward:
5′-GGTGAAGGTCGGAGTCAACG-3′ and reverse:
5′-CAAAGTTGTCATGGATGHACC-3′.
Plasmid construction and luciferase
reporter assay
TargetScan (www.targetscan.org), online software, predicted CDC42
was a target gene of miR-185 with binding site at 647-654 of its
3′UTR. In order to verify whether miR-185 interacts with CDC42,
double luciferase reporter assay was performed. The 3′UTR
oligonucleotide fragment of CDC42 was inserted into pcDNA3.1
plasmid vector (pcDNA3.1-CDC42-WT). Mutated the binding site from
5′-…UCUCUCC…-3′ to 5′-…AGAGAGG…-3′ and then inserted into pcDNA3.1
plasmid vector (pcDNA3.1-CDC42-MUT). The effectiveness of cloning
was detected by sequencing.
HuH-7 cells at 80% confluence were seeded in 6-well
plates and cultured overnight before transfection. miR-185 mimic or
negative control (NC) and pcDNA3.1-CDC42-WT or pcDNA3.1-CDC42-MUT
were co-transfected into HCC HuH-7 cells using Lipofectamine 3000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Then the
transfected cells were cultured at 37°C with 5% CO2 for
48 h and harvested for analysis.
Transfection
The influence of miR-185 to cell migration and
invasion, was investigated with miR-185 mimic and inhibitor were
used to overexpress or knock down miR-185. Similarly, we used small
interfering RNA (siRNA) to interfere with the expression of CDC42,
and detected the effect of miR-185 through CDC42 for cell migration
and invasion.
HCC cells were seeded into a 6-well plate and
cultivated overnight to make sure that cells adhered to the wall.
The medium was replaced before transfection, and the specific
plasmids were transfected in HCC cells by Lipofectamine 3000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
Statistical analysis
Experimental results were demonstrated using SPSS
20.0 software package (IBM Corp., Armonk, NY, USA). The differences
between the groups were calculated using t-test or Dunnetts,
Fishers after one-way ANOVA test. In addition, χ2 test
was used to compare the expression of miR-185 and the
clinicopathological features of HCC patients. The Kaplan-Meier
method with log-rank test was used to calculate the survival rates.
A total of 63 cases of patients were divided into two groups
[miR-185(+) and miR-185(−)] according to the miR-185 expression.
The effects of the miR-185 expression on overall survival (OS) and
disease-free survival (DFS) (in months) were analyzed using the
multivariate Cox regression method. For all tests, P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-185 expression is significantly
low and correlates with CDC42 in HCC
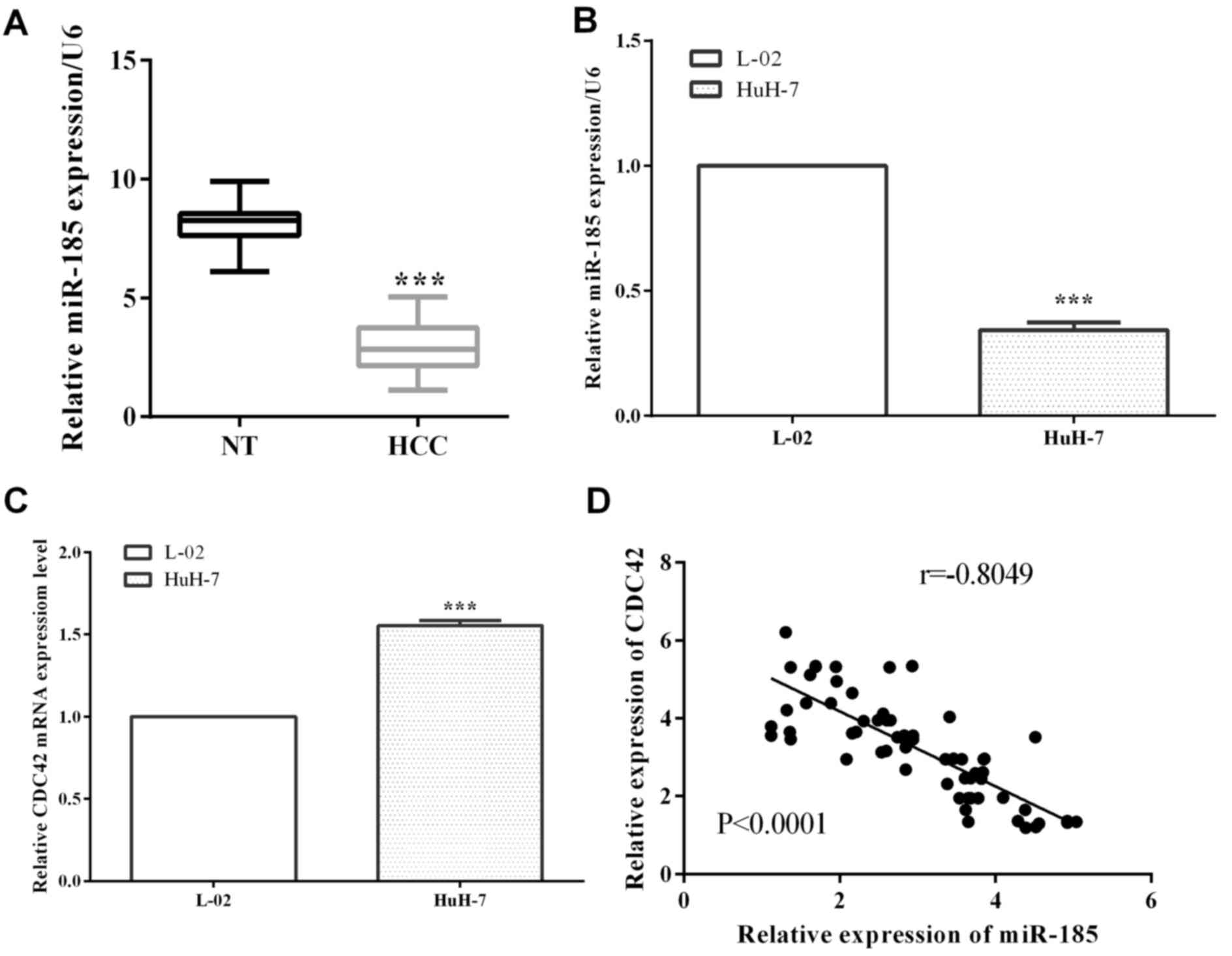
miR-185 and CDC42 expression level of the 63 paired
HCC and non-carcinomatous tissues was detected by RT-qPCR. For HCC
tissues, the expression of miR-185 was reduced significantly
compared with non-carcinomatous tissues (P<0.0001) (Fig. 1A). Similarly, miR-185 expression in
HCC cells HuH-7 was decreased in contrast to normal HCC L-02 cells
(P<0.0001), as shown in Fig. 1B.
On the contrary, CDC42 was overexpressed in HCC cell lines HuH-7
vs. normal liver L-02 cells (P<0.0001) (Fig. 1C). miR-185 was correlated with CDC42
in HCC (r=−0.8049, P<0.0001) (Fig.
1D).
miR-185 inhibits migration and
invasion and miR-185 deficiency predictes poor prognosis of HCC
patients
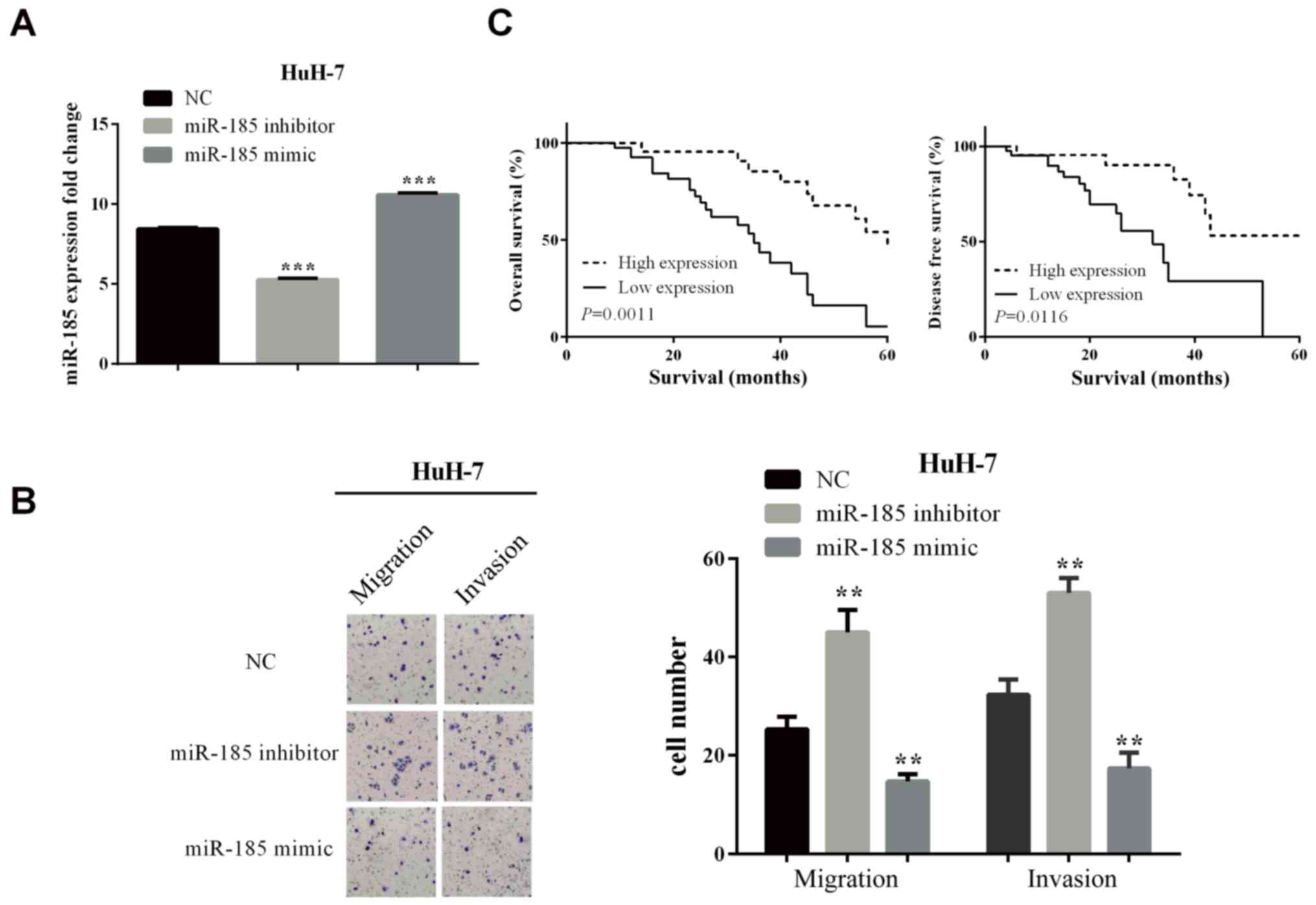
In order to test whether miR-185 influenced the
progress of HCC, we used mimic or inhibitor to overexpress or knock
down the expression of miR-185 in HuH-7 cells. The overexpression
(P-value was <0.0001) or knockdown (P-value was <0.0001)
effects are shown in Fig. 2A.
Transwell assay was used to detect the influence of
miR-185 on migration and invasion. As shown in Fig. 2B, in knockdown of miR-185, the cells
number that moved to the lower chamber was increased in HuH-7
(P-values for migration and invasion are 0.0029 and 0.0011,
respectively) cells, which illustrated the ability of migration and
invasion to increase. On the contrary, in the overexpression of
miR-185 inhibitor, the cell number was reduced in HuH-7 (P=0.0033
and 0.0042 for migration and invasion) cells.
miR-185 was significantly related to tumor size
(P=0.033), TNM stage (P=0.019), lymph node metastasis (P=0.023), as
shown in Table I. OS and DFS of
patients in miR-185(−) group was lower than that in miR-185(+)
group (log-rank P=0.0011 and 0.0116) based on Kaplan-Meier, as
shown in Fig. 2C. The results
revealed that miR-185 deficiency predicted poor prognosis in
HCC.
 | Table I.miR-185 expression and
clinicopathological features in 63 paired HCC. |
Table I.
miR-185 expression and
clinicopathological features in 63 paired HCC.
|
|
| miR-185
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | Cases (n=63) | High (%) | Low (%) | P-valuea |
|---|
| Sex |
|
|
| 0.782 |
| Male | 30 | 11 (36.7) | 19 (63.3) |
|
|
Female | 33 | 11 (33.3) | 22 (66.7) |
|
| Age (years) |
|
|
| 0.679 |
| ≤60 | 28 | 9 (32.1) | 19 (67.9) |
|
|
>60 | 35 | 13 (37.1) | 22 (62.9) |
|
| Tumor size (mm) |
|
|
| 0.029a |
| ≤5.0 | 25 | 13 (52.0) | 12 (48.0) |
|
|
>5.0 | 38 | 9 (28.9) | 27 (71.1) |
|
| TNM stage |
|
|
| 0.035a |
|
I–II | 26 | 13 (50.0) | 13 (50.0) |
|
|
III–IV | 37 | 9 (24.3) | 28 (75.7) |
|
| Local invasion |
|
|
| 0.077 |
|
T1-T2 | 24 | 12 (48.0) | 13 (52.0) |
|
|
T3-T4 | 39 | 10 (26.3) | 28 (73.7) |
|
| Lymph node
metastasis |
|
|
| 0.040a |
|
Positive | 33 | 8 (24.2) | 26 (75.8) |
|
|
Negative | 30 | 14 (46.7) | 15 (53.3) |
|
miR-185 mediates the expression of
CDC42 by direct targeting
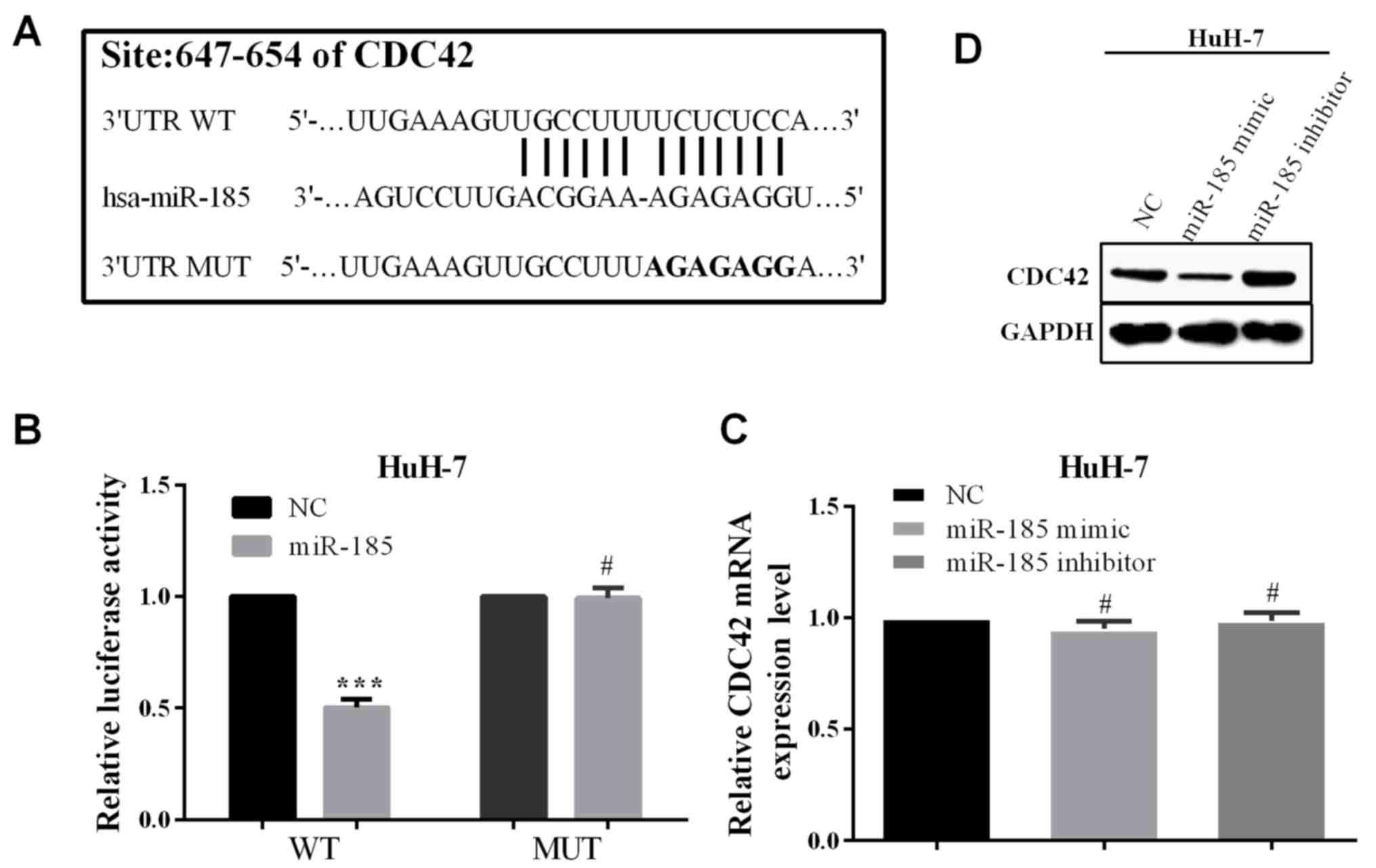
CDC42 was predicted to be a potential target by
online software TargetScan (http://www.targetscan.org/vert_71/), which was
reported to be related with tumor progression. The potential
binding site of CDC42 for miR-185 is located at 647-654 at 3′UTR.
Mutated the binding site from 5′-…UCUCUCC…-3′ to 5′-…AGAGAGG…-3′,
as shown in Fig. 3A. To verify the
association between CDC42 and miR-185, we co-transfected miR-185 or
negative control and pcDNA3.1-CDC42-WT or pcDNA3.1-CDC42-MUT into
HCC cells HuH-7. In co-transfection of miR-185 and
pcDNA3.1-CDC42-WT, the relative luciferase activity was reduced
compared with co-transfected negative control and pcDNA3.1-CDC42-WT
(P<0.0001 of HuH-7). Whereas, co-transfected miR-185 and
pcDNA3.1-CDC42-MUT had little change compared with NC, and the
P-value of HuH-7 was 0.8105 (Fig.
3B). The results show that miR-185 can directly bind to the
3′UTR of CDC42. Previously it was shown that miR-185 can directly
bind to the 3′UTR of CDC42, and the expression of miR-185 and CDC42
had a negative correlation. Therefore, we hypothesized that miR-185
could regulate the expression of CDC42. Unfortunately, in
overexpressed or knocked down miR-185, the mRNA level of CDC42 had
almost no change in either HuH-7 (P=0.074 or 0.8190 for
overexpression or knockdown miR-185) cells. On the contrary, the
expression in protein level was reduced or increased significantly
in HuH-7 cells (Fig. 3C).
Interference of CDC42 partially blocks
the function of miR-185
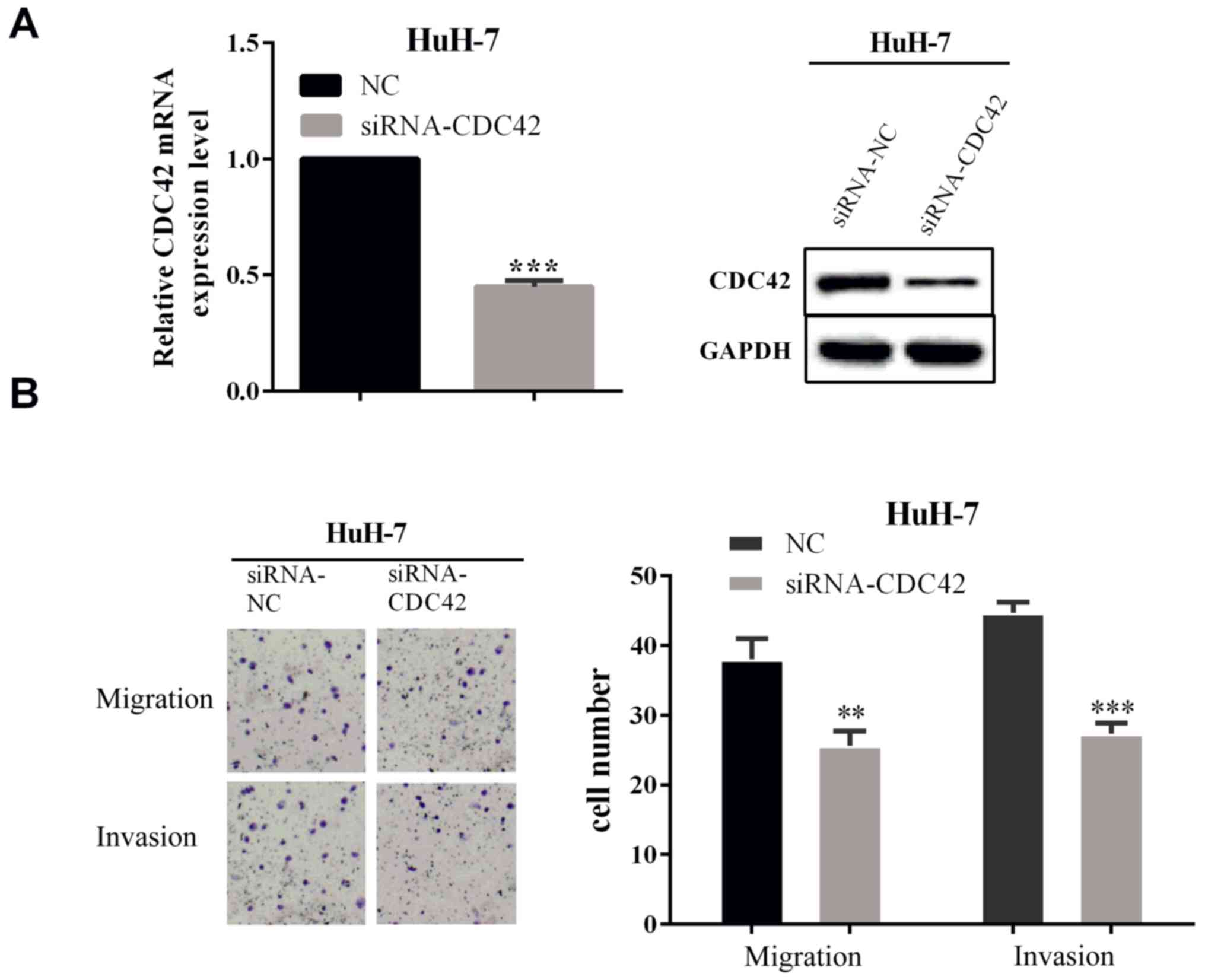
To verify whether miR-185 affected cell migration
and invasion by regulating CDC42 expression, we carried out a
rescue experiment, which directly interfered with the expression of
CDC42 to detect the ability of cell migration and invasion. The
interference effect was measured by RT-qPCR (P<0.0001) and WB as
shown in Fig. 4A. We calculated the
cell number of migration and invasion by Transwell assay. The
results showed that when interfered with CDC42, the cells number of
migration and invasion was reduced in HuH-7 (P=0.0043 and 0.0002)
cells, which illustrated that the interfered CDC42 expression could
reverse the partial function of miR-185.
Discussion
Primary liver cancer patients especially with HCC
are prone to metastasis and recurrence. It has been reported that
tumorigenesis was caused by oncogenes or loss of tumor inhibitors,
whereas, several of the molecular biological mechanisms remain
unknown (21). miRNAs are important
epigenetic regulators and usually act as oncogene or tumor
suppressor gene in progression of various cancers (22–28).
miR-185 induces cell cycle arrest at the G1 phase to suppress
proliferation in human non-small cell lung cancer cells (7). The expression of miR-185 in carcinoma
was significantly lower than that in normal tissues, including HCC,
colon cancer and prostate carcinoma (6,8,11). In this study, we discovered that
miR-185 is usually drownregulated in HCC and the average expression
level of miR-365 in tumor tissues was obviously lower than that in
paracancerous tissues.
To ivestigate the role of miR-185, we overexpressed
or knocked down miR-185 to detect the migration and invasion in
HCC. It was found that the level of miR-185 was correlated with
tumor migration and invasion. The ability of migration and invasion
were reduced in transfection with miR-185 mimic, conversely,
increased when the miR-185 inhibitor was transfected. Therefore,
miR-185 mimics suppressed HCC cell migration and invasion, whereas,
miR-215 inhibitor promoted cell migration and invasion.
miR-185 expression was negative correlated with
lymph node metastasis and low expression of miR-185 is associated
with poor prognosis in colon cancer and glioma patients (29). miR-185 was found to inhibit cell
invasion, which indicated that it may be a potential prognostic
marker in glioma (29). Previous
studies have revealed that miR-185 impedes cell migration and
invasion, suppressed tumor growth and increasing cisplatin
sensitivity by facilitating apoptosis, indicating it as a tumor
inhibiting factor (8,30,31). In
our study, we divided 63 patients into high expression group and
low expression group according to RNA levels, and we found that the
prognosis of high expression group was better.
In human colorectal cells miR-185 suppressed cell
proliferation through inhibiting CDC42 in transcription level
(11). In our study, we first
confirmed CDC42 was a direct target of miR-185 in HCC by RT-qPCR.
Furthermore, miR-185 was negatively correlated with CDC42, which
was highly expressed in carcinomatous tissues (32,33).
CDC42 is regulated by several miRNAs and genes in
HCC. For example, miR-137 was reported to inhibit cell
proliferation and metastasis by regulating CDC42 in HCC (13). In esophageal squamous cell carcinoma,
miR-107 targeting CDC42 suppress proliferation, migration and
invasion (33). Furthermore, Zhang
et al (34) and Xu et
al (32) found that Bif-1
(endophilin B1 or SH3GLB1) and HBx (Hepatitis B Virus X Protein)
promotes HCC cell proliferation, migration and inhibits apoptosis
via CDC42 expression and activity. To identify whether CDC42 is a
direct target of miR-365, we mutated the binding site of CDC42
3′UTR and performed the luciferase reporter assay. Trough
luciferase reporter assay, we confirmed that CDC42 was a direct
target of miR-185, which was the first time to propose that miR-185
targeted CDC42 in HCC. In addition, Liu et al suggested that
miR-185 targeted CDC42 and inhibited cell proliferation in
colorectal cells (11). Consistent
with these findings, in this study we found that CDC42 protein
expression was mediated by miR-185 and changed along with miR-185
in protein level, whereas miR-185 did not alter the CDC42 mRNA
level.
Furthermore, in order to confirm the biological role
of CDC42 in HCC, recovery experiment was carried out and it was
found that interference of CDC42 inhibited the migration and
invasion of HCC cells. Additionally, we found that depletion of
CDC42 reversed partial the function of miR-185.
In conclusion, our results suggest that miR-185 is
usually expressed at low level in HCC. Moreover, interference of
miR-185 promotes the ability of migration and invasion of HCC cells
by directly targeting CDC42. Thus, our findings displayed that
miR-185 may be developed to a potential diagnostic marker of HCC.
miR-185 was expressed at low level in HCC tissues and cell lines
indicating poor prognosis. miR-185 inhibited the ability of
migration and invasion through downregulation of the expression
level of CDC42 by direct targeting.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QZ contributed to the conception of the study and
wrote the manuscript. YC contributed significantly to perform the
experiment. KL contributed significantly to the analysis of the
data and helped in the writing of the manuscript. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Central Hospital of Zibo (Zibo, China) and informed consent of each
patient was received.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lauressergues D, Couzigou JM, Clemente HS,
Martinez Y, Dunand C, Becard G and Combier JP: Primary transcripts
of microRNAs encode regulatory peptides. Nature. 520:90–93. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Voinnet O: Origin, biogenesis, and
activity of plant microRNAs. Cell. 136:669–687. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Di Giacomo G, Koss M, Capellini TD,
Brendolan A, Popperl H and Selleri L: Spatio-temporal expression of
Pbx3 during mouse organogenesis. Gene Expr Patterns. 6:747–757.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lichtenauer UD, Duchniewicz M, Kolanczyk
M, Hoeflich A, Hahner S, Else T, Bicknell AB, Zemojtel T, Stallings
NR, Schulte DM, et al: Pre-B-cell transcription factor 1 and
steroidogenic factor 1 synergistically regulate adrenocortical
growth and steroidogenesis. Endocrinology. 148:693–704. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu C, Chen Z, Hu X, Wang L, Li C, Xue J,
Zhang P, Chen W and Jiang A: MicroRNA-185 downregulates androgen
receptor expression in the LNCaP prostate carcinoma cell line. Mol
Med Rep. 11:4625–4632. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Takahashi Y, Forrest AR, Maeno E,
Hashimoto T, Daub CO and Yasuda J: miR-107 and miR-185 can induce
cell cycle arrest in human non small cell lung cancer cell lines.
PLoS One. 18:e66772009. View Article : Google Scholar
|
|
8
|
Imam JS, Buddavarapu K, Lee-Chang JS,
Ganapathy S, Camosy C, Chen Y and Rao MK: MicroRNA-185 suppresses
tumor growth and progression by targeting the Six1 oncogene in
human cancers. Oncogene. 29:4971–4979. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Akçakaya P, Ekelund S, Kolosenko I,
Caramuta S, Ozata DM, Xie H, Lindforss U, Olivecrona H and Lui WO:
miR-185 and miR-133b deregulation is associated with overall
survival and metastasis in colorectal cancer. Int J Oncol.
39:311–318. 2011.PubMed/NCBI
|
|
10
|
Lu ZJ, Lu LG, Tao KZ, Chen DF, Xia Q, Weng
JJ, Zhu F, Wang XP and Zheng P: MicroRNA-185 suppresses growth and
invasion of colon cancer cells through inhibition of the
hypoxiainducible factor-2alpha pathway in vitro and in vivo. Mol
Med Rep. 10:2401–2408. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu M, Lang N, Chen X, Tang Q, Liu S,
Huang J, Zheng Y and Bi F: miR-185 targets RhoA and Cdc42
expression and inhibits the proliferation potential of human
colorectal cells. Cancer Lett. 301:151–160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu X, Li Y, Shen H, Li H, Long L, Hui L
and Xu W: miR-137 inhibits the proliferation of lung cancer cells
by targeting Cdc42 and Cdk6. FEBS Lett. 587:73–81. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu M, Lang N, Qiu M, Xu F, Li Q, Tang Q,
Chen J, Chen X, Zhang S, Liu Z, et al: miR-137 targets Cdc42
expression, induces cell cycle G1 arrest and inhibits invasion in
colorectal cancer cells. Int J Cancer. 128:1269–1279. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fritz G, Just I and Kaina B: Rho GTPases
are over-expressed in human tumors. Int J Cancer. 81:682–687. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zegers MM and Friedl P: Rho GTPases in
collective cell migration. Small GTPases. 5:e289972014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Arias-Romero LE and Chernoff J: Targeting
Cdc42 in cancer. Expert Opin Ther Targets. 17:1263–1273. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Stengel K and Zheng Y: Cdc42 in oncogenic
transformation, invasion, and tumorigenesis. Cell Signal.
23:1415–1423. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang L, Wang L and Zheng Y: Gene targeting
of Cdc42 and Cdc42GAP affirms the critical involvement of Cdc42 in
filopodia induction, directed migration, and proliferation in
primary mouse embryonic fibroblasts. Mol Biol Cell. 17:4675–4685.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chang CS, Huang SM, Lin HH, Wu CC and Wang
CJ: Different expression of apoptotic proteins between HBV-infected
and non-HBV-infected hepatocellular carcinoma.
Hepatogastroenterology. 54:2061–2068. 2007.PubMed/NCBI
|
|
20
|
Cooper AB, Wu J, Lu D and Maluccio MA: Is
autotaxin (ENPP2) the link between hepatitis C and hepatocellular
cancer? J Gastrointest Surg. 11:1628–1634. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Porta C, Larghi P, Rimoldi M, Totaro MG,
Allavena P, Mantovani A and Sica A: Cellular and molecular pathways
linking inflammation and cancer. Immunobiology. 214:761–777. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jiang C, Chen X, Alattar M, Wei J and Liu
H: MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis
of gastric cancer. Cancer Gene Ther. 22:291–301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Duan J, Zhang H, Qu Y, Deng T, Huang D,
Liu R, Zhang L, Bai M, Zhou L, Ying G, et al: Onco-miR-130 promotes
cell proliferation and migration by targeting TGFbetaR2 in gastric
cancer. Oncotarget. 7:44522–44533. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fabbri M, Garzon R, Cimmino A, Liu Z,
Zanesi N, Callegari E, Liu S, Alder H, Costinean S,
Fernandez-Cymering C, et al: MicroRNA-29 family reverts aberrant
methylation in lung cancer by targeting DNA methyltransferases 3A
and 3B. Proc Natl Acad Sci USA. 104:15805–15810. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He L, Thomson JM, Hemann MT,
Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe
SW, Hannon GJ, et al: A microRNA polycistron as a potential human
oncogene. Nature. 435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tang H, Wang Z, Liu X, Liu Q, Xu G, Li G
and Wu M: LRRC4 inhibits glioma cell growth and invasion through a
miR-185-dependent pathway. Curr Cancer Drug Targets. 12:1032–1042.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xiang Y, Ma N, Wang D, Zhang Y, Zhou J, Wu
G, Zhao R, Huang H, Wang X, Qiao Y, et al: miR-152 and miR-185
co-contribute to ovarian cancer cells cisplatin sensitivity by
targeting DNMT1 directly: a novel epigenetic therapy independent of
decitabine. Oncogene. 33:378–386. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li S, Ma Y, Hou X, Liu Y, Li K, Xu S and
Wang J: miR-185 acts as a tumor suppressor by targeting AKT1 in
non-small cell lung cancer cells. Int J Clin Exp Pathol.
8:11854–11862. 2015.PubMed/NCBI
|
|
32
|
Xu Y, Qi Y, Luo J, Yang J, Xie Q, Deng C,
Su N, Wei W, Shi D, Xu F, et al: Hepatitis B virus X protein
stimulates proliferation, wound closure and inhibits apoptosis of
HuH-7 cells via CDC42. Int J Mol Sci. 18:e5862017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sharma P, Saini N and Sharma R: miR-107
functions as a tumor suppressor in human esophageal squamous cell
carcinoma and targets Cdc42. Oncol Rep. 37:3116–3127. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang C, Liu F, Chen H, Li N, Luo Z, Guo
W, Huang D, Tang S, Wang H, Cheng S, et al: Bif-1 promotes tumor
cell migration and metastasis via Cdc42 expression and activity.
Clin Exp Metastasis. 34:11–23. 2017. View Article : Google Scholar : PubMed/NCBI
|