Introduction
Bladder cancer (BC), one of the most prevalent
cancers, arises from the epithelial lining of the urinary bladder,
and is mainly caused by smoking and mutation (1). In current years, the standard treatment
of invasive BC is radical cystectomy, although the therapeutic
strategies have improved (2), the
5-year survival was 62% due to high rate of metastasis and invasion
(3,4).
Therefore, looking for the tumor markers for early diagnosis is
particularly important for treatment of bladder tumors.
CAMP-response element binding protein (CREB1), a
transcription factor, regulates gene transcription through
phosphorylation and dephosphorylation. CREB1, as a proto-oncogenic
transcription factor promotes expression of its target genes taking
part in metabolism and DNA repair (5). Aberrant expression of CREB1 was reported
to be connected with variety of cancers, including gastric,
colorectal, ovarian and breast cancers (6–9). In
colorectal cancer, Ye et al discovered that CREB1 was
significantly upregulated and promoted cell migration and invasion
(10). Similar findings were reported
by Wang et al (11) knockdown
of CREB1 inhibited cell proliferation and motility in prostate
cells. Therefore, we strongly believe that CREB1 could play
important roles in BC.
microRNAs (miRNAs) are a class of non-coding RNA
sequences with 22–28 nucleotides, which could inhibit gene
expression at post-transcriptional level by binding to the
3′-untranslated region (3′-UTR) of target mRNA (12,13).
miRNAs are reported to be involved in several biological processes
in BC, including miR-1, miR-126, miR-202 and miR-149 (14–17). There
are several miRNAs binding to CREB1 and regulating the expression
of CREB1 in cancers, including miR-590, miR-1224, miR-205 and
miR-122 (5,18–20).
miR-122, a novel microRNA, was predicted to be downregulated in
many cancers including BC (21). In
hepatocellular carcinoma, miR-122 inhibited epithelial-mesenchymal
transition through snail1 and snail2 (22). Maierthaler et al discovered
that miR-122 was a prognostic marker in colorectal cancer (23). Similar finding were reported by Wang
et al that miR-122 inhibited tumor growth and angiogenesis
by targeting VEGFC in BC (24). In
addition, Rao et al discovered that miR-122 inhibited
proliferation and invasion by targeting CREB1 in gastric cancer
(20). Considering these functions,
to the best of our knowledge, we first propose that miR-122
regulated CREB1 expression and mediated cell proliferation and
invasion in BC.
Patients and methods
Patients and clinical samples
A collection of 47 BC tissues as well as
corresponding healthy tissue samples (5 cm away from the tissues)
were obtained from patients who underwent surgery at China-Japan
Union Hospital of Jilin University from 2015 to 2017. All samples
were snap-frozen in liquid nitrogen and saved in −80°C after
resection. None of the patients had medication or radiation
therapy. This study was approved by the Ethics Committee of
China-Japan Union Hospital of Jilin University (Changchun, China)
and obtained written informed consent from all the patients.
Cell lines and cell culture
Human BC cell lines T24, UM-UC-3 and J82 and normal
bladder cells SV-HUC-1 were obtained from the American Type Culture
Collection (ATCC; Rockville, MD, USA). Cells were maintained in
RPMI-1640 with 10% FBS (both from Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) then cultured in an incubator at 37°C with
5% CO2.
RNA isolation and RT-qPCR
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was employed to isolate RNAs, which including
miRNA from the cultured cells and tissues. OneStep
PrimeScript® cDNA Synthesis kit (Takara Biotechnology
Co., Ltd., Dalian, China) was applied to perform
reverse-transcription and synthesize first cDNA chain. Fast
SYBR-Green Master Mix and TaqMan microRNA assay kits were employed
to perform the RT-qPCR for CREB1 and miR-122, respectively, using
ABI PRISM7900 Sequence Detection System (all from Applied
Biosystems; Thermo Fisher Scientific, Inc.). GAPDH and U6 were
utilized as internal reference for CREB1 and miR-122, respectively.
The specific stem-loop RT primers were used for reverse
transcription reaction as follows: miR-122:
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAAACACC-3′; U6:
5′-AACGCTTCACGAATTTGCGT-3′; CREB1: F: CTTTTCTCCGGAACACAGATTTC; R:
GATTTGCCAAGTGGGAGGGA; GAPDH: F: 50-CACTCCTCCACCTTTGA-30, R:
50-CCACCACCCTGTTGCTG-3′. The thermocycling parameters were 95°C for
3 min and 40 cycles of 95°C for 15 sec followed by 60°C for 30 sec.
The quantification of CREB1 or miR-122 mRNA levels was through
measuring Cq values and normalized using the 2−ΔΔCq
method (25).
Protein extraction and western
blotting
The cells were washed with cold PBS buffer and
extracted using RIPA lysis buffer with protease inhibitor (Beyotime
Institute of Biotechnology, Shanghai, China), followed by
centrifugation for 20 min at 4°C with 12,000 × g. The supernatants
were collected to determine the protein concentration using BCA
reagent kit (Beijing Solarbio Science & Technology Co., Ltd.,
Beijing, China). The protein lysates (50 µg/lane) were fractionated
using 10% SDS-PAGE and transferred to a PVDF membrane (Bio-Rad
Laboratories, Inc., Hercules, CA, USA); The membrane was blocked
using 5% non-fat dried milk for 1 h at room temperature. The blots
were incubated with anti-CREB1 rabbit polyclonal antibody
(dilution, 1:1,000; cat. no. SAB4300519; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) and anti-GAPDH (dilution, 1:3,000; cat. no.
G5262; Sigma-Aldrich; Merck KGaA) at 4°C overnight and anti-rabbit
antibody (1:3,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA)
for 2 h at room temperature, which was normalized by GAPDH. Signal
detection employed ECL detection system (Thermo Fisher Scientific,
Inc., Waltham, MA, USA).
Cell proliferation assay
The cell proliferative ability was measured by Cell
Counting Kit-8 assay (CCK-8; Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan). A total of 100 µl T24 or J82 cells
(2×103 cells/well) were seeded in 96-well plates and
cultured for 24, 48, 72 or 96 h, respectively. Then 10 µl of CCK-8
solution was added in each well and incubated at 37°C for 1 h.
Microplate Reader (Epoch; BioTek Instruments, Inc., Winooski, VT,
USA) was applied to determine the absorbance at 490 nm.
Transwell assay
Tanswell chambers (Corning Costar, Beijing, China)
with 8 µm pore size membranes were employed to assess invasive
ability. Transwell chamber in 24-plate well with Matrigel (Clontech
Laboratories, Inc., Mountainview, CA, USA) was coated. T24 or J82
cells with a density of 5×104 were added in the upper
chamber in 200 µl medium without FBS. Whereas, 500 µl normal medium
with 15% FBS was added in the lower chamber for use as a
chemoattractant. The cells were incubated for 24 h, and the
unattached cells were removed using cotton swab. The invaded cells
were fixed with methanol and then stained using 1% crystal violet;
and cell counting was carried out under the microscope BX51 Olympus
(Shenzhen, China).
Transfection
All vectors, including miR-122 mimic, miR-122
inhibitor, pcDNA3.1-CREB1 and luciferase reporter plasmids were
purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China).
miR-122 mimic or inhibitor were applied to overexpress or knockdown
miR-122, while siRNA-CREB1 and pcDNA3.1-CREB1 were employed to
knock down or overexpress CREB1. The sequences of miR-122
mimic/inhibitor and negative control (NC) were:
5′-UGGAGUGUGACAAUGGUGUUUG-3′; 5′-CAAACACCAUUGUCACACUCCA-3′; and
5′-UUCUCCGAACGUGUCACGUTT-3′. Scrambled nucleotide sequences were
the NC of miR-184 inhibitor and miR-184 mimic.
Before transfection, T24 or J82 cells were seeded in
6-well plate and cultured overnight. A total of 4 µg vectors and 8
µl Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was mixed in 1.5 ml microcentrifuge tube and then
let stand for 20 min; then mixed and added into cells and cultured
at 37°C for 48 h.
Plasmid construction and luciferase
reporter assay
3′-UTR fragment of CREB1 mRNA, containing the
putative miR-122 binding sequence was amplified by PCR and cloned
into pmirGlo luciferase reporter vector (named pmirGlo-CREB1-WT;
WT). QuikChange Multi Site-Directed Mutagenesis kit (Agilent
Technologies, Inc., Santa Clara, CA, USA) was used for the
site-directed mutagenesis of CREB1 3′-UTR (pmirGlo-CREB1-MUT; MUT)
with WT as template.
For luciferase reporter assays, T24 cells were
seeded into 6-well plate, miRNA-122 and WT or MUT reporter plasmid
were transiently co-transfected using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). After transfection
for 48 h, luciferase activity was measured using a dual-luciferase
assay system (Promega Corporation, Madison, WI, USA) according to
the manufacturer's instruction, which was normalized by
renilla-luciferase activity.
Statistical analysis
The data are presented as mean ± standard deviation.
All statistical analyses were performed with SPSS 16.0 software
(SPSS, Inc., Chicago, IL, USA). Two-tailed Student's t-test and
one-way ANOVA followed by Tukey's post hoc test were employed to
analyze two groups and three or more groups, respectively. Pearsons
test analysis was applied to analyze the correlations between
miR-122 and CREB1 mRNA expression. P<0.05 was considered to
indicate a statistically significant difference.
Results
CREB1 is upregulated in BC tissues and
cells
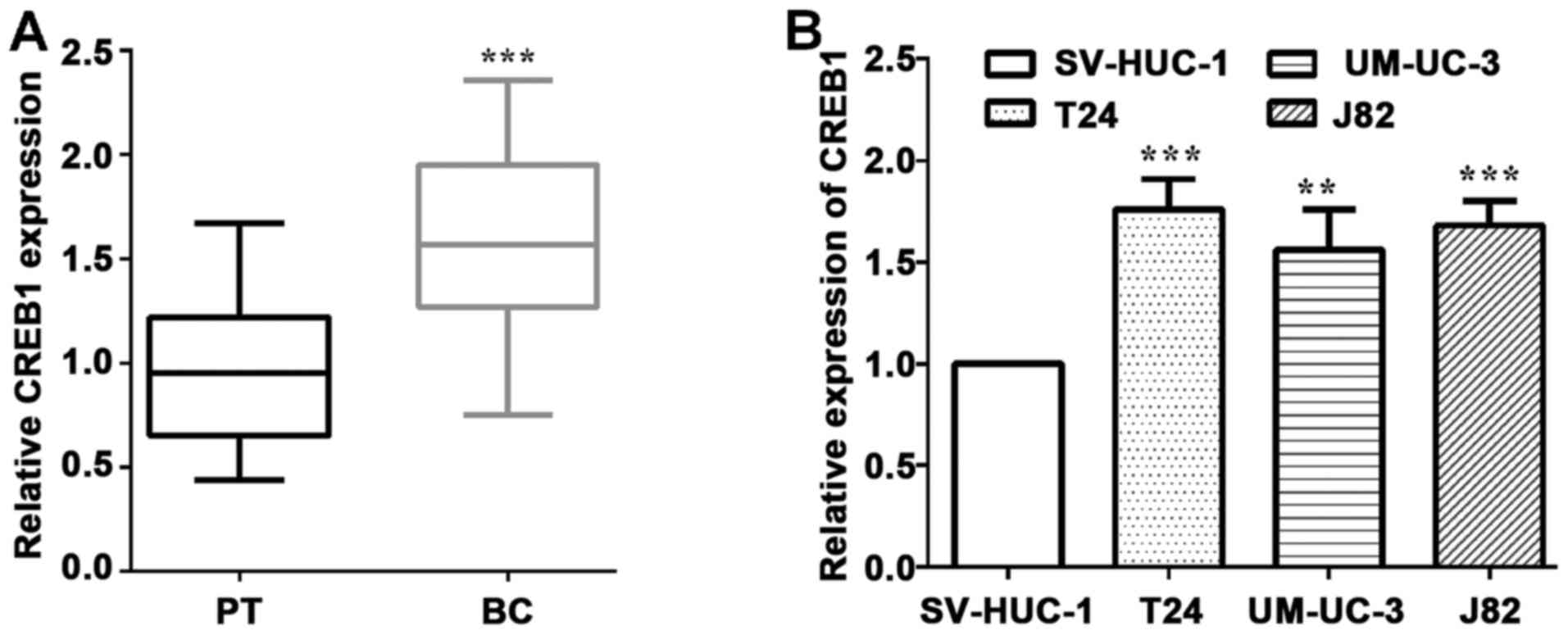
To validate the expression of CREB1 in BC, we
employed RT-qPCR to examined CREB1 expression in clinical specimens
and cells lines. As shown in Fig. 1A,
49 paired of BC and corresponding healthy tissues were collected
and measured the mRNA level of CREB1. As expected, CREB1 expression
in BC tissues was significantly higher than that in the
corresponding healthy tissues (P<0.0001) (Fig. 1A). Furthermore, CREB1 was expressed at
prominently higher levels in bladder cancer patients with tumor
(P=0.030), TNM stage (P=0.014), lymph node metastasis (P=0.028) and
the expression of miR-122 (P=0.014), while it had tendency to have
association with invasion (P=0.053) (Table I). We measured the mRNA levels of
CREB1 in three human BC cells and a normal bladder cell, and found
that CREB1 was upregulated in BC T24 (P=0.0009), UM-UC-3 (P=0.0083)
and J82 cells (P=0.0006), compared to the normal SV-HUC-1 cells
(Fig. 1B).
 | Table I.CREB1 expression and
clinicopathological features in 47 BC. |
Table I.
CREB1 expression and
clinicopathological features in 47 BC.
|
|
| CREB1 expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | Cases (n=47) | Low (%) (n=22) | High (%) (n=25) | P-valuea |
|---|
| Sex |
| Male | 27 | 13 (48.1) | 14 (51.9) | 0.831 |
|
Female | 20 | 9 (45.0) | 11 (55.0) |
|
| Age (years) |
|
<50 | 22 | 10 (45.6) | 12 (54.4) | 0.861 |
| ≥50 | 25 | 12 (48.0) | 13 (52.0) |
|
| Tumor size (mm) |
| ≤5.0 | 22 | 14 (63.6) | 8 (36.4) | 0.030a |
|
>5.0 | 25 | 8 (32.0) | 17 (68.0) |
|
| TNM stage |
| I–II | 21 | 14 (66.7) | 7 (33.3) | 0.014a |
|
III–IV | 26 | 8 (30.8) | 18 (67.2) |
|
| Lymph-node
metastasis |
| 0–2 | 24 | 15 (62.5) | 9 (37.5) | 0.028a |
|
>2 | 23 | 7 (30.4) | 16 (69.6) |
|
| Invasion |
| No | 25 | 15 (60.0) | 10 (40.0) | 0.053 |
|
Yes | 22 | 7 (31.8) | 15 (68.2) |
|
| miR-122 |
| Low
expression | 26 | 8 (30.8) | 18 (69.2) | 0.014a |
| High
expression | 21 | 14 (66.7) | 7 (33.3) |
|
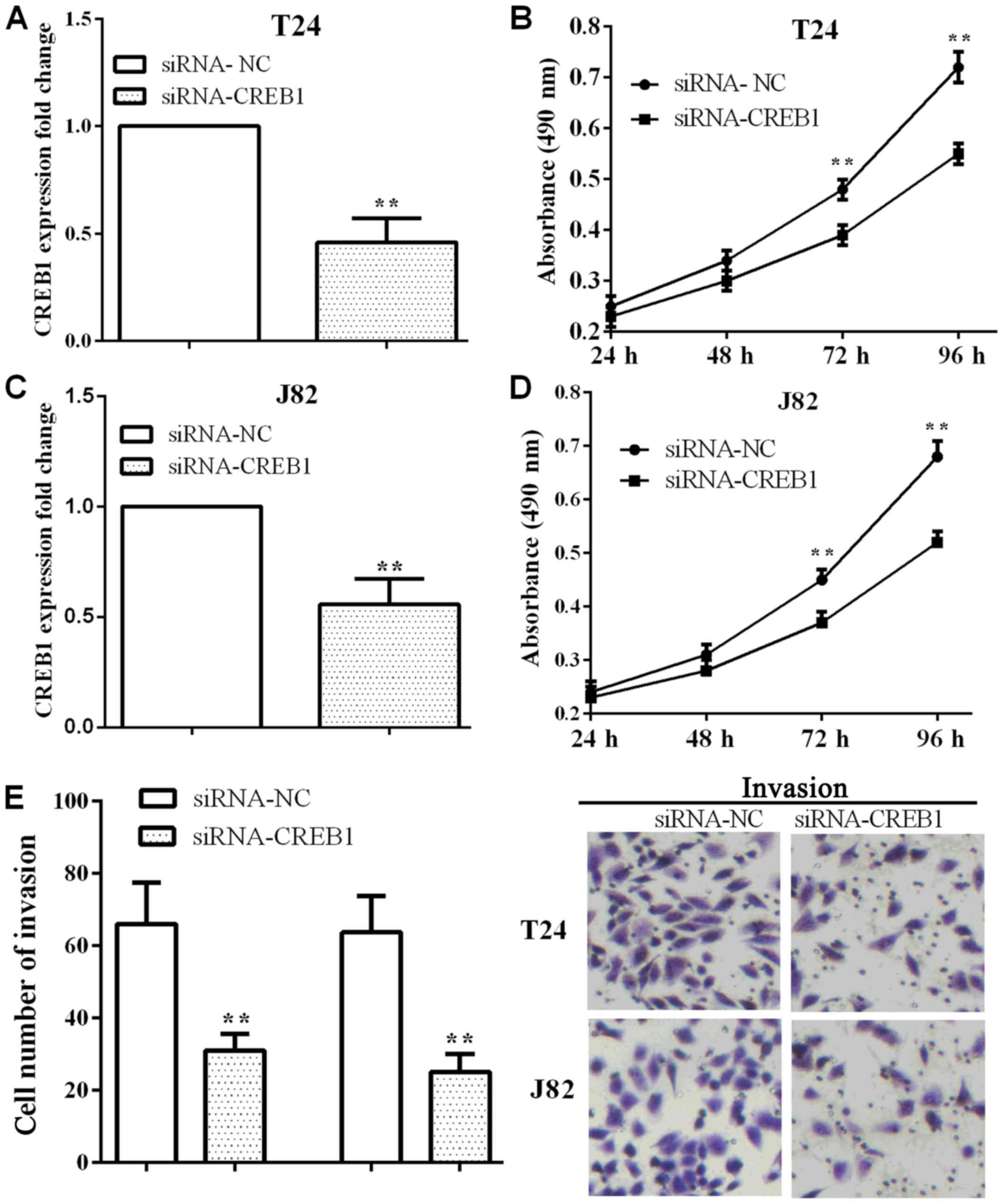
Knockdown of CREB1 inhibits cell
proliferation and invasion
To explore the impact of CREB1, the cell
proliferative and invasive abilities were measured after knockdown
CREB1 in T24 cells. siRNA-CREB1 was applied to knockdown CREB1, and
the CREB1 mRNA level was reduced in T24 (P=0.0010) and J82
(P=0.0026) cells as shown in Fig. 2A and
C. When CREB1 was knocked down, the absorbance was reduced in
T24 and J82 cells at 72 h (P=0.0053 and 0.0080) and 96 h (P=0.0012
and 0.0015) (Fig. 2B and D). On the
other hand, the number of invasion was reduced (P=0.0081 and
0.0039) either in T24 or J82 cells (Fig.
2E), which suggested that knockdown CREB1 inhibited cell
proliferation an invasion in BC T24 and J82 cells.
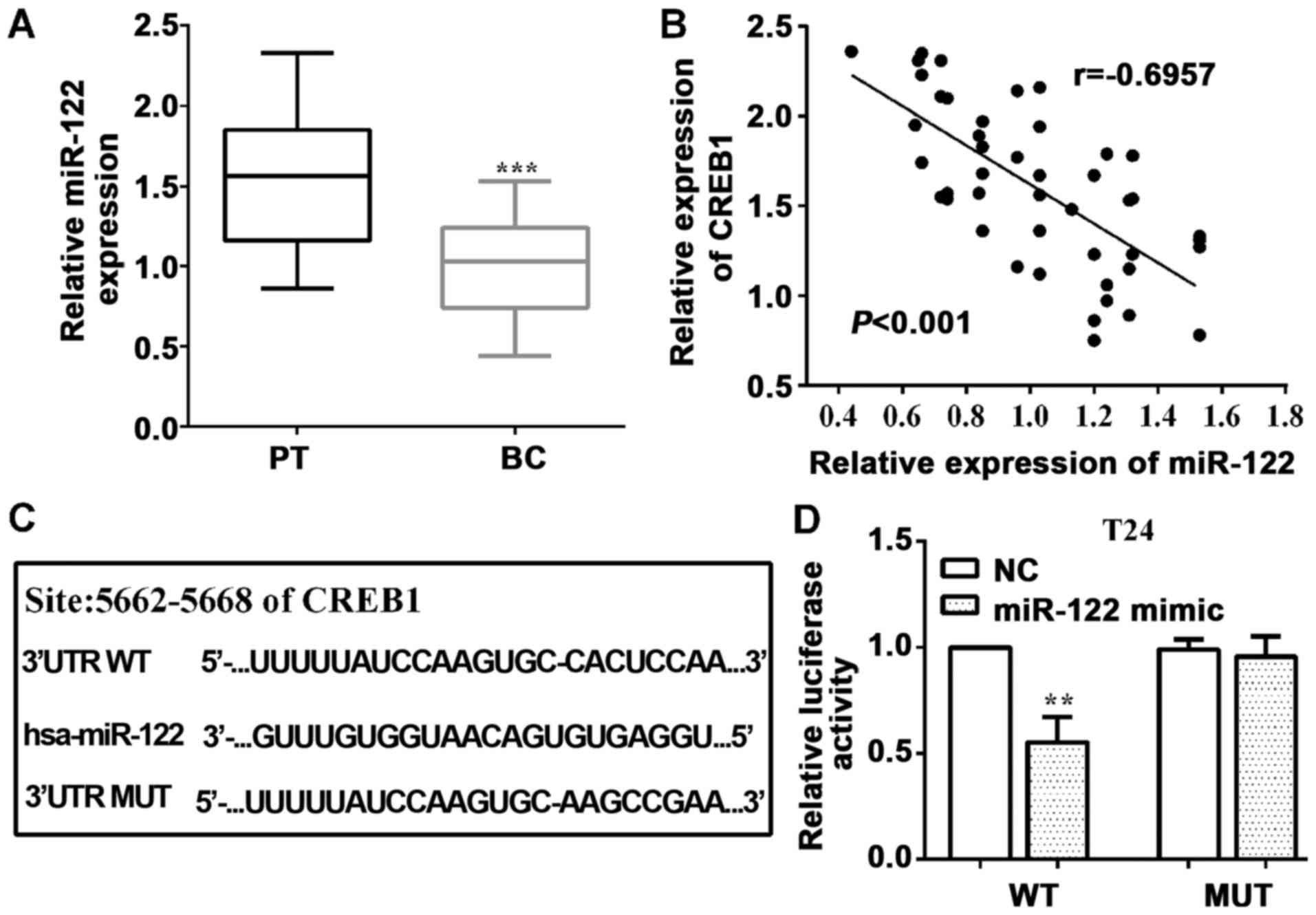
miR-122 is downregulated and targeted
to CREB1 in BC
As we detected the effect of CREB1 on cell
proliferation an invasion, we considered what impacted CREB1 and
then influenced cell proliferative an invasive abilities.
Therefore, we predicted microRNAs by TargetScan, and we found
miR-122 was upstream of CREB1. Then, we determined miR-122 level
employing RT-qPCR and found that miR-122 was downregulated
(P<0.0001) in BC tissues vs. healthy tissues (Fig. 3A). Furthermore, we analyzed the
expression of CREB1 and miR-122 in BC tissue samples, and
discovered that they had a negative association between them
(P<0.0001, r=−0.6957) (Fig.
3B).
In addition, we mutated the binding sequences of
miR-122 on CREB1 3′-UTR from 5′-CACUCCA-3′ (WT) to 5′-AAGCCGA-3′
(MUT), which were inserted into pmirGlo vector, as shown in
Fig. 3C. To confirm miR-122 direct
targeting to CREB1, luciferase reporter assay was performed. As
expected, the luciferase activity was reduced (P=0.0027) by
wild-type, while the mutant was not in T24 cells (P=0.6194)
(Fig. 3D).
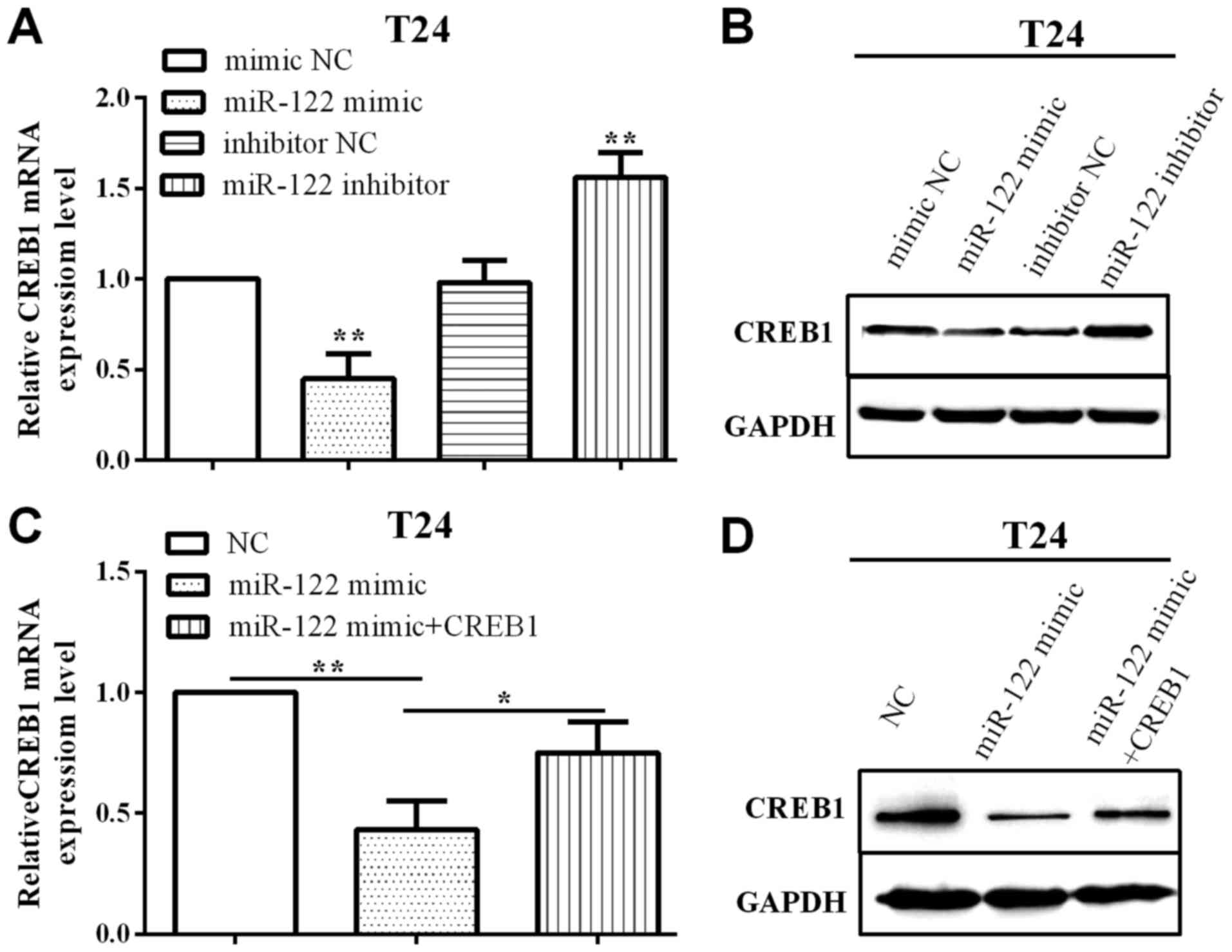
CREB1 is mediated by miR-122 and
regulates cell proliferation and invasion in BC
To evaluate the impact of miR-122 on CREB1, we
employed miR-122 mimic and inhibitor to overexpress or knock down
miR-122 and measured the CREB1 expression in T24 cells. As shown in
Fig. 4A and B, when transfected with
miR-122 mimic, both CREB1 miRNA and protein levels were decreased
(P=0.0024), whereas increased (P=0.0055) when transfected with
miR-122 inhibitor.
In addition, when overexpressed CREB1 was used pcDNA
3.1-CREB1, CREB1 expression was increased (P=0.0351), based on the
reduction by miR-122 evaluated in T24 cells by RT-qPCR and western
blotting (Fig. 4C and D).
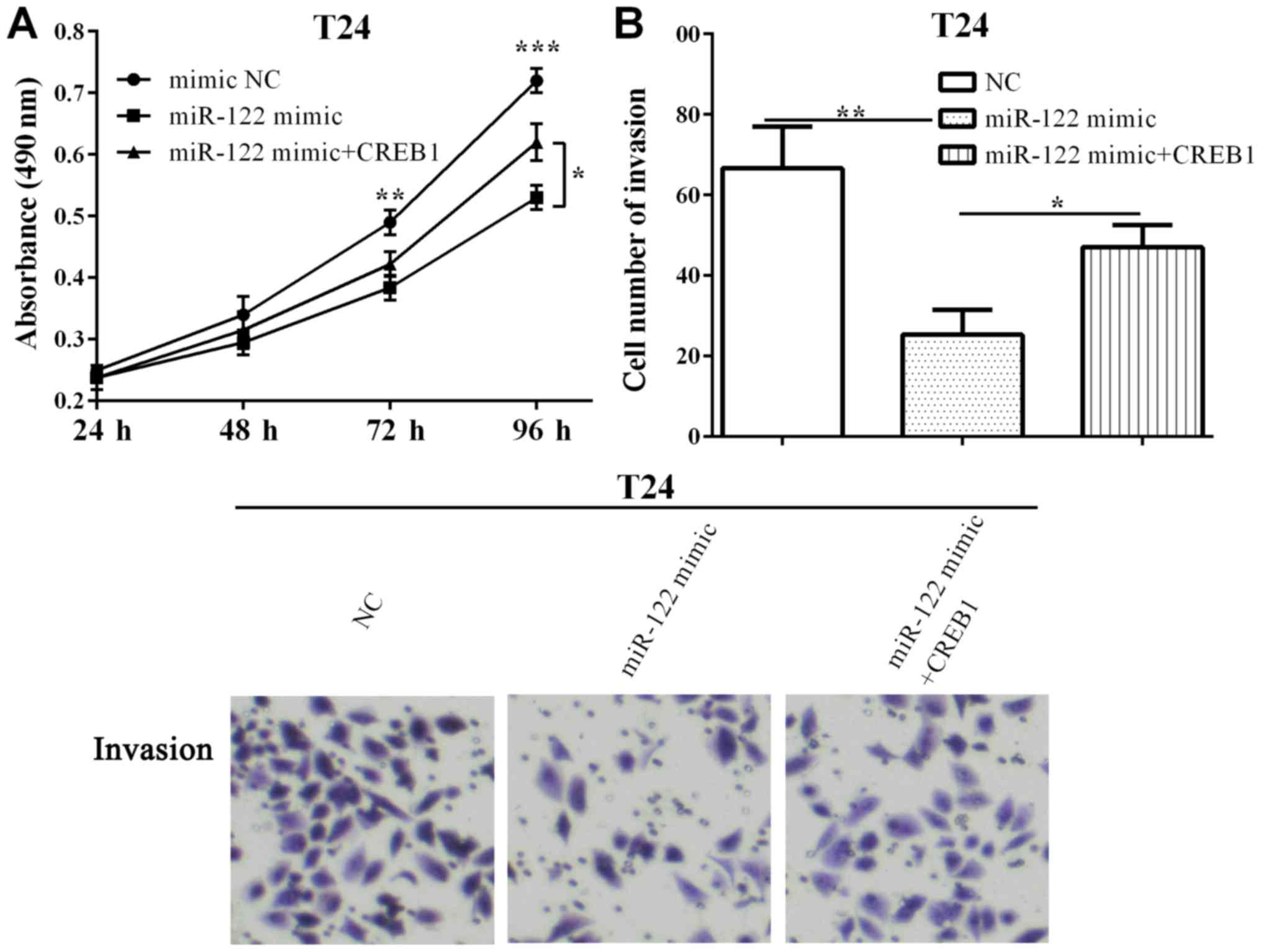
Furthermore, the proliferative ability was decreased
(P=0.0029 and 0.0003) when transfected with miR-122 after 72 h and
96 h, and was reversed (P=0.0124) by re-expressing CREB1 at 96 h
(Fig. 5A). Additionally, we
determined the invasive ability that CREB1 could counteract
(P=0.0105) and the inhibitory (P=0.0039) effect of miR-122 on cell
invasion in T24 cells (Fig. 5B).
Discussion
BC is one of the most prevalent cancers, which
arises from the epithelial lining of the urinary bladder (1). Moreover, although the therapeutic
strategies have improved, the 5-year survival is 62% due to high
rate of metastasis and invasion (3,4). Thus,
identifying new targets for the development of effective
therapeutics for bladder tumors is urgent. In our study, we found
that CREB1 was upregulated in bladder tissues and T24, UM-UC-3 and
J82 cells, while miR-122 was upregulated and had a negative
correlation with CREB1. Knockdown of CREB1 inhibited T24 and J82
cell proliferative and invasive capacities. In addition, CREB1 was
directly targeted by miR-122 in BC and regulated its expression. We
discovered that CREB1 could reverse the function of miR-122
partially on cell proliferation and invasion in T24 cells.
CREB1 acted as a proto-oncogenic transcription
factor, promoted gene transcription through phosphorylation and
dephosphorylation (5). CREB1 acted as
predictor of prostate cancer recurrence and a critical driver of
pro-survival, cell cycle and metabolic transcription programs
(26). CREB1 was upregulated and
promoted cell proliferation, invasion and acted as an independent
prognostic factor in various cancers (10,11). Even
in BC, CREB1 was involved in EMT (27), but the underlying molecular mechanisms
are still elusive. Our findings are consistent with the above
findings, CREB1 was upregulated in BC tissues and cell lines.
Further investigation in our present research found similar results
that CREB1 promoted BC T24 and J82 cell proliferation and invasion
using MTT and Transwell assays, indicating that CREB1 can be an
oncogene in BC. The data uncovered that the proliferation and
invasion activity were significantly suppressed in T24 cells
transfected with siRNA-CREB1 as compared with the NC group.
However, the biological mechanism is unclear. The results were
consistent with the findings of Shabestari et al in human
pre-B acute lymphoblastic leukemia cells (28). However, due to the limitation of
conditions we did not do IHC to evaluate the expression of
CREB1.
miRNAs are non-coding RNAs, which inhibited gene
expression at post-transcriptional level through targeting the
3′-untranslated region (3′-UTR) of target mRNA (12,13).
miR-122 was predicted to downregulate and inhibit cell
proliferation, invasion and EMT in many cancers (22–24). In
gastric cancer, CREB1 was a direct target gene of miR-122 and
miR-122 regulated the expression of CREB1 (20). Consistent with previous findings of
Wang et al (24), we
discovered that miR-122 was downregulated in BC. Moreover,
consistent with Rao et al (20), miR-122 had a negative correlation with
CREB1 in BC tissues, which, to the best of our knowledge, was the
first time to propose the connection between CREB1 and miR-122 in
BC. We hypothesized that CREB1 may ablate the inhibitory effects of
miR-122 on cell proliferation and invasion in BC based on these
results. In order to prove this hypothesis, we examined the rescue
experiments in BC T24 cells. We found that the overexpression of
miR-122 inhibited cell proliferation and invasion in T24 cells, and
CREB1 partially reversed the role of miR-122 on cell proliferation
and invasion in BC. In the present study, we first proposed the
relationship between CREB1 and miR-122; and we used rescue
experiments to verify that miR-122 regulated cell proliferation and
invasion through targeting CREB1, which was the novelty of the
study.
In conclusion, we demonstrated that CREB1 acts as an
oncogene in BC by reducing cancer growth and invasion. Moreover, we
indicated that CREB1 has an inverse correlation with miR-122 and
the expression of CREB1 by miR-122 in T24 cells. The newly
identified CREB1 may provide further insight into the progression
of BC and offers a promising therapeutic target for the treatment
of BC. The newly identified miR-122/CREB1 axis provides a novel
insight into the pathogenesis of BC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LG performed the data analyses and wrote the
manuscript. MY contributed significantly to analysis and manuscript
preparation. YW contributed to the conception and design of the
study. All authors have read and approved the final study.
Ethics approval and consent to
participate
The Ethics Committee of China-Japan Union Hospital
of Jilin University (Changchun, China) approved the study, and
informed consent was obtained by all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Knowles MA: Molecular pathogenesis of
bladder cancer. Int J Clin Oncol. 13:287–297. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Choueiri TK and Raghavan D: Chemotherapy
for muscle-invasive bladder cancer treated with definitive
radiotherapy: Persisting uncertainties. Nat Clin Pract Oncol.
5:444–454. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stein JP, Lieskovsky G, Cote R, Groshen S,
Feng AC, Boyd S, Skinner E, Bochner B, Thangathurai D, Mikhail M,
et al: Radical cystectomy in the treatment of invasive bladder
cancer: Long-term results in 1,054 patients. J Clin Oncol.
19:666–675. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mayr R, Fritsche HM and Pycha A and Pycha
A: Radical cystectomy and the implications of comorbidity. Expert
Rev Anticancer Ther. 14:289–295. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tan X, Wang S, Zhu L, Wu C, Yin B, Zhao J,
Yuan J, Qiang B and Peng X: cAMP response element-binding protein
promotes gliomagenesis by modulating the expression of oncogenic
microRNA-23a. Proc Natl Acad Sci USA. 109:15805–15810. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gu L, Lu LS, Zhou DL and Liu ZC: UCA1
promotes cell proliferation and invasion of gastric cancer by
targeting CREB1 sponging to miR-590-3p. Cancer Med. 7:1253–1263.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu Y, Lang T, Jin B, Chen F, Zhang Y,
Beuerman RW, Zhou L and Zhang Z: Luteolin inhibits colorectal
cancer cell epithelial-to-mesenchymal transition by suppressing
CREB1 expression revealed by comparative proteomics study. J
Proteomics. 161:1–10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dimitrova N, Nagaraj AB, Razi A, Singh S,
Kamalakaran S, Banerjee N, Joseph P, Mankovich A, Mittal P, DiFeo
A, et al: InFlo: A novel systems biology framework identifies
cAMP-CREB1 axis as a key modulator of platinum resistance in
ovarian cancer. Oncogene. 36:2472–2482. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu J, Zou Z, Nie P, Kou X, Wu B, Wang S,
Song Z and He J: Downregulation of microRNA-27b-3p enhances
tamoxifen resistance in breast cancer by increasing NR5A2 and CREB1
expression. Cell Death Dis. 7:e24542016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ye Q, Su L, Chen D, Zheng W and Liu Y:
Astragaloside IV induced miR-134 expression reduces EMT and
increases chemotherapeutic sensitivity by suppressing CREB1
signaling in colorectal cancer cell line SW-480. Cell Physiol
Biochem. 43:1617–1626. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang S, Wang X, Li J, Meng S, Liang Z, Xu
X, Zhu Y, Li S, Wu J, Xu M, et al: c-Met, CREB1 and EGFR are
involved in miR-493-5p inhibition of EMT via AKT/GSK-3β/Snail
signaling in prostate cancer. Oncotarget. 8:82303–82313.
2017.PubMed/NCBI
|
|
12
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ying SY, Chang DC and Lin SL: The microRNA
(miRNA): Overview of the RNA genes that modulate gene function. Mol
Biotechnol. 38:257–268. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao L, Yan P, Guo FF, Liu HJ and Zhao ZF:
MiR-1-3p inhibits cell proliferation and invasion by regulating
BDNF-TrkB signaling pathway in bladder cancer. Neoplasma. 65:89–96.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhai X and Xu W: Long noncoding RNA ATB
promotes proliferation, migration and invasion in bladder cancer by
suppressing microRNA-126. Oncol Res. 2018.(Epub ahead of print).
https://doi.org/10.3727/096504018×15152072098476
View Article : Google Scholar
|
|
16
|
Zhang L, Xu J, Yang G, Li H and Guo X:
miR-202 inhibits cell proliferation, migration, and invasion by
targeting EGFR in human bladder cancer. Oncol Res. 2018.(Epub ahead
of print). https://doi.org/10.3727/096504018×15149787144385
View Article : Google Scholar
|
|
17
|
Yang D, Du G, Xu A, Xi X and Li D:
Expression of miR-149-3p inhibits proliferation, migration, and
invasion of bladder cancer by targeting S100A4. Am J Cancer Res.
7:2209–2219. 2017.PubMed/NCBI
|
|
18
|
Qian J, Li R, Wang YY, Shi Y, Luan WK, Tao
T, Zhang JX, Xu YC and You YP: MiR-1224-5p acts as a tumor
suppressor by targeting CREB1 in malignant gliomas. Mol Cell
Biochem. 403:33–41. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang J, Ma Y, Wang S, Chen F and Gu Y:
C/EBPα inhibits proliferation of breast cancer cells via a novel
pathway of miR-134/CREB. Int J Clin Exp Pathol. 8:14472–14478.
2015.PubMed/NCBI
|
|
20
|
Rao M, Zhu Y, Zhou Y, Cong X and Feng L:
MicroRNA-122 inhibits proliferation and invasion in gastric cancer
by targeting CREB1. Am J Cancer Res. 7:323–333. 2017.PubMed/NCBI
|
|
21
|
Pop-Bica C, Gulei D, Cojocneanu-Petric R,
Braicu C, Petrut B and Berindan-Neagoe I: Understanding the role of
non-coding RNAs in bladder cancer: From dark matter to valuable
therapeutic targets. Int J Mol Sci. 18:E15142017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jin Y, Wang J, Han J, Luo D and Sun Z:
MiR-122 inhibits epithelial-mesenchymal transition in
hepatocellular carcinoma by targeting Snail1 and Snail2 and
suppressing WNT/β-cadherin signaling pathway. Exp Cell Res.
360:210–217. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Maierthaler M, Benner A, Hoffmeister M,
Surowy H, Jansen L, Knebel P, Chang-Claude J, Brenner H and
Burwinkel B: Plasma miR-122 and miR-200 family are prognostic
markers in colorectal cancer. Int J Cancer. 140:176–187. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Y, Xing QF, Liu XQ, Guo ZJ, Li CY and
Sun G: miR-122 targets VEGFC in bladder cancer to inhibit tumor
growth and angiogenesis. Am J Transl Res. 8:3056–3066.
2016.PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sunkel B, Wu D, Chen Z, Wang CM, Liu X, Ye
Z, Horning AM, Liu J, Mahalingam D, Lopez-Nicora H, et al:
Integrative analysis identifies targetable CREB1/FoxA1
transcriptional co-regulation as a predictor of prostate cancer
recurrence. Nucleic Acids Res. 44:4105–4122. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu X, Zhu Y, Liang Z, Li S, Xu X, Wang X,
Wu J, Hu Z, Meng S, Liu B, et al: c-Met and CREB1 are involved in
miR-433-mediated inhibition of the epithelial-mesenchymal
transition in bladder cancer by regulating Akt/GSK-3/Snail
signaling. Cell Death Dis. 7:e20882016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shabestari RM, Safa M, Alikarami F, Banan
M and Kazemi A: CREB knockdown inhibits growth and induces
apoptosis in human pre-B acute lymphoblastic leukemia cells through
inhibition of prosurvival signals. Biomed Pharmacother. 87:274–279.
2017. View Article : Google Scholar : PubMed/NCBI
|