Introduction
Cholangiocarcinomas (CCAs), originating from the
epithelial cells of the biliary tract, are now divided into
intrahepatic CCA, perihilar CCA (PHCC) and distal CCA based on
anatomical location, according to a recent study (1). PHCC, accounting for ~50% of CCAs, is a
relatively rare type of carcinoma, but with aggressive
characteristics and a poor prognosis. Radical resection is the only
effective curative treatment at present. However, the resection
rate of PHCC is relatively low as the majority of patients are
diagnosed at an advanced stage of its insidious development
(2). Additionally, the highly
desmoplastic nature, extensive support by a rich tumor
microenvironment and profound genetic heterogeneity of PHCC make it
resistant to current adjuvant therapeutic strategies (1). Therefore, PHCC remains one of the most
difficult challenges for hepatobiliary surgeons (3). In addition, tumor markers, including
carbohydrate antigen 19-9 (CA 19-9) and carcinoembryonic antigen
(CEA), may be raised in PHCC, but these markers lack the
sensitivity required for diagnosis (4). Therefore, further investigations on the
underlying molecular mechanisms of PHCC progression, and
identifying more effective prognostic biomarkers and therapeutic
targets, are urgently required for improving the prognosis of PHCC
patients.
Recent improvements in genome-wide surveys and high
throughput transcriptome analysis have revealed that the human
genome contains only ~20,000 protein-coding genes, representing
<2% of the total genome (5,6). While 10-
to 20-fold more genomic sequence is transcribed to long non-coding
RNA (lncRNA) than to protein-coding RNA (7,8). lncRNAs
are functionally defined as transcripts >200 nucleotides (nt) in
length with no protein-coding potential. In recent years, an
increasing amount of scientific interest has been focused on
investigating the functions and mechanisms of lncRNAs (8–10).
Furthermore, it has been suggested that lncRNAs have fulfilled a
wide variety of regulatory roles at almost every stage of gene
expression, including transcriptional, post-transcriptional and
epigenetic levels (9). Furthermore,
accumulating evidence has indicated that lncRNAs serve essential
roles in cancer development and progression through various
regulatory pathways, hierarchies and networks (11,12).
Therefore, identification of cancer-associated lncRNAs, and
investigation of their molecular and biological functions, may shed
light on investigating the molecular biology of cancer.
GAPLINC is a recently discovered non-coding RNA and
has been revealed to serve essential roles in multiple types of
cancer. Hu et al (13)
reported that GAPLINC is involved in gastric cancer progression and
may serve as a biomarker. Yang et al (14) demonstrated that GAPLINC may promote
invasion in colorectal cancer by targeting snail family zinc finger
2 through binding with PTB-associated splicing factor and
non-POU-domain-containing octamerbinding, and may be a potential
therapeutic target of colorectal cancer. However, the role of
lncRNA GAPLINC in PHCC has yet to be identified.
The present study investigated the expression of
GAPLINC in PHCC cell lines and tissues. The association between
GAPLINC expression and PHCC clinicopathological features was also
investigated. Furthermore, high GAPLINC expression was revealed to
be associated with the prognosis of PHCC. GAPLINC may also promote
PHCC cell metastasis and proliferation in in vitro assays.
These results indicated that GAPLINC may be considered as a
potential prognostic marker and therapeutic target for patients
with PHCC.
Materials and methods
Patients and tissue specimens
A total of 96 patients with PHCC who had undergone
surgical resection at Hongqi Hospital of Mudanjiang Medical College
(Mudanjiang, China) between January 2006 and November 2012 were
included in the present study. The mean age of the patients was
60.6±9.1 years old, ranging from 41–82. A total of 57 male patients
and 39 female patients were involved. The tumor tissues and paired
adjacent non-cancerous tissues, isolated from >1 cm away from
the tumor margin, were collected. It was confirmed by a pathologist
at Hongqi Hospital of Mudanjiang Medical College that the adjacent
non-cancerous tissues did not contain any clear tumor cells. PHCC
tissues and adjacent non-cancerous tissues from each patient were
immediately frozen in liquid nitrogen, prior to being stored at
−80°C until subsequent RNA extraction. No local or systemic
anticancer treatments were administered to these patients prior to
surgery. The detailed patient information is presented in Table I. The last follow-up was terminated in
June 2016 and the mean follow-up period was 47.8 months (range,
3–100 months). Patients with two or more different malignancies
were excluded. Written informed consent was obtained from all
patients and the study was approved by the Ethics Committee of
Hongqi Hospital of Mudanjiang Medical College.
 | Table I.Association between GAPLINC expression
and clinicopathological parameters of perihilar
cholangiocarcinoma. |
Table I.
Association between GAPLINC expression
and clinicopathological parameters of perihilar
cholangiocarcinoma.
| Parameter | No. patients | GAPLINC
expression | P-value |
|---|
| Age, years |
|
| 0.572 |
|
<65 | 66 | 4.87±2.92 |
|
| ≥65 | 30 | 5.26±3.58 |
|
| Sex |
|
| 0.836 |
| Male | 57 | 4.94±3.15 |
|
|
Female | 39 | 5.07±3.14 |
|
| Tumor size, cm |
|
| 0.571 |
|
<3 | 45 | 4.80±3.07 |
|
| ≥3 | 51 | 5.16±3.19 |
|
| Differentiation
grade |
|
| 0.716 |
| Well +
moderately | 79 | 5.05±3.11 |
|
| Poorly
+ undifferentiated | 17 | 4.74±3.27 |
|
| T stage |
|
| 0.013 |
|
T1+T2 | 78 | 4.62±3.14 |
|
|
T3+T4 | 18 | 6.63±2.58 |
|
| N stage |
|
| <0.001 |
| N0 | 77 | 4.22±2.84 |
|
|
N1+N2 | 19 | 8.11±2.20 |
|
| M stage |
|
| 0.564 |
| M0 | 76 | 4.94±3.11 |
|
| M1 | 3 | 6.53±4.01 |
|
| TNM stage |
|
| <0.001 |
|
I+II | 65 | 4.02±2.87 |
|
|
III+IV | 31 | 7.04±2.65 |
|
Cell culture
The human normal biliary epithelial HIBEpiC cell
line, and the PHCC QBC939 and FRH0201 cell lines, were purchased
from the Cell Bank of Chinese Academy of Sciences (Shanghai,
China). Cells were cultured in Dulbecco's modified Eagle's medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) or
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.),
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) and 1% penicillin/streptomycin (Gibco; Thermo
Fisher Scientific, Inc.) at 37°C in a humidified incubator with 5%
CO2.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cell lines or frozen
tissue samples using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocols. DNase
I (RNase-free; Takara Biotechnology Co., Ltd., Dalian, China) was
used to prevent contamination of DNA and was then inactivated by
heat treatment. cDNA was reverse transcribed with the extracted
total RNA (2 µg) in a total volume of 20 µl using the Prime-Script
one step RT-PCR kit (Takara Biotechnology Co., Ltd.), the
conditions were as follows: Incubation for 5 min at 25°C followed
by 60 min at 42°C, and then terminatin the reaction by heating at
70°C for 5 min. The mRNA level of lncRNA GAPLINC was evaluated by
RT-qPCR, which was performed using a SYBR PrimeScript RT-PCR kit
(Takara Biotechnology Co., Ltd.) on an ABI7300HT instrument
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The PCR
thermocycling conditions were as follows: Initial denaturation at
95°C for 5 min, followed by 40 cycles of denaturation at 95°C for
30 sec, annealing at 50°C for 30 sec and extension at 72°C for 30
sec. Each experiment was performed in triplicate. Relative GAPLINC
expression level was normalized to GAPDH using the comparative
cycle threshold (2−ΔΔCq) method (15). The primer sequences were as follows:
GAPLINC forward, 5′-ACACACAGCAGCCTGGTTTC-3′ and reverse,
5′-ATGGCACAATCAGGGCTCTT-3′; and GAPDH forward,
5′-ACCCACTCCTCCACCTTTGAC-3′ and reverse,
5′-TGTTGCTGTAGCCAAATTCGTT-3′.
Transfection of cell lines
The GAPLINC sequence was synthesized according to
the full-length GAPLINC sequence (based on the GAPLINC sequence on
NCBI; http://www.ncbi.nlm.nih.gov/), prior
to being sub-cloned into a pcDNA3.1 vector (Genewiz, Inc., Beijing,
China). An empty pcDNA3.1 vector was used as the control. Small
interfering RNAs (siRNAs) specifically targeting GAPLINC
(siGAPLINC-1 and siGAPLINC-2), and a scramble RNA negative control
(si-NC) were purchased from Genewiz, Inc. PHCC cells were cultured
on 6-well plates to confluency and were transfected using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocols. At 48 h
post-transfection, the cells were used for in vitro
assays.
Transwell and matrigel assays
Migration of QBC939 cells and FRH0201 cells were
assessed using a Transwell assay conducted in a Transwell chamber
(Corning Incorporated, Corning, NY, USA). After transfection for 24
h, cells (2.5×104) were plated into the upper chamber
filled with serum-free RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.), while RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) was added to the lower chamber. Invasion of the
cells was examined using the Matrigel method. The Transwell chamber
was pre-coated with Matrigel (Corning Incorporated) prior to the
cells being plated, followed by incubation of the chamber at 37°C
for 30 min. After incubation for 24 h at 37°C, cells that had
migrated to the bottom surface of the membrane were fixed with 4%
paraformaldehyde for 30 min at room temperature, stained with
crystal violet solution for 30 min at room temperature. Finally,
stained cells were visualized under a light microscope
(magnification, ×100), and the number of cells counted in five
random fields of view were averaged.
Cell proliferation assay
Cells proliferation was monitored using WST-8 and a
Cell Counting kit-8 (CCK-8; Roche Diagnostics GmbH, Mannheim,
Germany), according to the manufacturer's protocols. Briefly, 2,000
cells were seeded onto 96-well plates and cell proliferation was
monitored every 24 h for 4 days using a CCK-8 assay. The number of
rate of cell proliferation was quantified by reading the absorbance
of reduced WST-8 at a wavelength 450 nm.
Statistical analysis
A two-tailed Student's t-test was used to compare
two different groups with parametric variables. One-way analysis of
variance was used to compare multiple groups. Survival curves were
plotted using the Kaplan-Meier method and were compared using the
log-rank test. To assess the relative risk for each factor,
univariate and multivariate Cox regression analyses were performed.
Cut-off scores to discriminate high and low GAPLINC expression
samples were selected by evaluating the receiver-operating
characteristic (ROC) curves. The point on the curve with the
shortest distance to the coordinate (0,1) was selected as the
threshold value. All tests were two-sided and P<0.05 was
considered to indicate a statistically significant difference.
Analysis was performed using the SPSS 19.0 software package (IBM
Corp., Armonk, NY, USA).
Results
Expression of GAPLINC is upregulated
in human PHCC
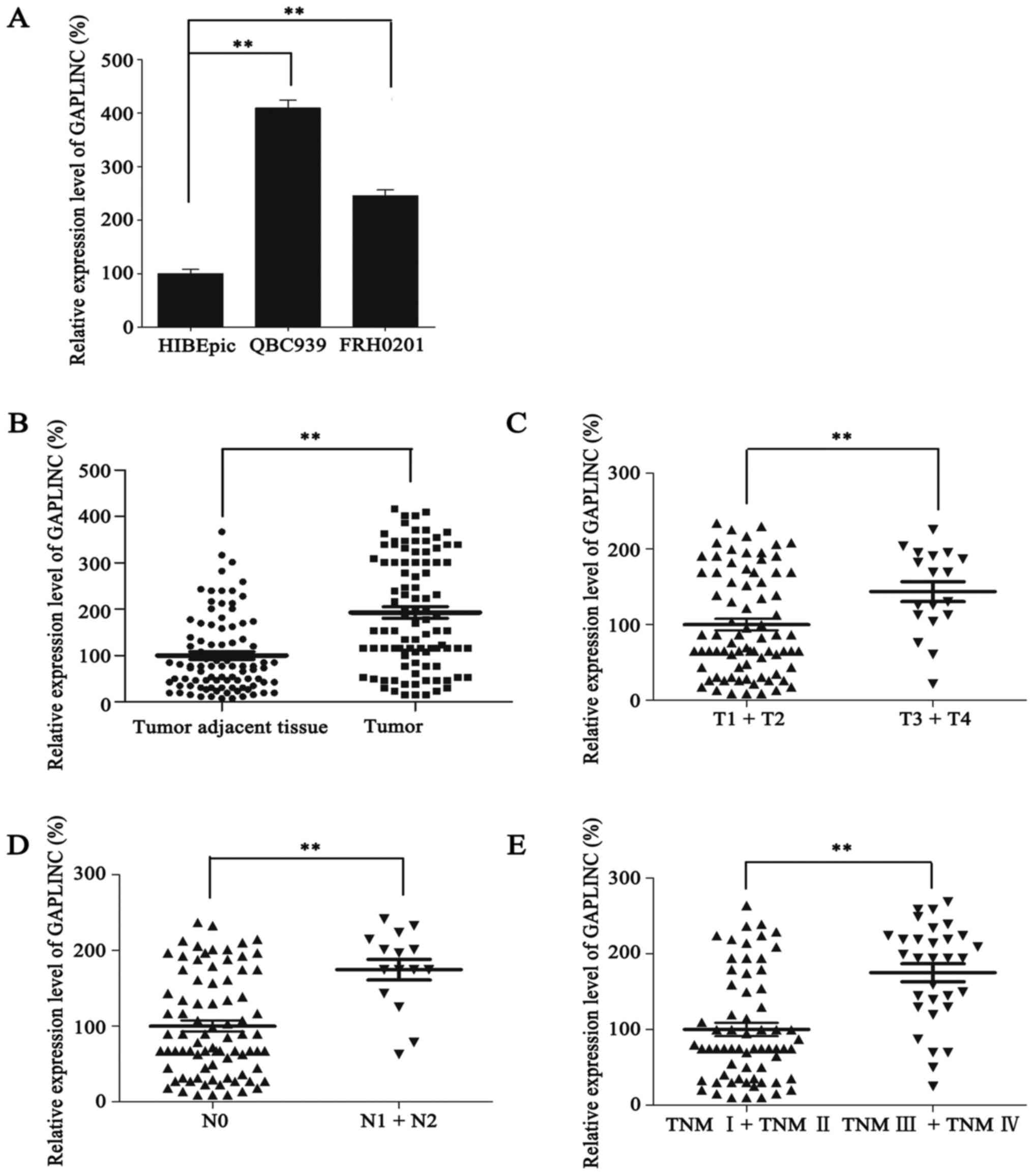
To identify the functional role of GAPLINC in PHCC,
RT-qPCR was performed to evaluate its expression in PHCC cell
lines. Notably, the PHCC QBC939 and FRH0201 cell lines both
exhibited higher expression levels of GAPLINC in comparison with
the HIBEpic cell line (Fig. 1A).
Furthermore, the expression of GAPLINC in 96 PHCC tissues and
paired adjacent non-cancerous tissues was detected. As a result,
GAPLINC was more highly expressed in 84.4% (81/96) PHCC tissues
compared with the adjacent non-cancerous tissues. Furthermore, the
mean expression level of GAPLINC in PHCC tissues was also much
higher than that in tumor adjacent tissues (1±0.80 vs. 1.93±1.21;
P<0.001; Fig. 1B). These results
suggested that GAPLINC may function as an oncogene in PHCC.
High GAPLINC expression promotes PHCC
progression
To extend the present understanding of the role of
GAPLINC in PHCC progression, further statistical analyses were
performed comparing GAPLINC expression with PHCC
clinicopathological characteristics. Patients with advanced T stage
(P=0.013), N stage (P<0.001) and TNM stage (P<0.001)
exhibited significantly higher GAPLINC expression, compared with
patients with earlier stages of disease (Fig. 1C-E; Table
I). However, no statistically significant associations were
observed between GAPLINC expression and other clinicopathological
parameters, including age, sex, tumor size, and M stage (P>0.05;
Table I). Taken together, these
results indicated that GAPLINC may promote PHCC progression.
Selection of a GAPLINC expression
cut-off value for prognosis prediction in PHCC using an ROC
curve
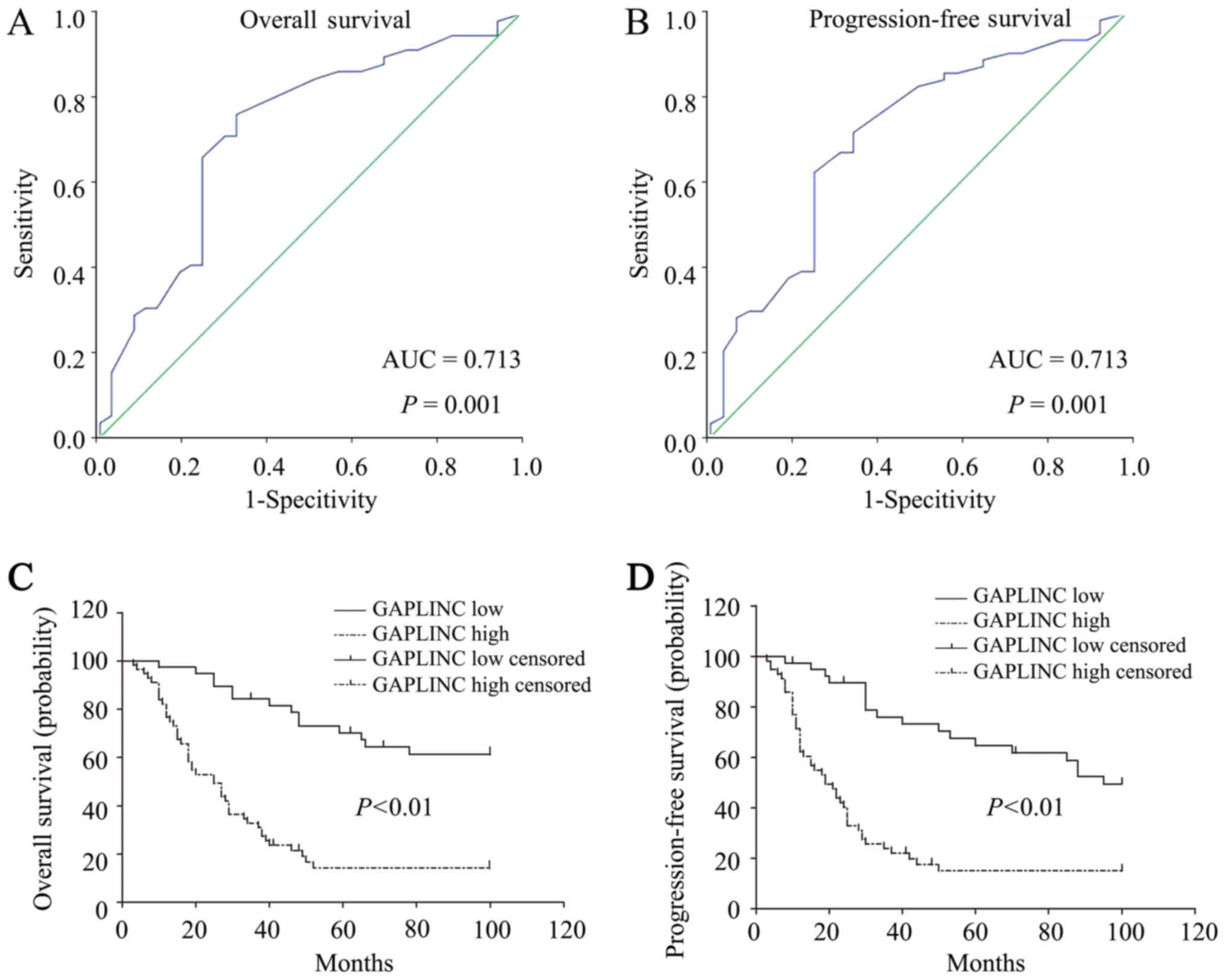
To better define the role of GAPLINC expression in
the prognosis prediction of patients with PHCC, the ROC curve was
employed to identify the optimal cut-off value for high and low
expression of GAPLINC. In order to use the ROC curve analysis, the
prognosis parameters were dichotomized: Overall survival (OS),
death due to PHCC or censored (lost to follow-up, alive or
succumbed to mortality from other causes); progression-free
survival (PFS), PHCC progressed or censored (lost to follow-up,
alive or succumbed to mortality from other causes). As demonstrated
in Fig. 2A and B, the area under the
ROC curve for OS and PFS was 0.713 (95% CI, 0.597–0.829), which
indicated that GAPLINC may serve as a valuable prognostic marker
for patients with PHCC (P<0.001). The cut-off value of the OS
and PFS for high and low GAPLINC expression was 3.10. Based on the
obtained cut-off value, the patients were classified into two
groups, the high GAPLINC expression group (n=57) and the low
GAPLINC expression group (n=39).
GAPLINC overexpression predicts a poor
prognosis in PHCC
To further verify the prognostic potential of
GAPLINC expression in PHCC, Kaplan-Meier analysis was performed,
which demonstrated that high GAPLINC expression was associated with
a lower OS rate (P<0.001; Fig.
2C). Furthermore, high GAPLINC expression was associated with a
poorer PFS (P<0.001; Fig. 2D).
Univariate analysis identified the following 4 risk factors for OS:
Differentiation (HR=0.306; 95% CI, 0.122–0.767; P=0.012), N stage
(HR=2.779; 95% CI, 1.554–4.970; P=0.001), TNM stage (HR=2.503; 95%
CI, 1.475–4.250; P=0.001) and GAPLINC expression level (HR=4.655;
95% CI, 2.514–8.621; P<0.001). Furthermore, the same 4 risk
factors were identified for PFS: Differentiation (HR=0.319; 95% CI,
0.137–0.743; P=0.008), N stage (HR=2.667; 95% CI, 1.496–4.754;
P=0.001), TNM stage (HR=2.302; 95% CI, 1.372–3.862; P=0.002) and
GAPLINC expression level (HR=3.839; 95% CI, 2.191–6.727;
P<0.001). Other clinicopathological features, including sex and
age, were not statistically significant prognosis factors (Table II). Furthermore, by performing
multivariate Cox regression analysis on these 4 factors, the
following 3 prognostic factors were revealed to be independent risk
factors for OS: Differentiation (HR=0.154; 95% CI, 0.054–0.437;
P<0.001), TNM stage (HR=3.464; 95% CI, 0.173–1.259; P=0.011) and
GAPLINC expression level (HR=4.470; 95% CI, 2.269–8.807;
P<0.001), and the same 3 factors were identified for PFS:
Differentiation (HR=0.167; 95% CI, 0.064–0.435; P<0.001), TNM
stage (HR=3.164; 95% CI, 1.240–8.072; P=0.016) and GAPLINC
expression (HR=3.804; 95% CI, 2.044–7.078; P<0.001; Table II). Taken together, these results
suggested that GAPLINC may be considered as a predictive factor for
poor survival and early tumor recurrence in patients with PHCC.
 | Table II.Multivariate analysis of
clinicopathological features for overall survival and
progression-free survival in patients with perihilar
cholangiocarcinoma. |
Table II.
Multivariate analysis of
clinicopathological features for overall survival and
progression-free survival in patients with perihilar
cholangiocarcinoma.
|
| Overall
survival | Progression-free
survival |
|---|
|
|
|
|
|---|
| Parameter | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Univariate
analysis |
|
|
|
|
|
|
| Age |
|
|
|
|
|
|
| ≥65
years vs. <65 years | 1.321 | 0.779–2.240 | 0.302 | 1.194 | 0.712–2.002 | 0.501 |
| Sex |
|
|
|
|
|
|
| Male
vs. Female | 1.033 | 0.614–1.738 | 0.901 | 0.963 | 0.583–1.593 | 0.884 |
|
Differentiation |
|
|
|
|
|
|
| Well +
Moderately vs. Poorly + Undifferentiated | 0.306 | 0.122–0.767 | 0.012 | 0.319 | 0.137–0.743 | 0.008 |
| Tumor size |
|
|
|
|
|
|
| ≥3 cm
vs. <3 cm | 1.125 | 0.673–1.125 | 0.653 | 1.066 | 0.651–1.745 | 0.801 |
| T stage |
| T3+T4
vs. T1+T2 | 1.712 | 0.922–3.179 | 0.089 | 1.516 | 0.822–2.795 | 0.183 |
| N stage |
|
|
|
|
|
|
| N1+N2
vs. N0 | 2.779 | 1.554–4.970 | 0.001 | 2.667 | 1.496–4.754 | 0.001 |
| M stage |
|
|
|
|
|
|
| M1 vs.
M0 | 3.149 | 0.402–4.691 | 0.275 | 2.043 | 0.270–5.475 | 0.489 |
| TNM stage |
|
|
|
|
|
|
|
(III+IV) vs. (I+II) | 2.503 | 1.475–4.250 | 0.001 | 2.302 | 1.372–3.862 | 0.002 |
| GAPLINC
expression |
|
|
|
|
|
|
| High
vs. Low | 4.655 | 2.514–8.621 | <0.001 | 3.839 | 2.191–6.727 | <0.001 |
| Multivariate
analysis |
|
|
|
|
|
|
|
Differentiation |
|
|
|
|
|
|
| Well +
Moderately vs. Poorly + Undifferentiated | 0.154 | 0.054–0.437 | <0.001 | 0.167 | 0.064–0.435 | <0.001 |
| N stage |
|
|
|
|
|
|
| N1+N2
vs. N0 | 0.466 | 0.173–1.259 | 0.132 | 0.530 | 0.200–1.406 | 0.202 |
| TNM stage |
|
|
|
|
|
|
|
(III+IV) vs. (I+II) | 3.464 | 1.332–9.006 | 0.011 | 3.164 | 1.240–8.072 | 0.016 |
| GAPLINC
expression |
|
|
|
|
|
|
| High
vs. Low | 4.470 | 2.269–8.807 | <0.001 | 3.804 | 2.044–7.078 | <0.001 |
GAPLINC may facilitate the metastasis
and proliferation of PHCC cells
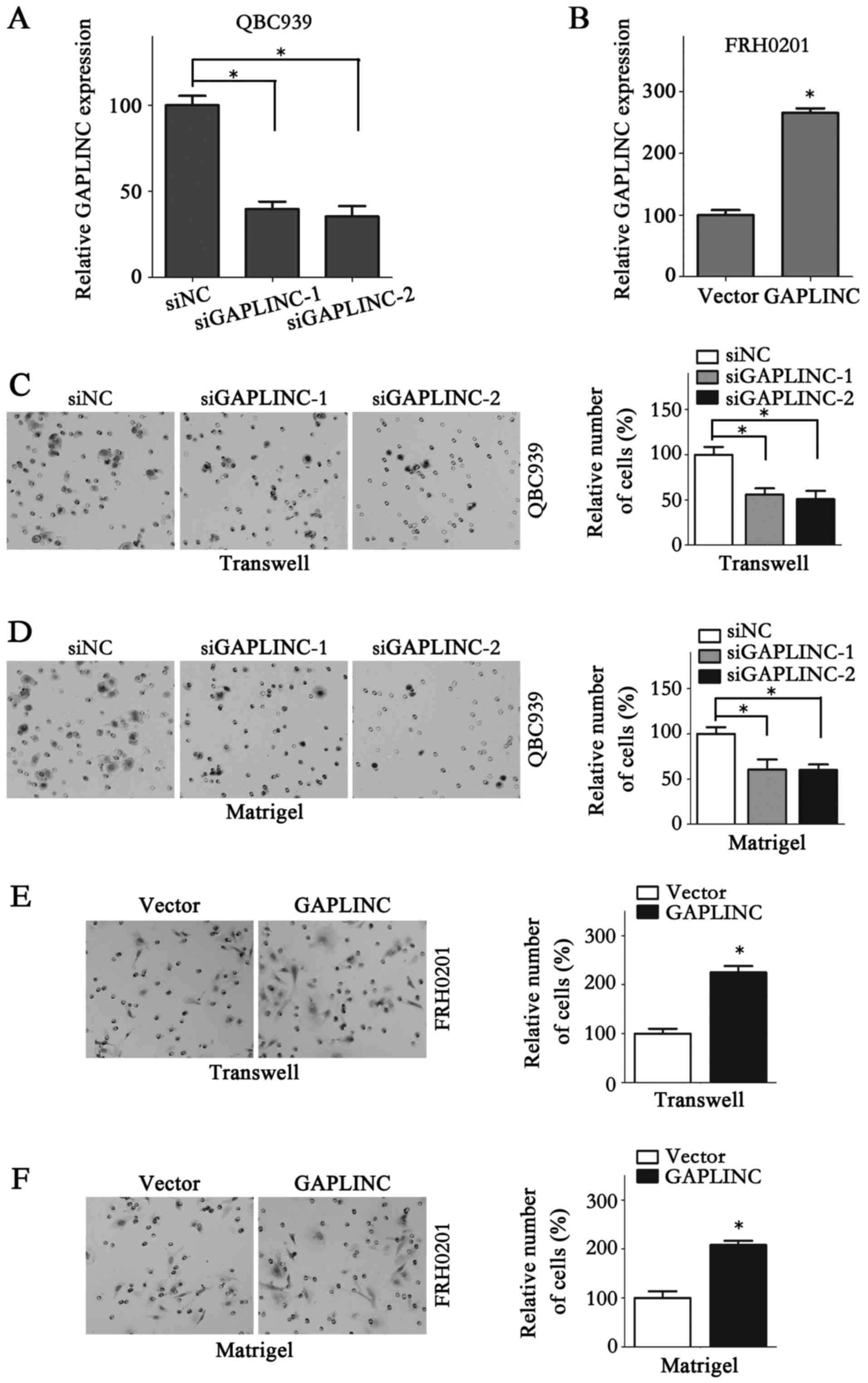
To further verify the function of GAPLINC in PHCC,
GAPLINC was knocked down and overexpressed. siNC, siGAPLINC-1 and
siGAPLINC-2 were used in QBC939 cells, while vector and GAPLINC
were used in FRH0201 cells, as QBC939 cells exhibit a comparatively
higher GAPLINC expression than FRH0201 (Fig. 1A). Therefore, GAPLINC silencing in
QBC939 cells and ectopic expression of GAPLINC in FRH0201 cells
would be more reliable for examining the functional role of GAPLINC
in PHCC cells. The expression of GAPLINC in QBC939 cells and
FRH0201 cells following transfection was determined using RT-qPCR
(Fig. 3A and B). The results of the
Transwell (Fig. 3C and D) and
Matrigel assays demonstrated that GAPLINC interference in QBC939
cells repressed the cell migration and invasion abilities markedly.
Accordingly, GAPLINC ectopic expression in FRH0201 cells markedly
increased metastasis (Fig. 3E and F).
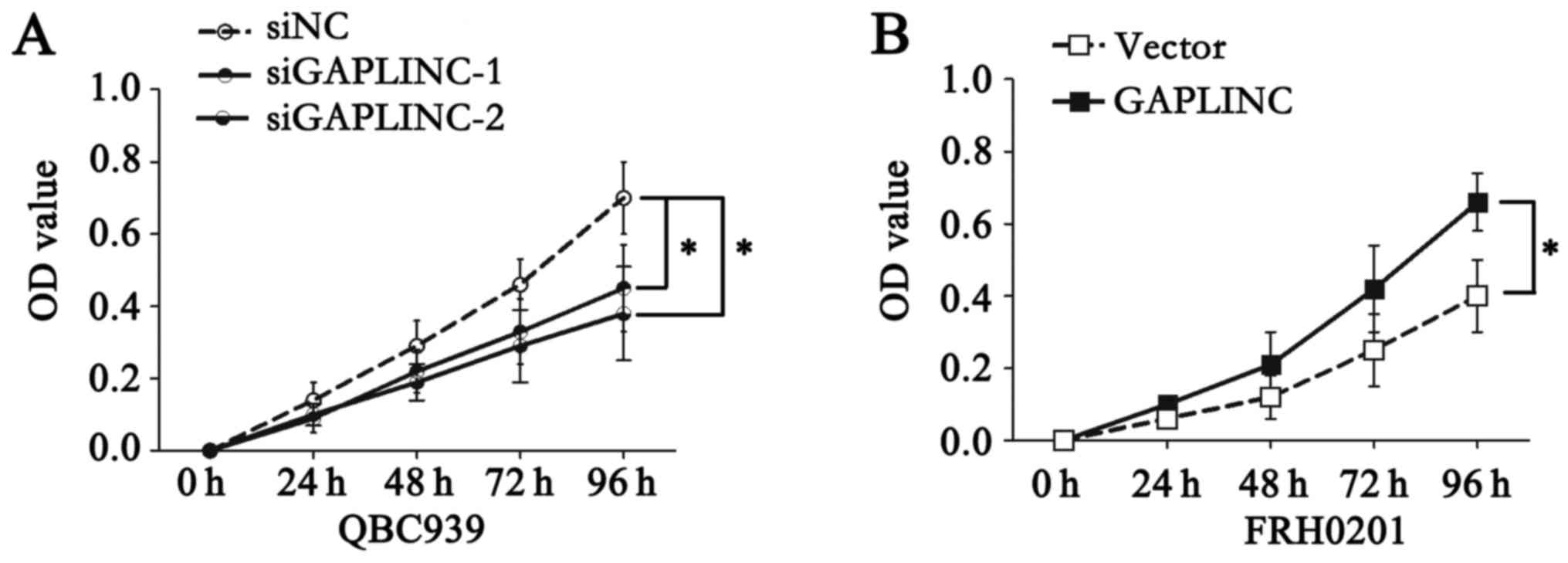
QBC939 cells with GAPLINC deficiency exhibited much lower
proliferation ability in a CCK-8 assay (Fig. 4A), while GAPLINC upregulation in
FRH0201 cells markedly promoted proliferation (Fig. 4B). These results indicated that GALINC
may promote the metastasis and proliferation of PHCC cells.
Discussion
Previous studies have focused on the diagnosis and
management of PHCC, and numerous advances have been made in recent
decades (3,16). However, standard adjuvant therapeutic
strategies remain unavailable for patients with PHCC. Furthermore,
due to the relatively low incidence rate, alterations in
classification, diagnosis and treatment strategies and the highly
aggressive nature of the disease, the vast majority of published
studies on PHCC are statically underpowered, nonrandomized,
restricted to short-term follow-up or demonstrate a poor response
rate (16). GAPLINC was selected for
the present study as it has been demonstrated to promote the
progression of gastric and colorectal cancer, and to be associated
with cancer prognosis (13,14). PHCC also originates from the
epithelium cancer, as with gastric and colorectal cancer. However,
the functional role of GAPLINC in PHCC has yet to be elucidated.
The present study collected 96 PHCC specimens and the prognosis of
these patients was determined. Clinicopathological features of
these patients were also reviewed and statistical analysis was
performed to verify the functional role of GAPLINC in the
progression of PHCC. lncRNA GAPLINC overexpression was revealed to
promote tumor progression by being associated with advanced T, N
and TNM stages of PHCC. Furthermore, ROC curves were constructed
and the optimal cut-off value of GAPLINC expression in the
prognostic prediction of PHCC patients was defined. Based on the
cut-off value, the patients were divided into a high GAPLINC
expression group (57/96, 59.4%) and a low GAPLINC expression group
(39/96, 40.6%). The percentage of patients with PHCC exhibiting a
high expression of GAPLINC was relatively higher compared than the
number reported in patients with colorectal cancer (85/180, 47.2%)
and gastric cancer (45/90, 50.0%) (13,14). The
log-rank test revealed that high GAPLINC expression predicted lower
OS and PFS rates. Furthermore, univariate and multivariate Cox
regression analyses revealed that high GAPLINC expression was an
independent risk factor for lower OS and PFS rates in patients with
PHCC, indicating that GAPLINC may be considered as a survival and
recurrence biomarker in patients with PHCC.
The number of lncRNAs identified in recent years has
markedly increased, so too has the number of lncRNAs involved in
cancer biology. Certain lncRNAs, including GAS5 (17,18),
PTENP1 (19), HOTAIR (20,21),
MALAT1 (22–24) and PCAT1 (25,26), have
been revealed to be associated with various types of cancer.
However, studies on the role of lncRNAs in PHCC are relatively rare
(27–29). Wang et al (27) revealed that lncRNA H19 and HULC,
upregulated by oxidative stress, may regulate CCA cell migration
and invasion by targeting interleukin 6 and C-X-C chemokine
receptor type 4 (CXCR4) via competing endogenous RNA patterns of
sponging let-7a/let-7b and microRNA (miR)-372/miR-373,
respectively. Ma et al (28)
demonstrated that the expression levels of carbamoyl phosphate
synthase 1 (CPS1) and its long non-coding RNA, CPS1-intronic
transcript 1, were increased in intrahepatic CCA tissues and cell
lines. Additionally, overexpression of CPS1 promoted tumor
proliferation and was associated with poor liver function and a
poorer prognosis in patients with intrahepatic CCA. Tan et
al (29) indicated that lncRNA
metastasis-associated lung adenocarcinoma transcript 1 may interact
with miR-204 to modulate PHCC proliferation, migration and invasion
by targeting CXCR4. These studies have provided novel insight into
the roles of lncRNAs in the diagnosis and treatment of PHCC.
Therefore, the clinical significance of other lncRNAs, including
H19, HULC and CPS1-IT1 has been verified in previous studies. The
present study aimed to define the role of GAPLINC in PHCC and
therefore, the expression status of other lncRNAs was not
determined in the present study. The results of the present study
demonstrated that GAPLINC is upregulated in PHCC cell lines and
patients tissues, and that there is an association between GAPLINC
expression and PHCC clinicopathological characteristics, OS and
PFS. Furthermore, high GAPLINC expression was demonstrated to be an
independent risk factor for PHCC, indicating that GAPLINC may also
be an effective therapeutic target for patients with PHCC. In
vitro assays further revealed that GAPLINC promoted the
metastasis and proliferation of PHCC cells.
Several different types of molecules have been
suggested to be potential diagnostic markers or therapeutic targets
for patients with cancer. However, few of them have been commonly
used in clinical practice due to relatively low specificity and/or
sensitivity. In the present study, the clinical samples were staged
based on the TNM staging criteria, a commonly used method for
evaluating tumor development in clinical practice at present. No
previous studies have verified that the expression levels of
existing tumor biomarkers are significantly associated with the
PHCC stage. CEA and CA 19-9 are widely used in the diagnosis of
PHCC, but recent studies have reported that they lack sufficient
specificity and sensitivity (4).
Therefore, the clinical samples were not further stained using
these tumor markers in the present study. However, the association
between GAPLINC expression and certain commonly used tumor markers
require further investigation in future studies. lncRNAs may be
detected in human plasma, which is a non-invasive way to achieve
early diagnosis compared with diagnosing using protein-coding
genes, which usually requires invasive manipulation to obtain
clinical samples. Certain lncRNAs, including H19, have been
revealed to have strong potential as blood biomarkers for the
diagnosis of cancer (30).
Furthermore, lncRNAs exhibit cell-specific expression patterns to a
greater degree than mRNA and usually possess an evolutionarily
conserved function and secondary structure, making lncRNAs more
appropriate for use as diagnostic and prognostic markers.
In conclusion, the present study confirmed that the
expression of GAPLINC was upregulated in PHCC tissues compared with
expression in paired adjacent non-cancerous tissues. Furthermore,
GAPLINC overexpression may promote the development of PHCC. In
addition, high GAPLINC expression predicts a poor OS rate and an
early recurrence in patients with PHCC, and serves as an
independent risk factor for PHCC prognosis. Further analysis
demonstrated that GAPLINC may promote the metastasis and
proliferation of PHCC cells. All these results indicated that
GAPLINC may be a prognostic maker and therapeutic target in
PHCC.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XW performed the in vitro assays and the
RT-qPCR assays. JS and GL collected the PHCC tissues and performed
the statistical analysis. HG was involved in designing the study,
collection of tissues, analysis of the data, drafting the
manuscript, revising it critically for important intellectual
content, and gave final approval of the version to be published and
JW designed the present study.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients and the study was approved by the Ethics Committee of
Hongqi Hospital of Mudanjiang Medical College (Mudanjiang,
China).
Patient consent for publication
Written informed consent was obtained from all
patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Razumilava N and Gores GJ:
Cholangiocarcinoma. Lancet. 383:2168–2179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Khan SA, Davidson BR, Goldin RD, Heaton N,
Karani J, Pereira SP, Rosenberg WM, Tait P, Taylor-Robinson SD,
Thillainayagam AV, et al: Guidelines for the diagnosis and
treatment of cholangiocarcinoma: An update. Gut. 61:1657–1669.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhimin G, Noor H, Jian-Bo Z, Lin W and Jha
RK: Advances in diagnosis and treatment of hilar
cholangiocarcinoma-a review. Med Sci Monit. 19:648–656. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Patel T: Cholangiocarcinoma-controversies
and challenges. Nat Rev Gastroenterol Hepatol. 8:189–200. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
ENCODE Project Consortium, . Birney E,
Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH,
Weng Z, Snyder M, Dermitzakis ET, et al: Identification and
analysis of functional elements in 1% of the human genome by the
ENCODE pilot project. Nature. 447:799–816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li X, Wu Z, Fu X and Han W: lncRNAs:
Insights into their function and mechanics in underlying disorders.
Mutat Res Rev Mutat Res. 762:1–21. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cheetham SW, Gruhl F, Mattick JS and
Dinger ME: Long noncoding RNAs and the genetics of cancer. Br J
Cancer. 108:2419–2425. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Evans JR, Feng FY and Chinnaiyan AM: The
bright side of dark matter: lncRNAs in cancer. J Clin Invest.
126:2775–2782. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Serviss JT, Johnsson P and Grander D: An
emerging role for long non-coding RNAs in cancer metastasis. Front
Genet. 5:2342014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu Y, Wang J, Qian J, Kong X, Tang J, Wang
Y, Chen H, Hong J, Zou W, Chen Y, et al: Long noncoding RNA GAPLINC
regulates CD44-dependent cell invasiveness and associates with poor
prognosis of gastric cancer. Cancer Res. 74:6890–6902. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang P, Chen T, Xu Z, Zhu H, Wang J and He
Z: Long noncoding RNA GAPLINC promotes invasion in colorectal
cancer by targeting SNAI2 through binding with PSF and NONO.
Oncotarget. 7:42183–42194. 2016.PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Howell M and Valle JW: The role of
adjuvant chemotherapy and radiotherapy for cholangiocarcinoma. Best
Pract Res Clin Gastroenterol. 29:333–343. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mourtada-Maarabouni M, Pickard MR, Hedge
VL, Farzaneh F and Williams GT: GAS5, a non-protein-coding RNA,
controls apoptosis and is downregulated in breast cancer. Oncogene.
28:195–208. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xing Z, Lin A, Li C, Liang K, Wang S, Liu
Y, Park PK, Qin L, Wei Y, Hawke DH, et al: lncRNA directs
cooperative epigenetic regulation downstream of chemokine signals.
Cell. 159:1110–1125. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Poliseno L, Salmena L, Zhang J, Carver B,
Haveman WJ and Pandolfi PP: A coding-independent function of gene
and pseudogene mRNAs regulates tumour biology. Nature.
465:1033–1038. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li L, Liu B, Wapinski OL, Tsai MC, Qu K,
Zhang J, Carlson JC, Lin M, Fang F, Gupta RA, et al: Targeted
disruption of Hotair leads to homeotic transformation and gene
derepression. Cell Rep. 5:3–12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gutschner T, Hämmerle M and Diederichs S:
MALAT1-a paradigm for long noncoding RNA function in cancer. J Mol
Med (Berl). 91:791–801. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Prensner JR, Iyer MK, Balbin OA,
Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso
CS, Kominsky HD, et al: Transcriptome sequencing across a prostate
cancer cohort identifies PCAT-1, an unannotated lincRNA implicated
in disease progression. Nat Biotechnol. 29:742–749. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Prensner JR, Chen W, Han S, Iyer MK, Cao
Q, Kothari V, Evans JR, Knudsen KE, Paulsen MT, Ljungman M, et al:
The long non-coding RNA PCAT-1 promotes prostate cancer cell
proliferation through cMyc. Neoplasia. 16:900–908. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang WT, Ye H, Wei PP, Han BW, He B, Chen
ZH and Chen YQ: LncRNAs H19 and HULC, activated by oxidative
stress, promote cell migration and invasion in cholangiocarcinoma
through a ceRNA manner. J Hematol Oncol. 9:1172016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma SL, Li AJ, Hu ZY, Shang FS and Wu MC:
Co-expression of the carbamoyl-phosphate synthase 1 gene and its
long non-coding RNA correlates with poor prognosis of patients with
intrahepatic cholangiocarcinoma. Mol Med Rep. 12:7915–7926. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tan X, Huang Z and Li X: Long non-coding
RNA MALAT1 interacted with miR-204 to modulates human Hilar
cholangiocarcinoma proliferation, migration and invasion by
targeting CXCR4. J Cell Biochem. 118:3643–3653. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gan L, Xu M, Zhang Y, Zhang X and Guo W:
Focusing on long noncoding RNA dysregulation in gastric cancer.
Tumour Biol. 36:129–141. 2015. View Article : Google Scholar : PubMed/NCBI
|