Introduction
Head and neck squamous cell carcinoma (HNSCC) is one
of the most prevalent cancer types globally (1). Although there has been extensive
progress in HNSCC treatment, the overall survival rate remains at
33.24%, and numerous patients exhibit metastatic recurrence and
drug resistance eventually (2). Thus,
exploring novel targets and associated mechanisms that inhibit or
facilitate HNSCC progression is an urgent task for ameliorating
HNSCC treatment.
MicroRNAs (miRNAs/miRs) are a type of non-coding RNA
(18–24 nucleotides in length) that regulate mRNA expression by
binding to the 3′untranslated regions (UTR) of target mRNAs
(3). Increasingly, evidence has
revealed that miR-204 is downregulated in breast cancer (4), glioma (5),
gastric cancer (6) and acute myeloid
leukemia (7). However, to the best of
our knowledge, no information is available regarding the function
of miR-204 in HNSCC progression. One previous study has
demonstrated that Brd4 overexpression may decrease cyclin-dependent
kinase inhibitor 1B (p27) mRNA stability or transcription in
neuroendocrine tumor cells (8).
Additionally, the p27 protein has been demonstrated to serve a
critical function in inhibiting tumor progression (9). Nevertheless, whether the Brd4/p27
regulatory route is involved in HNSCC progression has not yet been
reported, to the best of our knowledge. Thus, it may be
hypothesized that miR-204 enhances p27 mRNA stability and thus
inhibits HNSCC development via targeting Brd4. The potential
mechanism of this effect may be the involvement of the Brd4/p27
pathway.
In the present study, the function of miR-204 in
modulating cyclin-dependent kinase inhibitor 1B (p27) mRNA
stability in HNSCC was investigated.
Materials and methods
HNSCC clinical samples and cell
culture
A total of 23 pairs of HNSCC paraffin-embedded
tissue samples were randomly selected from the Affiliated Hospital
of Jining Medical College (Jining, China) between October 2014 and
November 2016. The thickness of these tissue samples was 4–7 µm,
and were used for detecting miR-204 and Brd4 levels via reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
assay. Written informed consent from all patients and ethical
approval from the Hospital Ethic Review Committee was obtained.
HNSCC cell lines SCC25, Cal27, SCC4, HN12, HN13 and FaDu were
purchased from the Type Culture Collection of the Chinese Academy
of Sciences (Shanghai, China). All cell lines were maintained in
Dulbecco's modified Eagle Medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) in a
humidified atmosphere with 5% CO2 at 37°C.
Bioinformatics
TargetScan v6.2 (http://www.targetscan.org/; August 23, 2017) was used
to predict miRNAs that could potentially bind to Brd4 3′UTR. In
this software, gene name ‘Brd4’ and ‘ENST00000263377.2’ were used
as the search terms.
miRNA, plasmids and transfection
miR-204 mimics/inhibitors and the associated
negative control (NC) were synthesized by Guangzhou RiboBio Co.,
Ltd. (Guangzhou, China). Brd4 coding sequences were cloned into the
pcDNA 3.1 vector (plasmid no. 70219, Addgene, Inc., Cambridge, MA,
USA) and the construct was verified using DNA sequencing, which is
denoted as Brd4-CDS. Lipofectamine 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc) was used for miR-204 mimics (50 nm),
inhibitor (50 nm) and plasmid (2.5 µg) transfection. A total of 48
h after transfection, the expression of genes was detected using
RT-qPCR, and the protein expression of genes was tested using
western blotting. The sequences of the primers used are included in
Table I.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Gene | Sequence (5′ to
3′) |
|---|
| Brd4-CDS forward
(Spel) |
CCCAAGCTTATGTCTGCGGAGAGCGGCCCTGGGA |
| Brd4-CDS reverse
(Xbal) |
CGCGGATCCTCAGAAAAGATTTTCTTCAAATATT |
| GAPDH forward |
CGGAGTCAACGGATTTGGTCGTAT |
| GAPDH reverse |
AGCCTTCTCCATGGTGGTGAAGAC |
| Brd4-RT-qPCR
forward |
CAGTGACAGTTCGACTGATGACTC |
| Brd4-RT-qPCR
reverse |
TTTCCTTTTGTGCTTTTCTTTTTT |
| p27-RT-qPCR
forward |
CTCCAAGACAAACAGCGGAAAATC |
| p27-RT-qPCR
reverse |
GTCTGTGTGCAGGCCATGACATCT |
Cell proliferation assay
FaDu cells with or without miR-204 overexpression
were seeded in 96-well plates at 3,000 cells/well and incubated at
37°C the day prior to transfection. The cell growth rate was
evaluated using a Cell Counting Kit-8 assay (Beyotime Institute of
Biotechnology, Haimen, China) according to the manufacturer's
protocol.
Cell cycle assay
Following transfection for 48 h, FaDu cells were
starved using FBS-free DMEM medium for 12 h and harvested,
re-suspended in 4°C 70% ethanol and stored overnight at −20°C. Then
the cell cycle assays were performed using a cell cycle detection
kit (Beyotime Institute of Biotechnology) according to the
manufacturer's protocol. The percentage of cells in each phase was
analyzed using ModFit LT 4.0 software (Verity Software House, Inc.,
Topsham, ME, USA).
Cell apoptosis assay
Cell apoptotic rate was determined using Annexin
V-fluorescein isothiocyanate (FITC) and propidium iodide (PI)
(Beyotime Institute of Biotechnology) with flow cytometry analysis
(BD Biosciences, San Jose, CA, USA). Briefly, subsequent to
transfection for 48 h, 5×105 FaDu cells were harvested
and washed with ice-cold PBS for one time. Then cells were double
stained with Annexin V-FITC (50 nM) and PI (50 nM) for 10 min at
room temperature, and washed with ice-cold PBS twice following the
manufacturer's protocol. 10,000 cells were collected and FlowJo v10
software (FlowJo LLC, Ashland, OR, USA) was used to analyze the
data, which were expressed as cell percentage.
Luciferase reporter assays
For miRNA target validation, pMIR-Report vector
(cat. no. AM5795; Thermo Fisher Scientific, Inc.) was used to
introduce the fragment of Brd4 3′UTR containing the wild-type (WT)
or the mutant (MUT) binding sites for miRNA-204, referred to as
Luc-Brd4-WT and Luc-Brd4-MUT, respectively. A total of
1×106 FaDu cells were co-transfected with Luc-Brd4-WT or
Luc-Brd4-MUT and miR-204 mimics or NC using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). A total of 48 h later, cells were lysed with
Reporter lysis buffer (cat. no. E397A; Promega Corporation,
Madison, WI, USA) and luciferase activity was measured with VivoGlo
Luciferin kit (cat. no. P1041; Promega Corporation) using a
luminometer (Thermo Fisher Scientific, Inc.). β-gal was utilized to
normalize the transfection efficiency.
RT-qPCR
Total RNA was extracted from FaDu cells using
TRizol® reagent (Life Technologies; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocols. The
first strand cDNA was synthesized using M-MLV reverse transcriptase
(Promega Corporation, Madison, WI, USA) and using an oligo(dT) 18
primer (5′-TTTTTTTTTTTTTTTTT-3′). The thermocycler conditions were
30°C for 10 min, 42°C for 1 h and 95°C for 10 min. RT-qPCR was
performed on an ABI Prism 7500 Sequence Detector (Applied
Biosystems; Thermo Fisher Scientific, Inc.) with the SYBR-Green
RT-qPCR kit (Takara Biotechnology Co., Ltd., Dalian, China)
according to the manufacturer's protocol. The thermocycler
conditions were 94°C for 5 min, followed by 94°C for 40 sec, 60°C
for 30 sec and 72°C for 1 min, for 40 cycles, and then 72°C for 10
min. For the analysis of mRNA expression, all primers used are
included in Table I. GAPDH served as
a reference gene. For miRNA expression detection, the miR-204
primer and U6 primer were purchased from Shanghai GenePharma Co.,
Ltd. (Shanghai, China). U6 was used as an internal reference gene.
Relative quantification was calculated using the 2−ΔΔCq
method (10).
Western blotting
FaDu cells were lysed using RIPA buffer (cat. no.
P0013B, Beyotime Institute of Biotechnology) and the concentration
of protein was determined utilizing a BCA Protein Assay kit
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). A total of 30 µg
protein was separated using 10% SDS-PAGE and transferred onto
polyvinylidene difluoride membranes. The membranes were blocked
with 5% non-fat milk at room temperature for 1 h, and then
incubated with primary antibodies against Brd4 (dilution 1:1,000;
cat. no. ab75898), p27 (dilution 1:5,000; cat. no. ab32034) and
β-actin (dilution 1:10,000; cat. no. ab8226) at 4°C overnight,
which were purchased from Abcam (Cambridge, MA, USA). Following
washing three times with tris-buffered saline with 0.5% Tween-20,
the membranes were incubated with horseradish peroxidase-conjugated
secondary goat anti-rabbit (cat. no. A0208) and goat anti-mouse
antibody (cat. no. A0216) (both Beyotime Institute of
Biotechnology; and both dilution, 1:5,000) at room temperature for
1 h. The membranes were subsequently developed using an ECL system
(Thermo Fisher Scientific, Inc.). β-actin was used as an internal
control.
mRNA stability assays
FaDu cells were performed on this experiment.
miR-204 was induced by transfection with miR-204 mimics for 48 h.
Then, de novo RNA synthesis was blocked at 37°C for 2, 4 and
6 h by the addition of 5 µg/ml of actinomycin D (ActD; Apexbio,
Houston, TX, USA) into DMEM. Total RNA was harvested at the 2, 4
and 6 h, and mRNA expression was detected using RT-qPCR as
aforementioned. The mRNA half-life was determined by comparing to
the mRNA levels prior to the addition of ActD.
Statistical analysis
GraphPad Prism 7.0 (GraphPad Software Inc., La
Jolla, CA, USA) was used to analyze the data. All data were
obtained from at least three independent experiments (n≥3) and
presented as the mean ± standard deviation. Datasets with only two
groups were analyzed using an unpaired student's t-test.
Differences between multiple groups were analyzed using one-way
analysis of variance followed by a Tukey-Kramer post-hoc test, and
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-204 is downregulated and
negatively associated with Brd4 expression levels in HNSCC tumor
samples
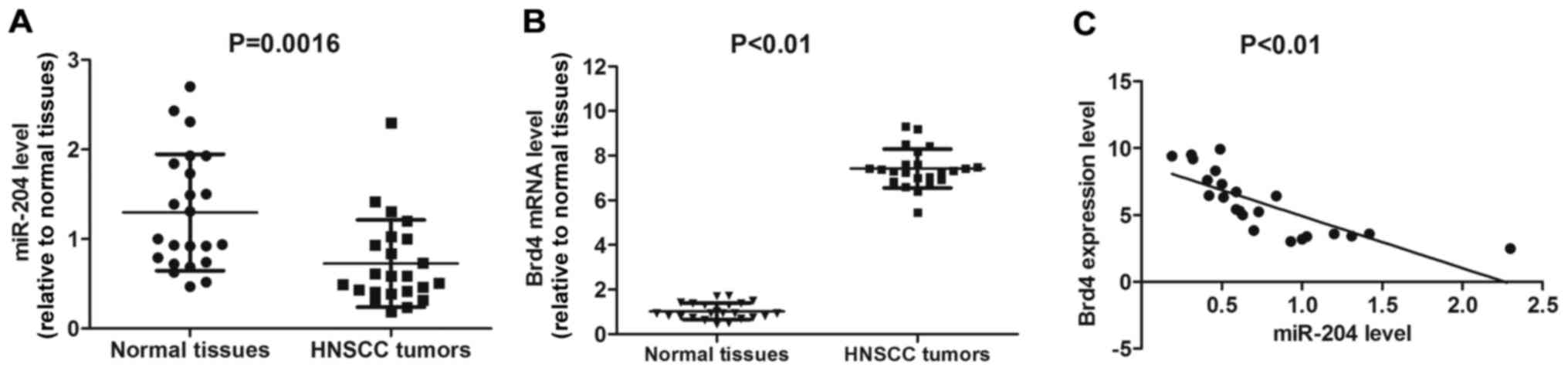
RT-qPCR was performed to detect the expression
levels of miR-204 in HNSCC tumor and adjacent normal tissues. The
levels of miR-204 were significantly downregulated in HNSCC tumor
types compared with the normal adjacent tissues (P=0.0016; Fig. 1A). In contrast, Brd4 mRNA expression
level was significantly higher in HNSCC tumor types compared with
normal tissues (P<0.01; Fig. 1B).
Interestingly, the miR-204 expression level was significantly
negatively associated with Brd4 expression in HNSCC tissues
(P<0.01; Fig. 1C). These results
indicate that miR-204 may hold a suppressive function in HNSCC
tumor types, which is associated with Brd4 expression.
Effects of miR-204 on HNSCC cell
proliferation, cycle and apoptosis
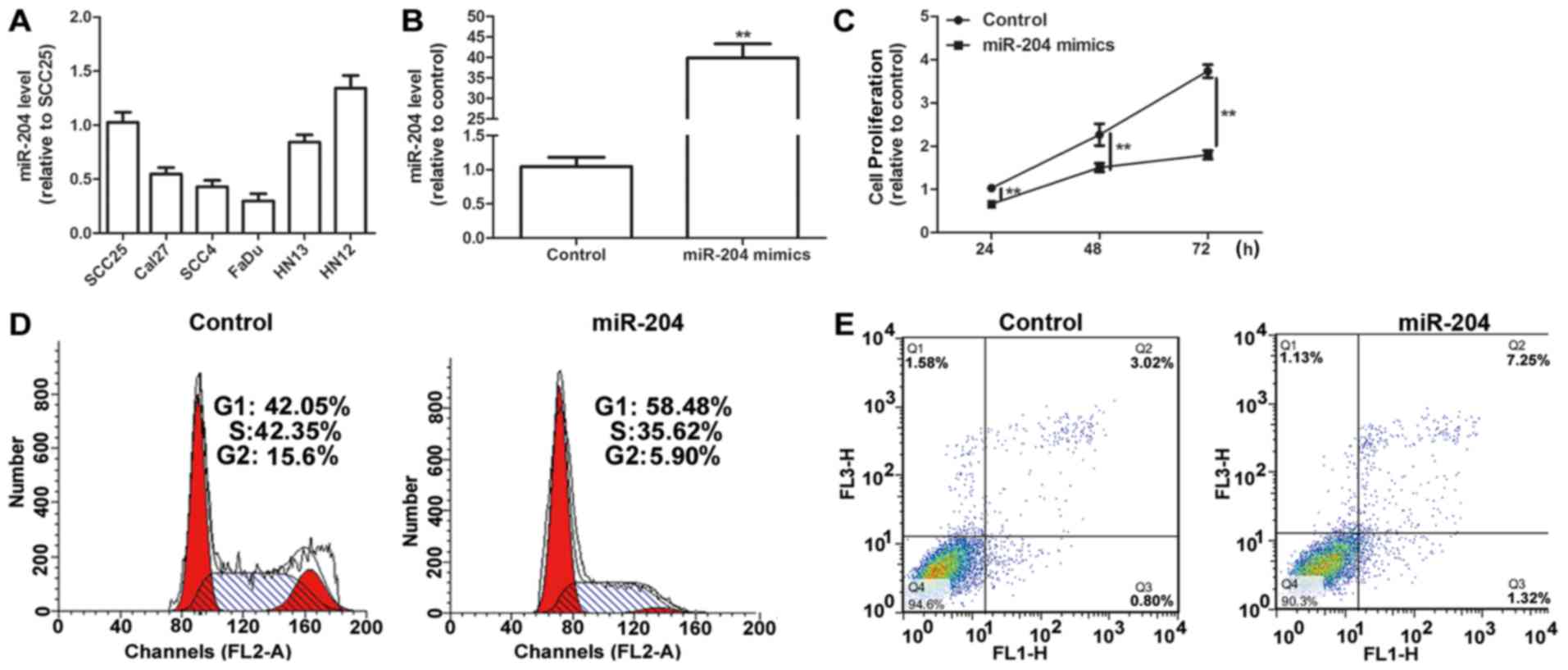
Initially, the expression levels of miR-204 in seven
HNSCC cell lines (SCC25, Cal27, SCC4, HN12, HN13 and FaDu) were
examined, and the lowest level of miR-204, compared with all other
cell lines, was observed in FaDu cells (Fig. 2A). Then, miR-204 expression levels
were upregulated using miR-204 mimics transfection in FaDu cells.
Transfection with miR-204 mimics significantly upregulated miR-204
level in FaDu cells compared with the control (P<0.01; Fig. 2B). A cell proliferation assay revealed
that the upregulation of miR-204 expression levels significantly
inhibited cell proliferation in HNSCC cells compared with the
control (P<0.01; Fig. 2C).
Additionally, a cell cycle assay indicated that the ectopic
expression of miR-204 promoted cell cycle arrest in G1/S phase
compared with the control (Fig. 2D).
Additionally, miR-204 overexpression was demonstrated to enhance
cell apoptosis in HNSCC cells compared with the control (Fig. 2E).
Brd4 is potential target of
miR-204
TargetScan (version 6.2, http://www.targetscan.org/) was used to predict
potential targets of miR-204 and it was revealed that Brd4 may be a
potential target with two putative miR-204 binding sites within its
3′UTR (Fig. 3A). Brd4 expression was
measured in SCC25, Cal27, SCC4, HN12, HN13 and FaDu cells via
RT-qPCR and western blotting. It was revealed that there were
higher levels of mRNA expression of Brd4 in FaDu cells compared
with in any other cells (Fig. 3B and
C). Fig. 3D revealed that the
co-transfection of FaDu cells with miR-204 mimics and the
Luc-Brd4-WT constructs resulted in a significant reduction in
luciferase activity compared with transfection with NC (P<0.05).
Conversely, no significant effect on the luciferase activity of
Luc-Brd4-MUT was observed following miR-204 upregulation,
suggesting that Brd4 is a direct target of miR-204. RT-qPCR
experiments further confirmed that upregulation of miR-204
significantly decreased Brd4 mRNA expression levels in HNSCC cells
(P<0.01; Fig. 3E), and western
blotting experiments produced similar results (Fig. 3F). In summary, it was confirmed that
Brd4 is a potential target of miR-204 in FaDu cells.
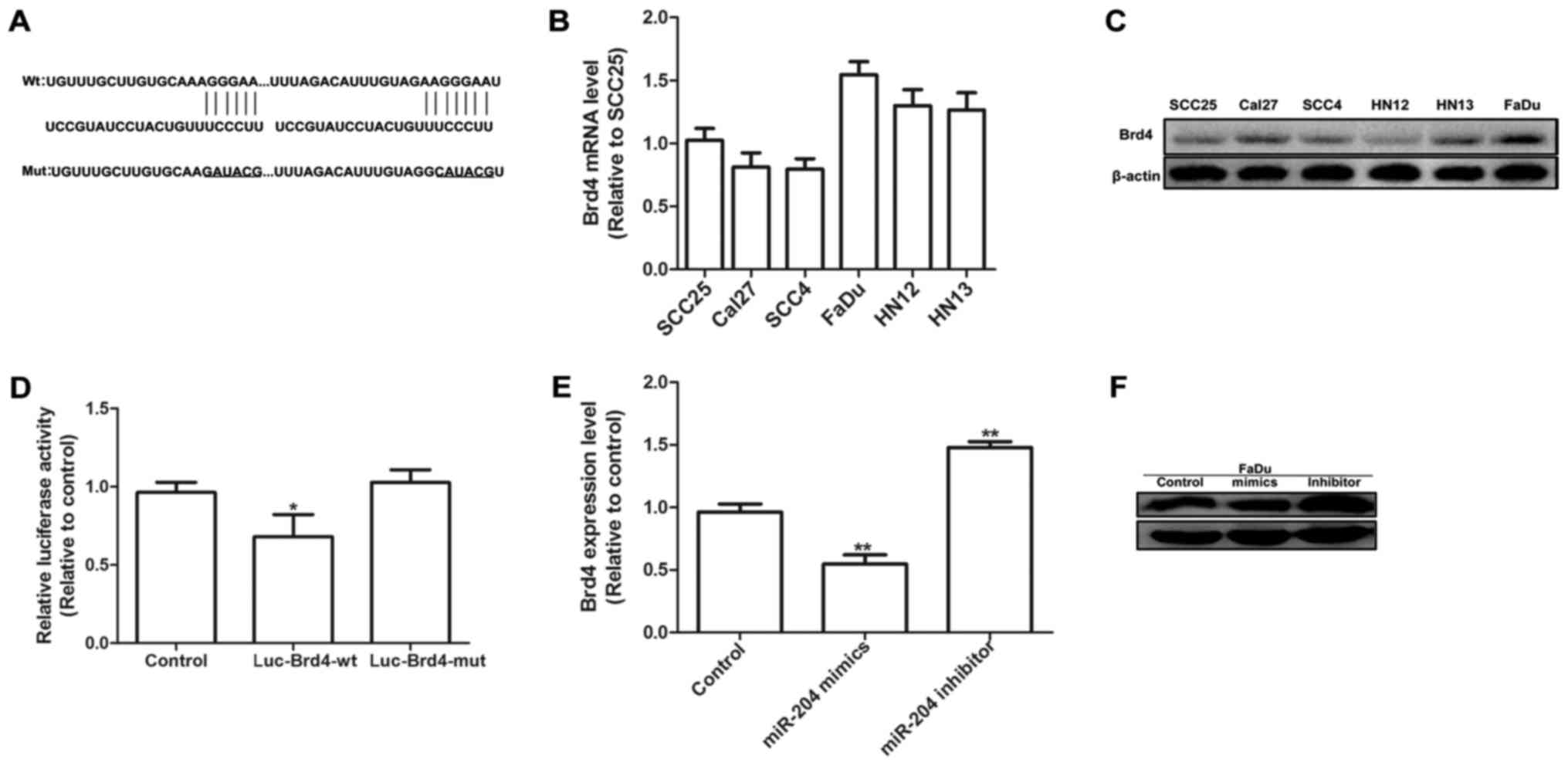 | Figure 3.Brd4 is potential target of miR-204.
(A) Predicted binding sites for miR-204 in the 3′-untranslated
region of human Brd4 mRNA (wild type and mutant). Brd4 mRNA
expression levels and protein expression were detected in various
head and neck squamous cell carcinoma cell lines (SCC25, Cal27,
SCC4, HN12, HN13 and FaDu) by (B) RT-qPCR and (C) western blotting.
(D) FaDu cells were co-transfected with Luc-Brd4-WT or
Luc-Brd4-MUT, miR-204 mimics and β-gal control plasmid for 48 h,
then luciferase activity was measured and normalized to β-gal
activity. *P<0.05 vs. control. (E) RT-qPCR and (F) western
blotting results of Brd4 expression in FaDu cells following
transfection with miR-204 mimics for 48 h. Data were presented as
the mean ± standard deviation. **P<0.01 vs. control. miR,
microRNA; Brd4, bromodomain-containing protein 4; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; Luc,
luciferase; WT, wild type; MUT, mutant. |
miR-204 enhances p27 mRNA stability
via targeting Brd4
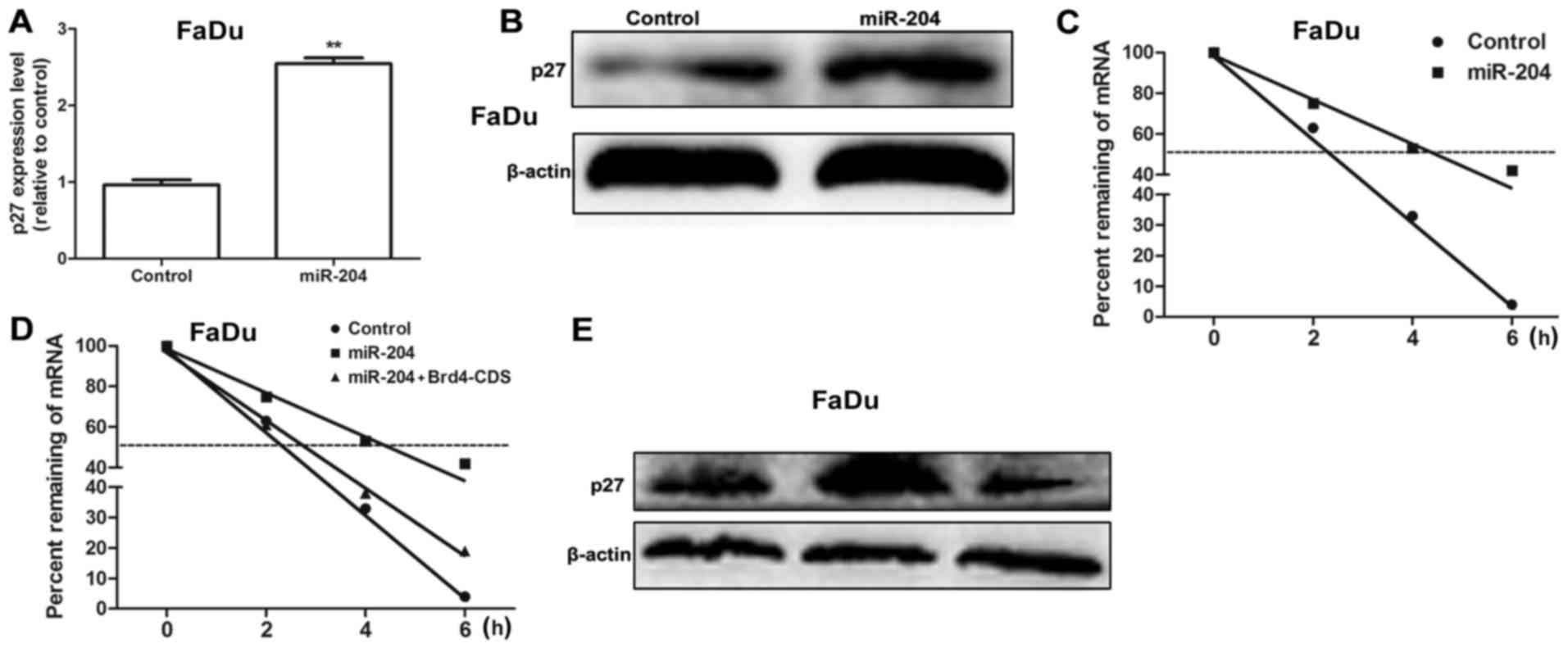
It was further examined whether miR-204 may affect
the expression of anti-proliferative factors including p27. As
expected, transfection with miR-204 mimics significantly increased
p27 mRNA expression levels (P<0.01; Fig. 4A) and protein expression in FaDu cells
(Fig. 4B), which indicates that
miR-204 modulates p27 expression at the transcriptional level. To
determine whether miR-204 influences p27 mRNA stability, HNSCC
cells were transfected with miR-204 for 48 h, followed by ActD
treatment. Fig. 4C demonstrated that
transfection with miR-204 mimics delayed the reduction of p27 mRNA.
As Brd4 may bind to the acetylated histones in the chromatin,
resulting in the suppression of gene transcription (11), it was hypothesized that miR-204
enhances p27 mRNA stability by targeting Brd4. When FaDu cells were
co-transfected with miR-204 mimics with Brd4 overexpression plasmid
(Brd4-CDS), Brd4 overexpression attenuated or even reversed the
promotive effect of miR-204 on p27 mRNA stability and expression
(Fig. 4D and E). Overall, the results
of the present study reveal that miR-204 may enhance p27 mRNA
stability through the use of Brd4.
miR-204 exerts its effects on HNSCC
cell proliferation, cycle and apoptosis partially through Brd4
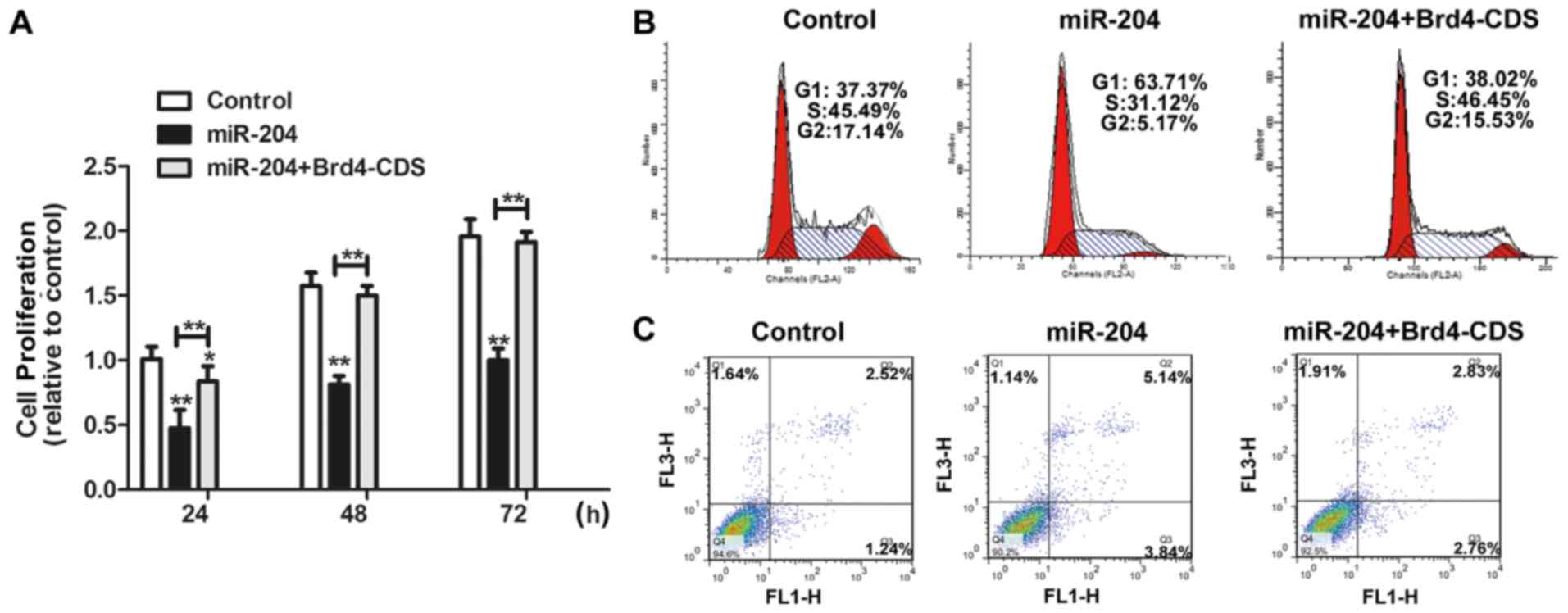
Further investigations were conducted into whether
the functional effects of miR-204 are dependent on Brd4 expression.
Brd4-CDS was co-transfected with miR-204 mimics in FaDu cells. As
presented in Fig. 5A and B,
overexpression of Brd4 significantly counteracted the inhibitory
effects of miR-204 on cell proliferation (P<0.05) and
counteracted the inhibitory effects of miR-204 on the percentage of
cells in the G1/S phase. Furthermore, the promotive effects of
miR-204 on cell apoptosis were decreased by Brd4 overexpression
(Fig. 5C). Therefore, these results
demonstrate that the inhibitory effects that miR-204 exerts on
HNSCC progression is partially dependent on Brd4 expression.
Discussion
miR-204 is known as a multifunctional miRNA involved
in tumor development (12–14). However, to the best of our knowledge,
the functions of miR-204 in HNSCC have never been reported.
The present study focuses on the functions of
miR-204 on HNSCC cell proliferation and apoptosis. The clinical
tissue samples revealed that miR-204 was downregulated, however
Brd4 expression was upregulated in HNSCC tumor tissues, and their
expression were negatively associated. The targeted association
between miR-204 and Brd4 was confirmed using a luciferase reporter
assay, RT-qPCR and western blotting. As Brd4 may decrease p27 mRNA
stability or transcription in neuroendocrine tumor cells (8), it was further investigated whether p27
participates in HNSCC progression regulated by miR-204 and the
results of the present study revealed that the Brd4/p27 pathway
regulated by miR-204 serves an essential role in HNSCC
progression.
To the best of our knowledge, the present study is
the first to demonstrate the function of miR-204 in HNSCC
progression. However, as miR-204 may exert inhibitory effects on
tumor angiogenesis and metastasis (4,15), it is
additionally interesting to note whether miR-204 exerts inhibitory
functions on HNSCC metastasis and angiogenesis. Notably, Brd4 was
initially identified as a potential target of miR-204 and was
involved in the p27 mRNA stability regulated by miR-204 in HNSCC.
Future experiments should focus on whether the miR-204/Brd4
regulatory route exists in other tumor types. Although the Brd4
inhibitor IBET has been demonstrated to exert inhibitory functions
in the development of various tumor types (8,16,17), exploring the functions of miR-204 may
provide another clue to develop novel Brd4 inhibitors. In summary,
the results of the present study offer novel insight into the
function of miR-204 in HNSCC progression, suggesting use as a
compelling biomarker or therapeutic target for HNSCC
progression.
Acknowledgements
The authors would like to thank Professor Kaixue
Wang from Shanghai Jiaotong University (Shanghai, China) for
critically reviewing this work.
Funding
Funding information is not applicable.
Availability of data and materials
All data generated or analyzed during the current
study are included in this published article.
Authors' contributions
CW and YZ designed the study. YZ and GC analyzed the
data. CW, YZ, DZ and YW performed the experiments. CW wrote the
manuscript.
Ethics approval and consent to
participate
Written informed consent from all patients and
ethical approval from was obtained the Hospital Ethic Review
Committee.
Patient consent for publication
The manuscript declared that the patients have
provided written informed consent for the publication of any
associated data and accompanying images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Datta J, Islam M, Dutta S, Roy S, Pan Q
and Teknos TN: Suberoylanilide hydroxamic acid inhibits growth of
head and neck cancer cell lines by reactivation of tumor suppressor
microRNAs. Oral Oncol. 56:32–39. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li X, Zheng L, Zhang F, Hu J, Chou J, Liu
Y, Xing Y and Xi T: STARD13-correlated ceRNA network inhibits EMT
and metastasis of breast cancer. Oncotarget. 7:23197–23211.
2016.PubMed/NCBI
|
|
4
|
Zheng L, Li X, Gu Y, Lv X and Xi T: The
3′UTR of the pseudogene CYP4Z2P promotes tumor angiogenesis in
breast cancer by acting as a ceRNA for CYP4Z1. Breast Cancer Res
Treat. 150:105–118. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ye ZN, Liu JP, Wu LY, Zhang XS, Zhuang Z,
Chen Q, Lu Y, Liu CG, Zhang ZH, Zhang HS, et al: Downregulation of
miR-204 expression correlates with poor clinical outcome of glioma
patients. Hum Pathol. 63:46–52. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Canu V, Sacconi A, Lorenzon L, Biagioni F,
Lo Sardo F, Diodoro MG, Muti P, Garofalo A, Strano S, D'Errico A,
et al: MiR-204 down-regulation elicited perturbation of a gene
target signature common to human cholangiocarcinoma and gastric
cancer. Oncotarget. 8:29540–29557. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Butrym A, Rybka J, Baczynska D, Tukiendorf
A, Kuliczkowski K and Mazur G: Low expression of microRNA-204
(miR-204) is associated with poor clinical outcome of acute myeloid
leukemia (AML) patients. J Exp Clin Cancer Res. 34:682015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang L, Matkar S, Xie G, An C, He X, Kong
X, Liu X and Hua X: BRD4 inhibitor IBET upregulates p27kip/cip
protein stability in neuroendocrine tumor cells. Cancer Biol Ther.
18:229–236. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shao J, Li S, Palmqvist L, Fogelstrand L,
Wei SY, Busayavalasa K, Liu K and Liu VM: p27(KIP1) and PTEN
cooperate in myeloproliferative neoplasm tumor suppression in mice.
Exp Hematol Oncol. 5:172016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Filippakopoulos P, Qi J, Picaud S, Shen Y,
Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et
al: Selective inhibition of BET bromodomains. Nature.
468:1067–1073. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shi L, Zhang B, Sun X, Lu S, Liu Z, Liu Y,
Li H, Wang L, Wang X and Zhao C: MiR-204 inhibits human NSCLC
metastasis through suppression of NUAK1. Br J Cancer.
111:2316–2327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yin Y, Zhang B, Wang W, Fei B, Quan C,
Zhang J, Song M, Bian Z, Wang Q, Ni S, et al: miR-204-5p inhibits
proliferation and invasion and enhances chemotherapeutic
sensitivity of colorectal cancer cells by downregulating RAB22A.
Clin Cancer Res. 20:6187–6199. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zeng J, Wei M, Shi R, Cai C, Liu X, Li T
and Ma W: MiR-204-5p/Six1 feedback loop promotes
epithelial-mesenchymal transition in breast cancer. Tumour Biol.
37:2729–2735. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun Y, Yu X and Bai Q: miR-204 inhibits
invasion and epithelial-mesenchymal transition by targeting FOXM1
in esophageal cancer. Int J Clin Exp Pathol. 8:12775–12783.
2015.PubMed/NCBI
|
|
16
|
Gao X, Wu X, Zhang X, Hua W and Zhang Y,
Maimaiti Y, Gao Z and Zhang Y: Inhibition of BRD4 suppresses tumor
growth and enhances iodine uptake in thyroid cancer. Biochem
Biophys Res Commun. 469:679–685. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hu Y, Zhou J, Ye F, Xiong H, Peng L, Zheng
Z, Xu F, Cui M, Wei C, Wang X, et al: BRD4 inhibitor inhibits
colorectal cancer growth and metastasis. Int J Mol Sci.
16:1928–1948. 2015. View Article : Google Scholar : PubMed/NCBI
|