Introduction
Anaplastic lymphoma kinase (ALK) gene
rearrangements are present in 3-5% of patients with non-small cell
lung cancer (NSCLC). Research has revealed the promising clinical
activity of ALK-tyrosine kinase inhibitors (TKIs) in the treatment
of patients with NSCLC who contain ALK rearrangements.
Crizotinib was the first ALK-TKI approved for patients with
ALK rearrangement-positive NSCLC. Recently, two
second-generation ALK-TKIs (alectinib and ceritinib) have been
approved for the treatment of ALK rearrangement-positive
NSCLC in Japan. Although these inhibitors exhibited significant
clinical responses, almost all patients treated with them developed
resistance. Some studies have reported that the resistance
mechanisms in patients with ALK rearrangement-positive NSCLC
comprised ALK gene alterations, such as ALK point
mutations and copy-number gains (1,2), bypass
signaling activation through the activation of other oncogenes
(3,4),
and SCLC transformation (5,6). However, to date, no study has reported a
correlation between tumor markers and SCLC transformation in both
EGFR-mutant and ALK rearrangement-positive NSCLC.
Here, we report a case of a patient with SCLC transformation after
alectinib treatment who exhibited an elevation of
pro-gastrin-releasing peptide precursor (ProGRP) and
neuron-specific enolase (NSE) levels, suggesting these to be
predictive of SCLC transformation during the development of
resistance to ALK-TKIs.
Case report
A 62-year-old former smoker male (38 pack-year
history) with contralateral lung metastases presented with lung
adenocarcinoma in clinical stage T1aN3M1a. The patient provided
informed consent. He underwent biopsy of the left supraclavicular
lymph node, and both immunohistochemistry (IHC) and fluorescence
in situ hybridization (FISH) revealed adenocarcinoma with
ALK rearrangement. Accordingly, he received cytotoxic
chemotherapy (carboplatin, pemetrexed, and bevacizumab), to which
his best response was a stable disease. After 4 months of the
treatment initiation, his drug regimen was changed to 300 mg
alectinib two times daily, which is the approved dosage in Japan,
due to the disease progression. Eventually, he attained complete
radiological remission. After 1 year of alectinib treatment, brain
magnetic resonance imaging revealed a new central nervous system
(CNS) metastasis. Accordingly, he underwent stereotactic
radiotherapy (SRT) for the isolated CNS lesion and continued
receiving alectinib beyond the CNS progression. After 2 years of
the alectinib initiation, computed tomography (CT) revealed a new
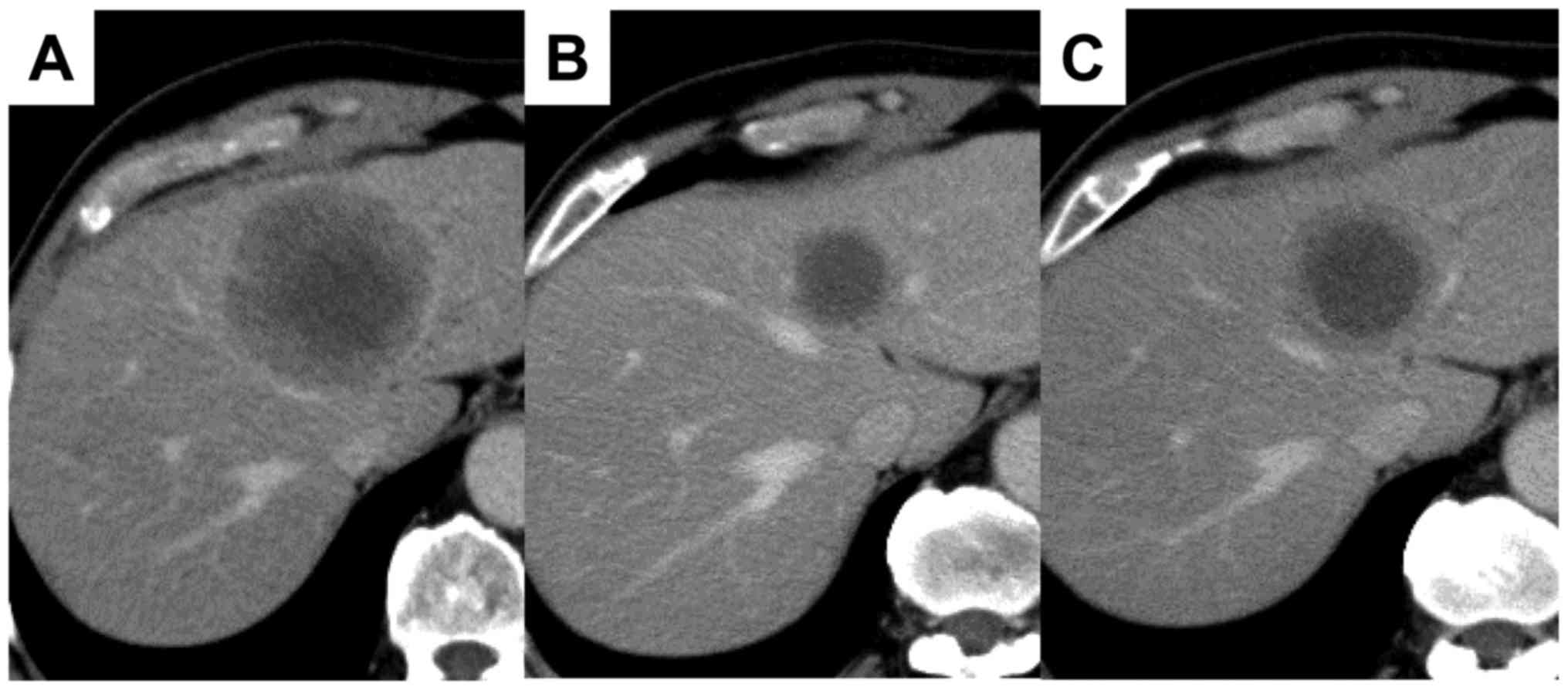
mass (55×60 mm) in the left lobe of the liver (Fig. 1A). The pathological finding
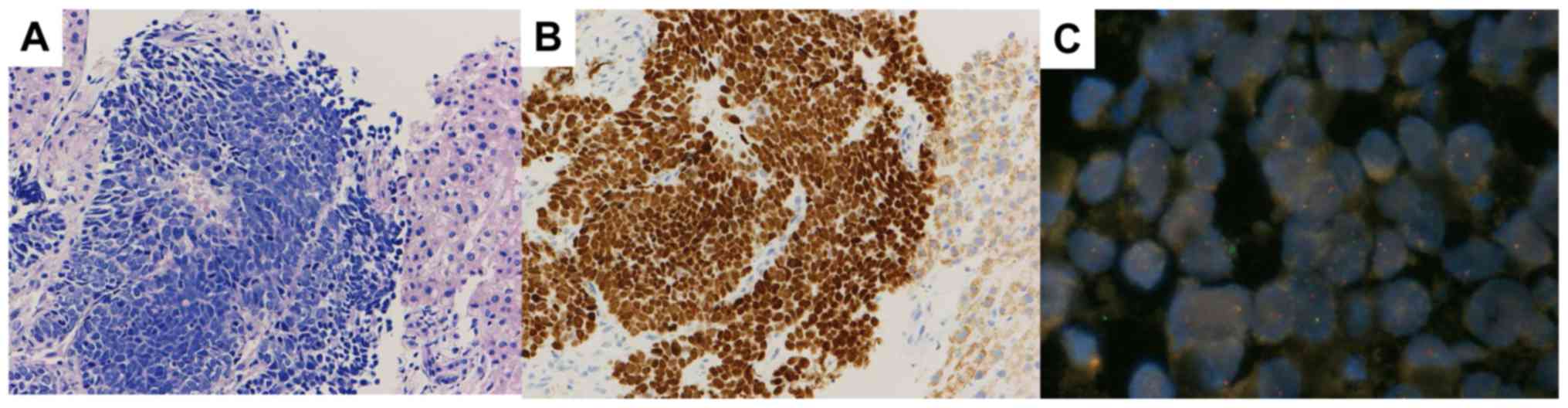
corroborated SCLC transformation (Fig.
2A). In addition, the detection of the ALK overexpression and
rearrangement with IHC and FISH confirmed SCLC transformation in
this specimen (Fig. 2B and C). During
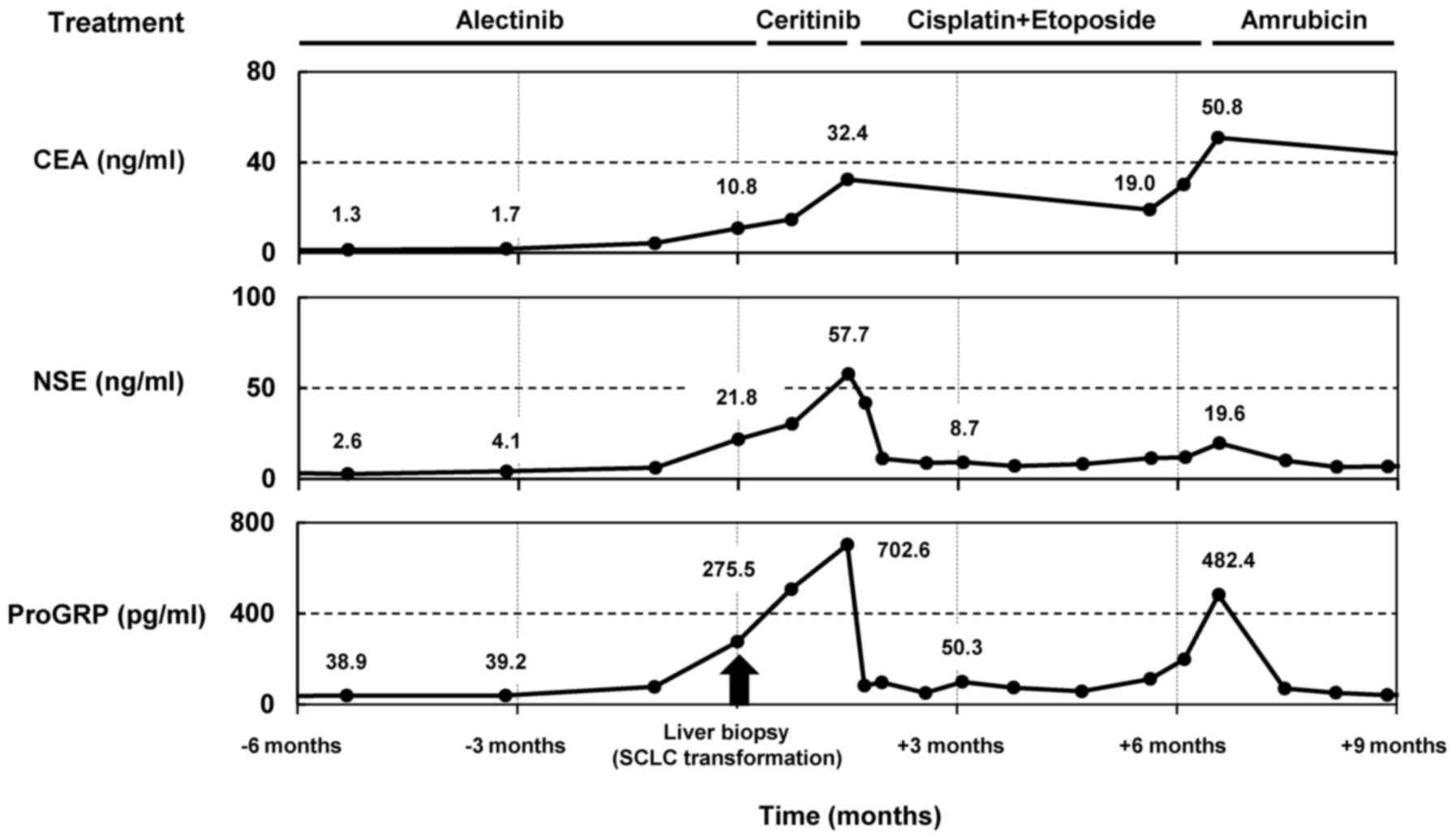
rebiopsy, serum ProGRP, NSE, and carcinoembryonic antigen (CEA)
levels were 275.5 pg/ml, 21.8 ng/ml and 10.8 ng/ml, respectively
(serum ProGRP and CEA levels upon ALK rearrangement-positive
NSCLC diagnosis were 56.3 pg/ml and 4.2 ng/ml, respectively).
Accordingly, he temporally received ceritinib after alectinib
treatment; however, serum ProGRP, NSE, and the size of liver
metastasis continued to increase. Thus, we switched to cisplatin
and etoposide. After four cycles, CT revealed a partial response of
the liver lesion to the treatment (Fig.
1B). After 3 months, liver metastasis reprogressed (Fig. 1C). Hence, we treated the patient with
three more regimens [amrubicin (AMR), nivolumab, and irinotecan],
but the tumor progressed. After 4 years of the treatment
initiation, the patient died due to disease progression.
Discussion
This case report retrospectively analyzed the serial
tumor markers CEA, NES, and ProGRP because the diagnosis of
ALK rearrangement-positive NSCLC in a patient (Fig. 3). Three months before SCLC
transformation was confirmed by a liver biopsy, serum CEA, NES, and
ProGRP levels were within normal limits. However, after SCLC
transformation, serum ProGRP and NSE levels correlated with tumor
response to chemotherapy for SCLC and were significantly beneficial
for monitoring tumor progression. Thus, this study revealed that
tumor markers, such as ProGRP and NSE, can predict SCLC
transformation at the time of developing resistance to
ALK-TKIs.
Apparently, ProGRP and NSE are two of the tumor
markers of SCLC that are preferentially used to diagnose diseases.
Typically, the diagnostic specificity of NSE is rather limited
owing to a relatively high false-positive rate. Conversely, the
diagnostic sensitivity and specificity of ProGRP in SCLC are higher
compared to those in NSE (7). To
date, no study has investigated whether tumor markers for SCLC
could be predictive markers for SCLC transformation in not only
EGFR-mutant NSCLC but also ALK rearrangement-positive
NSCLC. To the best of our knowledge, only one case study of SCLC
transformation in EGFR-mutant NSCLC reported the elevation
of serum ProGRP and NSE levels. However, whether tumor makers could
be predictive for the diagnosis of SCLC transformation, due to the
confirmed pathological diagnosis at autopsy, remains unclear.
Hence, our case is the first to report that both ProGRP and NSE
could be useful predictive markers for SCLC transformation.
Research has identified several ALK-TKI resistance
mechanisms in patients with ALK rearrangement-positive
NSCLC, and each resistance mechanism exhibits a different
sensitivity to other ALK inhibitors in ALK
rearrangement-positive NSCLC (8).
Thus, identifying the resistance mechanism that remains sensitive
to other ALK inhibitors could be essential in selecting appropriate
ALK inhibitors as subsequent ALK-TKIs. In addition, non-invasive
liquid biopsies and cell-free DNA (cfDNA) genotyping have been
developed to detect these resistance mechanisms efficiently because
multiple tumor biopsies are related to both risks and discomfort
(6,9–12).
However, liquid biopsies are limited in the diagnosis of
histological transformation. Hence, we anticipate that combining
ProGRP and specific cfDNA for the detection of ALK resistance
mutation could be more useful. Indeed, in our case, both ProGRP and
NSE levels were elevated at the time of developing resistance to
alectinib, suggesting that we could predict SCLC transformation
before rebiopsy. Further investigation of the correlation between
tumor markers ProGRP and NSE and SCLC transformation is
warranted.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YO, TY and TH collaborated in the conception and
design of the study. YO, TY, TU, YI and TH acquired the data. YO,
TY TU YM and TH performed data analysis. All authors were involved
in writing the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
The patient was informed that the data from his case
would be submitted for publication and provided consent.
Competing interests
Dr Hida obtained research grants from Ono
Pharmaceutical, Novartis Pharma, Chugai Pharmaceutical, Eli Lilly,
Taiho Pharmaceutical, AstraZeneca, Nippon Boehringer Ingelheim,
Pfizer, Bristol-Meyers Squibb, Clovis Oncology, Eisai, Takeda Bio,
Dainippon Sumitomo Pharma, Abbvie, MSD, Merck Serono, Kyowa Hakko
Kirin, Daiichi Sankyo, Servier, Kissei, Ignyta, and Astellas. He
has received personal fees from Ono Pharmaceutical, Novartis
Pharma, Chugai Pharmaceutical, Eli Lilly, Taiho Pharmaceutical,
AstraZeneca, Nippon Boehringer Ingelheim, Pfizer, MSD, Kissei,
Clovis Oncology, and Bristol-Meyers Squibb. All other authors have
no competing interests.
References
|
1
|
Katayama R, Khan TM, Benes C, Lifshits E,
Ebi H, Rivera VM, Shakespeare WC, Iafrate AJ, Engelman JA and Shaw
AT: Therapeutic strategies to overcome crizotinib resistance in
non-small cell lung cancers harboring the fusion oncogene EML4-ALK.
Proc Natl Acad Sci USA. 108:7535–7540. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Katayama R, Shaw AT, Khan TM,
Mino-Kenudson M, Solomon BJ, Halmos B, Jessop NA, Wain JC, Yeo AT,
Benes C, et al: Mechanisms of acquired crizotinib resistance in
ALK-rearranged lung cancers. Sci Transl Med. 4:120ra172012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sasaki T, Koivunen J, Ogino A, Yanagita M,
Nikiforow S, Zheng W, Lathan C, Marcoux JP, Du J, Okuda K, et al: A
novel ALK secondary mutation and EGFR signaling cause resistance to
ALK kinase inhibitors. Cancer Res. 71:6051–6060. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lovly CM, McDonald NT, Chen H,
Ortiz-Cuaran S, Heukamp LC, Yan Y, Florin A, Ozretić L, Lim D, Wang
L, et al: Rationale for co-targeting IGF-1R and ALK in ALK
fusion-positive lung cancer. Nat Med. 20:1027–1034. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fujita S, Masago K, Katakami N and Yatabe
Y: Transformation to SCLC after treatment with the ALK inhibitor
alectinib. J Thorac Oncol. 11:e67–e72. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ou SI, Lee TK, Young L, Fernandez-Rocha
MY, Pavlick D, Schrock AB, Zhu VW, Milliken J, Ali SM and Gitlitz
BJ: Dual occurrence of ALK G1202R solvent front mutation and small
cell lung cancer transformation as resistance mechanisms to second
generation ALK inhibitors without prior exposure to crizotinib.
Pitfall of solely relying on liquid re-biopsy? Lung Cancer.
106:110–114. 2017.PubMed/NCBI
|
|
7
|
Stieber P, Dienemann H, Schalhorn A,
Schmitt UM, Reinmiedl J, Hofmann K and Yamaguchi K:
Pro-gastrin-releasing peptide (ProGRP)-a useful marker in small
cell lung carcinomas. Anticancer Res. 19:2673–2678. 1999.PubMed/NCBI
|
|
8
|
Gainor JF, Dardaei L, Yoda S, Friboulet L,
Leshchiner I, Katayama R, Dagogo-Jack I, Gadgeel S, Schultz K,
Singh M, et al: Molecular mechanisms of resistance to first- and
second-generation ALK inhibitors in ALK-rearranged lung cancer.
Cancer Discov. 6:1118–1133. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Paweletz CP, Sacher AG, Raymond CK, Alden
RS, O'Connell A, Mach SL, Kuang Y, Gandhi L, Kirschmeier P, English
JM, et al: Bias-corrected targeted next-generation sequencing for
rapid, multiplexed detection of actionable alterations in cell-free
DNA from advanced lung cancer patients. Clin Cancer Res.
22:915–922. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lodrini M, Sprüssel A, Astrahantseff K,
Tiburtius D, Konschak R, Lode HN, Fischer M, Keilholz U, Eggert A
and Deubzer HE: Using droplet digital PCR to analyze MYCN and ALK
copy number in plasma from patients with neuroblastoma. Oncotarget.
8:85234–85251. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Johnson AC, Dô P, Richard N, Dubos C,
Michels JJ, Bonneau J and Gervais R: Identification of I1171N
resistance mutation in ALK-positive non-small-cell lung cancer
tumor sample and circulating tumor DNA. Lung Cancer. 99:38–40.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bordi P, Tiseo M, Rofi E, Petrini I,
Restante G, Danesi R and Del Re M: Detection of ALK and KRAS
mutations in circulating tumor DNA of patients with advanced
ALK-positive NSCLC with disease progression during crizotinib
treatment. Clin Lung Cancer. 18:692–697. 2017. View Article : Google Scholar : PubMed/NCBI
|