Introduction
Lung carcinoma has the highest incidence and
mortality rates worldwide (1,2). Lung carcinoma could be divided into
several subtypes, including small cell carcinoma, squamous cell
carcinoma, adenocarcinoma and large cell carcinoma and the last
three types are collectively called non-small cell lung cancer
(NSCLC), accounting for 80–85% of all the lung carcinomas (3). There are several incidence factors in
the pathogenesis of lung carcinoma, including air pollution,
smoking, occupation and eating habits (4,5).
Therefore, to clarify the molecular mechanisms of the tumorigenesis
and progression of lung carcinoma is necessary.
MicroRNAs (miRNAs) are a cohort of small non-coding
RNAs, which are ~22–28 oligonucleotides in length (6). miRNAs affect the cell process regulation
by binding to the 3′UTR of the mRNA of their target gene at
post-transcriptional level (7–14).
miR-23b, has been reported to have low expression and be involved
in multiple tumors, such as ovarian, bladder hepatocellular and
prostate cancers (15–18). In hepatocellular cancer, miR-23b
downregulates the expression of urokinase, c-Met and suppresses the
migration and epithelial-mesenchymal transition (EMT) (15,19).
Similar results have been reported in experimental autoimmune
encephalomyelitis, i.e., that miR-23b suppresses leukocyte
migration and pathogenesis (20). In
prostate cancer miR-23b represses proto-oncogene Src kinase and is
associated with cancer diagnosis and prognosis (16). However, the specific function and
regulatory mechanism of miR-202 in lung carcinoma progression has
not been reported.
Cyclin G1 (CCNG1), acts as a target of p53, is a
member of G-type cyclins located at chromosome 5q-32-q34, which has
six exons and constitutes of 259 amino acids (21,22). CCNG1
acts as a cell cycle regulator in human tumor cells such as
cervical carcinoma, hepatocellular carcinoma, breast cancer and
lung carcinoma (21,23–26). In
hepatocellular carcinoma, Wen et al revealed that CCNG1 may
act as a promising biomarker and contribute to the recurrence and
chemoresistance (24). Morever, some
miRNAs could interact with CCNG1 to affect tumor progression.
miR-1271 and miR-23b through targeting CCNG1 inhibit ovarian cancer
growth and progression (27,28).
In this study, we mainly investigated the
correlation of miR-23b and CCNG1 and the impact on lung carcinoma.
We measured the mRNA expression level of miR-23b and CCNG1 in lung
carcinoma tissues and lung cancer cells, using paracancerous
tissues and normal lung cell as internal reference, respectively.
We also explored the effects of changing miR-23b expression on the
cell proliferation ability of lung cancer cells A549 and NCI-H460.
Furthermore, we investigated the relationship between miR-23b or
CCNG1 expression and the survival of patients. The role of miR-23b
and CCNG1 in lung carcinoma was explored.
Materials and methods
Tissue specimens
According to WHO classification, 57 samples from
patients with lung carcinoma and paracancerous tissues were
collected from People's Hospital of Yan'an City (Yan'an, China)
from 2014 to 2016. All the specimens were frozen immediately after
surgery and stored at −80°C before RNA extraction and other tests.
No patient had received any therapy, including radiotherapy or
chemotherapy, before surgery. The complete clinicopathological
features of the patients with lung carcinoma (5 small cell
carcinoma, 24 squamous cell carcinoma, 19 adenocarcinoma and 9
large cell carcinoma) are described in Table I. Of this cohort, 28 patients were
diagnosed at advanced stage (stages III/IV) and all of these
patients did not suffer from distant metastasis at initial
diagnosis. Written informed consent was obtained from all patients
and the study was approved by the Ethics Committee of People's
Hospital of Yan'an City (Yan'an, China).
 | Table I.miR-23b expression and
clinicopathological features in 57 paired lung carcinoma
tissues. |
Table I.
miR-23b expression and
clinicopathological features in 57 paired lung carcinoma
tissues.
|
|
| miR-23b
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | Cases (n=57) | 26 High (%) | 31 Low (%) | P-value |
|---|
| Sex |
|
Male | 32 | 12 (37.5) | 20 (62.5) | 0.164 |
|
Female | 25 | 14 (56.0) | 11 (44.0) |
|
| Age (years) |
|
≤60 | 31 | 15 (48.4) | 16 (51.6) | 0.646 |
|
>60 | 26 | 11 (42.3) | 15 (57.7) |
|
| Tumor size
(mm) |
|
≤5.0 | 35 | 20 (57.1) | 15 (42.9) | 0.028a |
|
>5.0 | 22 | 6 (27.3) | 16 (72.7) |
|
| TNM stage |
|
I–II | 29 | 17 (58.6) | 12 (41.4) | 0.045a |
|
III–IV | 28 | 9 (32.1) | 19 (67.9) |
|
| Local invasion |
|
T1-T2 | 24 | 14 (58.3) | 10 (41.7) | 0.100 |
|
T3-T4 | 33 | 12 (36.4) | 21 (63.6) |
|
| Lymph node
metastasis |
|
0–2 | 30 | 18 (60.0) | 12 (40.0) | 0.022a |
|
>2 | 27 | 8 (29.6) | 19 (70.4) |
|
| Ki-67 |
|
<14% | 17 | 12 (70.6) | 5 (29.4) | 0.030a |
|
≥14% | 40 | 14 (35.0) | 26 (65.0) |
|
| CCNG1 |
|
Negative | 26 | 17 (65.4) | 9 (34.6) | 0.018a |
|
Positive | 31 | 9 (29.0) | 22 (71.0) |
|
Cell lines and culture conditions
Two human lung cancer cell lines (A549 and NCI-H460)
and normal lung cells (MRC-5) were obtained from the American Type
Culture Collection (ATCC, Rockville, MD, USA). All the cells were
maintained in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(FBS; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) cultured at
37°C in an atmosphere with 5% CO2.
RNA isolation and reverse
transcription quantitative PCR (RT-qPCR)
Total RNA or miRNA was isolated and extracted using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) or
miRcute Extraction and Separation of miRNAs kit (Tiangen Biotech
Co., Ltd., Beijing, China) according to the manufacturer's
instructions. To detect the expression of miRNA or mRNA, cDNA was
reverse-transcribed using PrimeScript™ II 1st strand cDNA synthesis
kit (Takara Biotechnology Co., Ltd., Dalian, China). Then RT-qPCR
was performed using the SYBR Premix kit or SYBR PrimeScript miRNA
RT-PCR kit (both from Takara Biotechnology Co., Ltd.). The
thermocycling conditions were 95°C for 3 min and 40 cycles of 95°C
for 15 sec followed by 60°C for 30 sec. Normalization was carried
out using glyceraldehyde-3-phosphate or dehydrogenase (GAPDH) and
U6 small nuclear RNA (U6). The relative expression levels of miRNA
and mRNA were calculated using 2−ΔΔCq method (29). All experiments were repeated at least
3 times. The primers were as follows: miR-23b forward,
5′-GGTGCTCTGGCTGCTTGG-3′ and reverse, 5′-GCCAAGGTCGTGGTTGCG-3′; U6
forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′; CCNG1 forward,
5′-GTTACCGCTGAGGAGCTGCAGTC-3′ and reverse,
5′-GCAGCCATCCTGGATGGATTCAG-3′; GAPDH forward,
5′-GGTGAAGGTCGGAGTCAACG-3′ and reverse,
5′-CAAAGTTGTCATGGATGHACC-3′.
Transfection
miR-23b mimic, miR-23b inhibitor and their negative
control (NC) were purchased from Guangzhou RiboBio Co., Ltd.
(Guangzhou, China). To assess the efficiency of miR-23b on cell
proliferation, we transfected miR-23b mimic or inhibitor into lung
cancer A549 and NCI-H460 cells, in order to overexpress or knock
down miR-23b expression, and normal control (NC) was included. To
detect the effect of miR-23b through CCNG1 for cell proliferation,
we used siRNA to interfere with the expression of CCNG1.
We seeded the lung cancer cells A549 and NCI-H460
into 6-well plates to cultivate overnight before transfection. The
plasmid vectors were transfected using Lipofectamine 3000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions.
Cell proliferation assay
Before the experiments, we seeded the lung cells
into 96-well plates with a density of 5×103 cells/well.
3-(4,5-Dimethyl-2-thia-zolyl)-2,5-diphenyl-2-H-tetrazolium bromide
(MTT; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) assay was
used to test the cell proliferative activity. After cultured for
24, 48, 72 or 96 h, 10 µl of MTT solution (5 mg/ml) were added in
each well. After cell incubattion with the MTT reagent at 37°C for
4 h, we added 150 µl dimethyl sulphoxide (DMSO), and measured the
absorbance at 490 nm on a microplate reader (BioTek Instruments,
Inc., Winooski, VT, USA). All experiments were repeated at least 3
times.
Transwell assay
The cell invasion assay was performed using
Transwell inserts (Millipore, Boston, MA, USA) coated with Matrigel
(BD Biosciences, Franklin Lakes, NJ, USA) on the upper surface.
Following the procedures described in the manufacturer's
instructions, 200 µl serum-free medium cell suspension containing
5×104 cells were added to the upper chamber of the
insert. Next, 500 µl RPMI-1640 medium with 15% FBS were added into
the lower compartment as a chemoattractant. After incubation at
37°C for 24 h, the cells on the upper surface of the membrane were
carefully removed using cotton swab and cells on the lower surface
were fixed at room temperature for 30 min with 100% methanol and
stained for 20 min at room temperature with 0.1% crystal violet.
Five visual fields of ×200 magnification of each insert were
randomly selected and counted under a light microscope (Olympus
Corp., Tokyo, Japan). Each experiment was performed in
triplicate.
Protein extraction and western
blotting
Total proteins were extracted from lung cancer cells
using RIPA lysis buffer, supplemented with PMSF (both from Beijing
Solarbio Science & Technology Co., Ltd., Beijing, China). After
centrifuged for 20 min at 4°C with 12,000 × g speed, the
concentration of protein was measured by BCA reagent kit (Beijing
Solarbio Science & Technology Co., Ltd.). Following
electrophoresis using 10% sodium dodecyl sulphate-polyacrylamide
gel electrophoresis (SDS-PAGE), the separated proteins were
transferred to the polyvinylidene fluoride membrane (PVDF;
Millipore). The concentration of the concentrated gel was 5%, and
the concentration of the separated gel was 10%. The membrane was
incubated with mouse anti-CCNG1 monoclonal antibody (1:1,000; cat.
no. WH0000900M1; Sigma-Aldrich; Merck KGaA) at 4°C overnight, with
GAPDH mouse antibody (1:3,000; cat. no. TA802519, ZSGB-BIO; OriGene
Technologies, Inc., Beijing, China) as internal control. A mouse
secondary antibody (1:4,000, cat. no. sc-516142; Santa Cruz
Biotechnology, Inc.) containing conjugated horseradish peroxidase
was used to incubate the membrane for 1 h at room temperature. The
proteins (CCNG1 and GAPDH) were evaluated by ECL western blotting
detection system (BestBio, Shanghai, China).
Plasmid construction and luciferase
reporter assay
TargetScan (www.targetscan.org) software predicted that CCNG1 was
a potential target gene of miR-23b with binding site located at
3′UTR of CCNG1. Therefore, we used double luciferase reporter assay
to verify whether miR-23b binds to the 3′UTR of CCNG1 mRNA. First,
we constructed the plasmid with CCNG1 3′UTR oligonucleotide
fragment inserted in (pcDNA3.1-CCNG1-WT). Then, we mutated miR-23b
binding sequences at CCNG1 3′UTR (pcDNA3.1-CCNG1-MUT) from
5′-…AAUGUGA…-3′ to 5′-…UUACACU…-3′. Effectiveness of constructs was
verified by sequencing.
miR-23b mimic or NC and pcDNA3.1-CCNG1-WT or MUT
were co-transfected into lung cancer A549 and NCI-H460 cells to
detect the luciferase reporter activity. Luciferase assay was
detected using Dual-Luciferase® Reporter Assay System
(Promega Corp., Madison, WI, USA) with Renilla luciferase as
normalization. Each experiment was performed in triplicate.
Statistical analysis
Experimental data are presented as mean ± standard
deviation (SD). Statistical analyses were performed using the SPSS
19.0 software (IBM Corp., Armonk, NY, USA). The differences between
groups were determined using Student's t-test and one-way ANOVA
followed by a Tukey's post hoc test. Pearson's χ2 test
was used to analyze the relationship between miR-23b expression and
the clinicopathological characteristics of lung cancer patients.
Pearson's correlation was performed to analyze the correlation
between the expression levels of miR-23b and CCNG1. Kaplan-Meier
method with log rank test were used to calculate the 5-year overall
survival (OS) and disease-free survival (DFS). P<0.05 indicates
a statistically significant difference.
Results
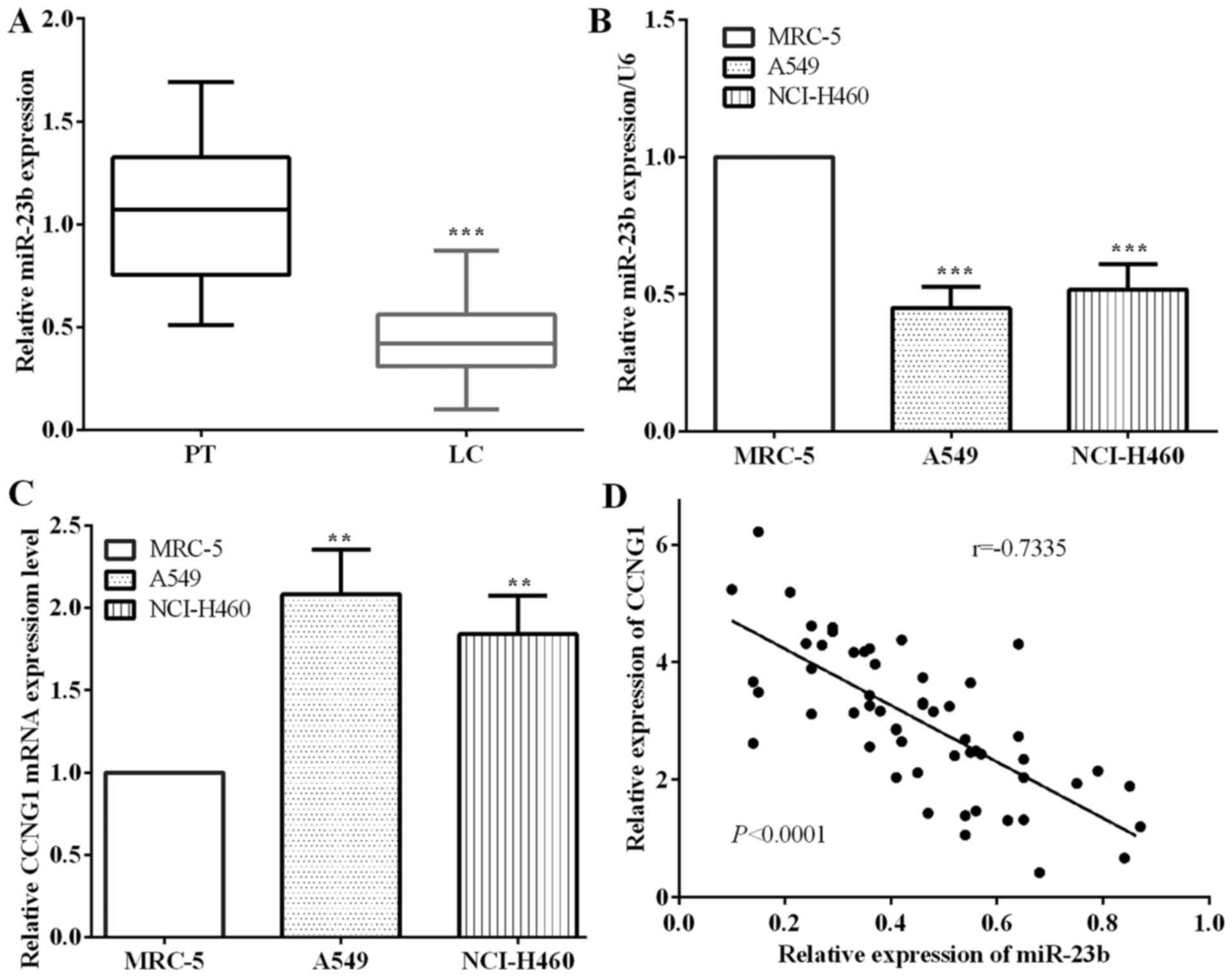
miR-23b expression is significantly
low and has negative correlation with CCNG1 in lung carcinoma
The expression of miR-23b and CCNG1 was examined by
RT-qPCR in cancerous and paracancerous tissues of 57 cases with
lung cancer. The results showed that the expression of miR-23b in
paracancerous tissues was obviously higher than that in cancerous
tissues (P<0.0001) (Fig. 1A).
There was no significant difference observed between the four lung
carcinoma subtypes. Moreover, we detected the expression of lung
cancer A549 and NCI-H460 cells and normal lung MRC-5 cells, and
found that the expression of miR-23b was significantly lower in
A549 (P=0.0002) and NCI-H460 (P=0.0008) cells compared with MRC-5
cells, which are similar results with those of tissues samples
(Fig. 1B).
Contrary to the results of miR-23b, the expression
of CCNG1 in lung cancer A549 (P=0.0024) and NCI-H460 cells
(P=0.0035) was significantly higher than that in normal lung cancer
MRC-5 cells (Fig. 1C). Furthermore,
the expression of miR-23b had negative correlation with CCNG1 in
lung carcinoma tissues as detected by Pearson's correlation test
(P<0.0001, r=−0.7335) (Fig.
1D).
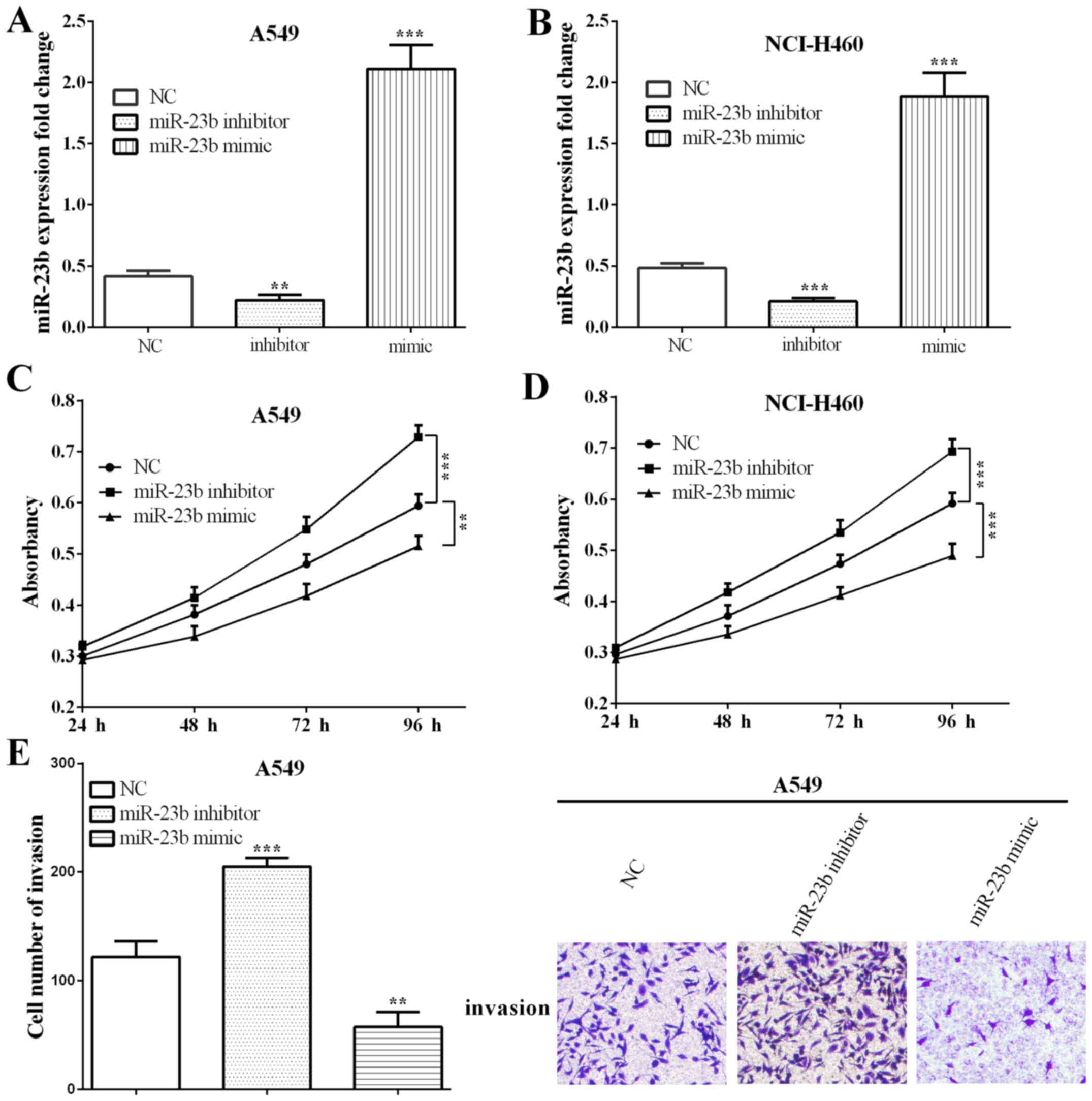
miR-23b inhibits proliferation and invasion of lung
carcinoma cells. To study the biological function of miR-23b on
proliferation and invasion, we overexpressed or knocked down
miR-23b using miR-23b inhibitor or mimic in lung cancer cells A549
(P=0.0056 and 0.0001) and NCI-H460 (P=0.0006 and 0.0002), and the
transfection efficiency was detected by RT-qPCR (Fig. 2A and B).
MTT assay was applied to detect the proliferation
after spreading cells for 24, 48, 72 and 96 h. As shown in Fig. 2C, when transfected with miR-23b
inhibitor in A549 cells, the ability of cell proliferation was
increased significantly (P=0.2039, 0.0553, 0.0046 and 0.0002) at 72
and 96 h. On the other hand, cell proliferation ability was reduced
when transfected with miR-23b mimic (P=0.5876, 0.0207, 0.0069 and
0.0021). Results were similar in NCI-H460 cells, with P-values
0.0319, 0.0156, 0.0064, 0.0007 and 0.0801, 0.0348, 0.0020 and
0.0006, respectively, when transfected with miR-23b inhibitor or
mimic (Fig. 2D).
To further explore the function of miR-23b in lung
carcinoma, Transwell assay was performed to detect the invasive
ability in altering miR-23b in A549 cells. Similar with
proliferation, Transwell assays showed that the invasive ability
was significantly increased (P=0.0010) in miR-202
inhibitor-transfected A549 cells, whereas the opposite results were
observed (P=0.0051) in miR-23b overexpressed cells (Fig. 2E).
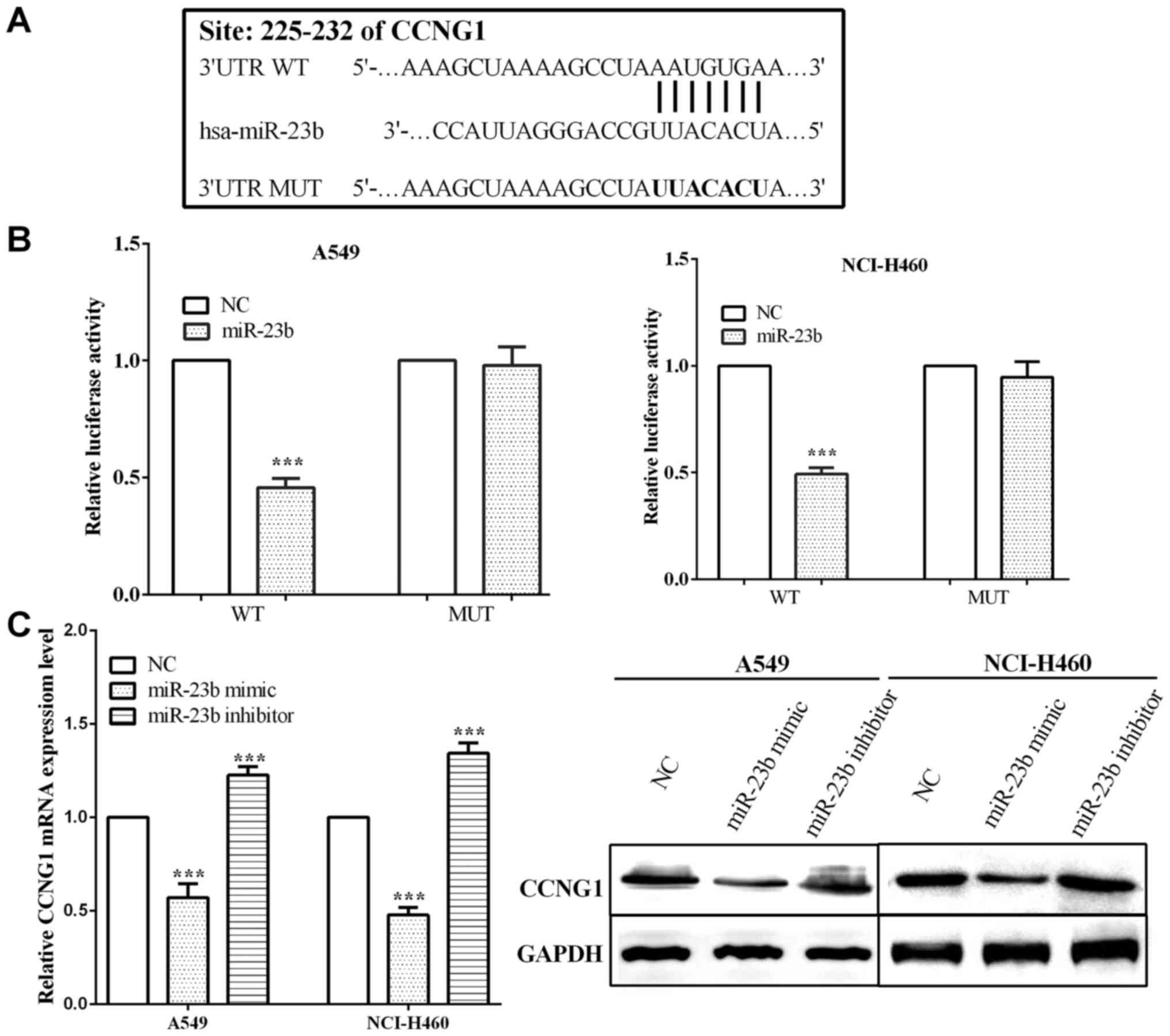
CCNG1 is a target of miR-23b and
mediated by miR-23b
TargetScan (http://www.targetscan.org/vert_71/), online software,
was utilized to predict potential target genes, and we found that
CCNG1 was a potential target gene of miR-23b. The potential binding
site of CCNG1 for miR-23b is located at 225–232 bp of its 3′UTR
mRNA (Fig. 3A). Luciferase reporter
assay was performed in order to test whether miR-23b interacted
with CCNG1. Firstly, we co-transfected two plasmid vectors,
respectively, containing CCNG1 3′UTR and miR-23b, and then detected
the change of luciferase activity. The binding sequences of CCNG1
3′UTR were mutated from 5′-…AAUGUGA…-3′ to 5′-…UUACACU…-3′, and the
luciferase activity was measured. We found that when co-transfected
with miR-23b and CCNG1 wild-type 3′UTR, the luciferase activity was
reduced significantly both in A549 (P<0.0001) and NCI-H460
(P<0.0001) cell lines. In contrast, when co-transfected with
miR-23b and CCNG1 mutant type 3′UTR, luciferase activity had almost
no change in both A549 (P=0.680) and NCI-H460 (P=0.278) cells
(Fig. 3B).
Furthermore, we transfected miR-23b mimic or
inhibitor to overexpress or knock down miR-23b, and detected the
change of CCNG1. As a result, when overexpressing miR-23b, the
expression of CCNG1 was reduced with P-values of A549 and NCI-H460
cells 0.0006 and <0.0001, respectively. On the contrary, in
knockdown of miR-23b, CCNG1 expression was increased significantly
both in mRNA (P=0.0010 and 0.0004 in A549 and NCI-H460,
respectively) and protein levels (Fig.
3C).
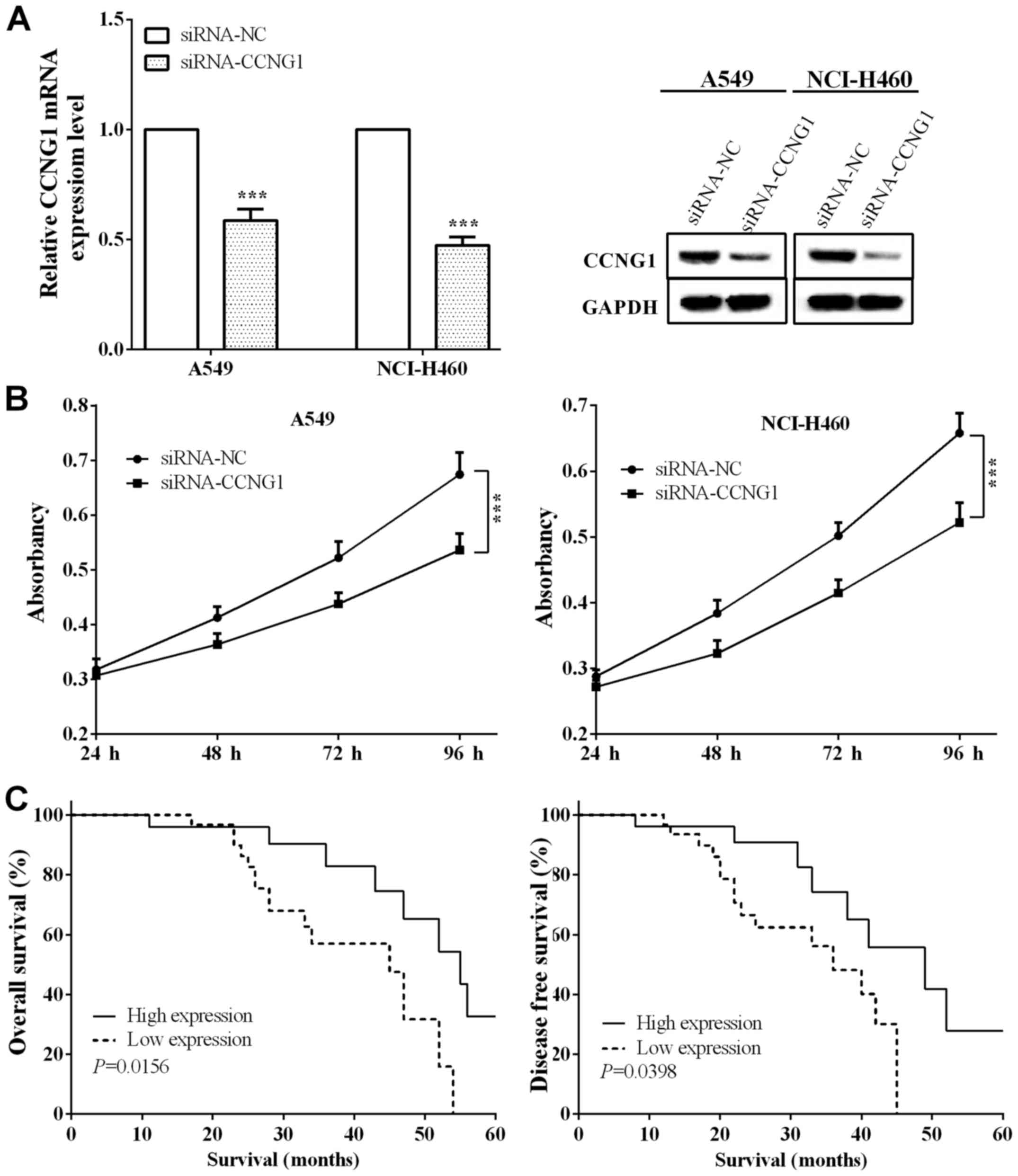
Interference of CCNG1 expression inhibits lung
cancer cell proliferation. In order to investigate the effect of
CCNG1 on cell proliferation, we disrupted the expression of CCNG1
in lung cancer cells A549 and NCI-H460. As Fig. 4A shows, RT-qPCR (P=0.0001 and
<0.0001, respectively, for A549 and NCI-H460) and western
blotting identified the interference results, respectively.
Proliferation ability was detected by MTT assay, and we found that
in disrupted CCNG1, the proliferation ability of A549 and NCI-H460
cells was significantly reduced (P=0.4422, 0.0399, 0.0157, 0.0088
and 0.1216, 0.0202, 0.0060, 0.0051, respectively) at 48, 72 and 96
h (Fig. 4B). The results revealed
that the interference of CCNG1 expression could inhibit the
proliferative ability of lung cancer cells.
Low expression of miR-23b in lung
carcinoma predicts poor prognosis
We divided the patients into miR-23b high expression
group [miR-23b(+)] and low expression group [miR-23b(−)] according
to the expression of miR-23b, with 26 and 31 cases, respectively.
The expression of miR-23b had negative correlation with tumor size
(P=0.028), TNM stage (P=0.045), lymph node metastasis (P=0.022),
Ki-67 (P=0.030) and CCNG1 (P=0.018), as shown in Table I. Kaplan-Meier was utilized to
calculate the OS and DFS of patients and we found that both OS and
DFS in miR-23b(−) group were significantly lower than miR-23b(+)
group (log-rank P=0.0156 and 0.0398), which expounded that lack of
miR-23b predicted poor prognosis in lung carcinoma (Fig. 4C).
Discussion
Lung carcinoma is the most common malignancy for men
and ranks second for women world-wide. Therefore, it is
particularly important to find molecular markers for the diagnosis
and prediction of lung carcinoma. miRNAs affect cell progression by
targeting the 3′UTR of the mRNA of their target gene (7–10,30,31).
miRNAs usually act as oncogenes or tumor suppressors in lung
carcinoma, including miR-23b, miR-221, miR-148b, miR-423 and many
other miRNAs (32). miR-23b was
reported to suppress cell proliferation, migration and inhibit EMT
in ovarian, bladder, hepatocellular, and prostate cancer (15–17,19,28).
However, the role of miR-23b in lung carcinoma has not been
reported. Therefore, the expression of miR-23b was tested in 57
paired lung carcinoma and paracancerous tissues, and we found that
miR-23b expression in lung carcinoma tissues was obviously lower
than paracancerous tissues, which is consistent with Shao et
al (3). Similar with the results
obtained in tissues, the expression of miR-23b in lung cancer cells
A549 and NCI-H460 was significantly lower than that in normal lung
MRC-5 cells.
Since miR-23b was downregulated in lung carcinoma,
we attempted to explore the molecular function of miR-23b in lung
carcinoma. We transfected miR-23b mimic or inhibitor to overexpress
or knockdown miR-23b, and then measured the proliferation ability
by MTT assay. Consistent with the results of Majid et al in
bladder cancer (17), we found that
miR-23b could inhibit lung cancer cell proliferation and invasion.
For Transwell assay, since we have verified cell proliferation
ability with two cell lines, according to Pan et al
(33), we believe that one cell line
could fully represent the entire experimental result.
We predicted that CCNG1 is a potential target of
miR-23b using TargetScan and it was verified in lung cancer cells
A549 and NCI-H460. CCNG1, a member of G-type cyclins, acted as cell
cycle regulator and was overexpressed in human tumor cells
especially in lung carcinoma. CCNG1 was up-expressed in lung
carcinoma and could increase cell sensitivity to radiotherapy to
promote cell death (24,34). Luciferase reporter assay was performed
to verify miR-23b binding to the 3′UTR of CCNG1, which was
consistent with the findings in ovarian cancer. Subsequently,
miR-23b expression was exogenously altered and the change of the
expression of CCNG1 was detected to determine miR-23b-regulated
CCNG1. The expression of CCNG1 was highly expressed in lung
carcinoma tissues and cells. miR-23b had negative correlation with
CCNG1 in lung carcinoma tissues. Furthermore, CCNG1 was interfered
to survey cell proliferation ability, and we found that CCNG1
destroyed the suppressed cell proliferation ability in lung cancer
cells.
It has been reported that low expression of miR-23b
in prostate cancer suggests poor prognosis (16). In order to explore the impact of
miR-23b on the survival of patients with lung cancer, we calculated
the OS and DFS of 57 lung cancer patients, and we discovered that
miR-23b low expression predicts shorter OS and DFS than high
expression. The results expound that miR-23b low expression
predicts poor prognosis in lung carcinoma. However, the limitation
of the study was that we did not perform the multivariate
analysis.
In conclusion, miR-23b expression was low in lung
carcinoma tissues and cell lines. miR-23b suppressed lung carcinoma
proliferation by targeting CCNG1 and predicted poor prognosis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XY contributed significantly to the analysis and
manuscript preparation; HH and ZZ contributed significantly in
performing the experiments and assisted in writing the manuscript;
WY performed the data analyses and wrote the manuscript; CX
assisted in performing the analysis with constructive discussions.
XC contributed to the conception of the study. All authors read and
approved the final study.
Ethics approval and consent to
participate
The Ethics Committee of People's Hospital of Yan'an
City (Yan'an, China) approved the research, and written informed
consent was obtained from all the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Siegel RL and Jemal A: Lung
cancer statistics. Adv Exp Med Biol. 893:1–19. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shao Y, Liang B, Long F and Jiang SJ:
Diagnostic microRNA biomarker discovery for non-small-cell lung
cancer adenocarcinoma by integrative bioinformatics analysis.
Biomed Res Int. 2017:25630852017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Butler LM, Montague JA, Koh WP, Wang R, Yu
MC and Yuan JM: Fried meat intake is a risk factor for lung
adenocarcinoma in a prospective cohort of Chinese men and women in
Singapore. Carcinogenesis. 34:1794–1799. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Paris C, Clement-Duchene C, Vignaud JM,
Gislard A, Stoufflet A, Bertrand O, Thiberville L, Grosdidier G,
Martinet Y, Benichou J, et al: Relationships between lung
adenocarcinoma and gender, age, smoking and occupational risk
factors: A case-case study. Lung Cancer. 68:146–153. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Christodoulatos GS and Dalamaga M:
Micro-RNAs as clinical biomarkers and therapeutic targets in breast
cancer: Quo vadis? World J Clin Oncol. 5:71–81. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Subtil FS, Wilhelm J, Bill V, Westholt N,
Rudolph S, Fischer J, Scheel S, Seay U, Fournier C, Taucher-Scholz
G, et al: Carbon ion radiotherapy of human lung cancer attenuates
HIF-1 signaling and acts with considerably enhanced therapeutic
efficiency. FASEB J. 28:1412–1421. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Flynt AS and Lai EC: Biological principles
of microRNA-mediated regulation: Shared themes amid diversity. Nat
Rev Genet. 9:831–842. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lauressergues D, Couzigou JM, Clemente HS,
Martinez Y, Dunand C, Bécard G and Combier JP: Primary transcripts
of microRNAs encode regulatory peptides. Nature. 520:90–93. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Voinnet O: Origin, biogenesis, and
activity of plant microRNAs. Cell. 136:669–687. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Di Giacomo G, Koss M, Capellini TD,
Brendolan A, Pöpperl H and Selleri L: Spatio-temporal expression of
Pbx3 during mouse organogenesis. Gene Expr Patterns. 6:747–757.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lichtenauer UD, Duchniewicz M, Kolanczyk
M, Hoeflich A, Hahner S, Else T, Bicknell AB, Zemojtel T, Stallings
NR, Schulte DM, et al: Pre-B-cell transcription factor 1 and
steroidogenic factor 1 synergistically regulate adrenocortical
growth and steroidogenesis. Endocrinology. 148:693–704. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Salvi A, Sabelli C, Moncini S, Venturin M,
Arici B, Riva P, Portolani N, Giulini SM, De Petro G and Barlati S:
MicroRNA-23b mediates urokinase and c-met downmodulation and a
decreased migration of human hepatocellular carcinoma cells. FEBS
J. 276:2966–2982. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Majid S, Dar AA, Saini S, Arora S,
Shahryari V, Zaman MS, Chang I, Yamamura S, Tanaka Y, Deng G, et
al: miR-23b represses proto-oncogene Src kinase and functions as
methylation-silenced tumor suppressor with diagnostic and
prognostic significance in prostate cancer. Cancer Res.
72:6435–6446. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Majid S, Dar AA, Saini S, Deng G, Chang I,
Greene K, Tanaka Y, Dahiya R and Yamamura S: MicroRNA-23b functions
as a tumor suppressor by regulating Zeb1 in bladder cancer. PLoS
One. 8:e676862013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li W, Liu Z, Chen L, Zhou L and Yao Y:
MicroRNA-23b is an independent prognostic marker and suppresses
ovarian cancer progression by targeting runt-related transcription
factor-2. FEBS Lett. 588:1608–1615. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cao J, Liu J, Long J, Fu J, Huang L, Li J,
Liu C, Zhang X and Yan Y: microRNA-23b suppresses
epithelial-mesenchymal transition (EMT) and metastasis in
hepatocellular carcinoma via targeting Pyk2. Biomed Pharmacother.
89:642–650. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Y, Han JJ, Liang XY, Zhao L, Zhang
F, Rasouli J, Wang ZZ, Zhang GX and Li X: miR-23b suppresses
leukocyte migration and pathogenesis of experimental autoimmune
encephalomyelitis by targeting CCL7. Mol Ther. 26:582–592. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Horne MC, Goolsby GL, Donaldson KL, Tran
D, Neubauer M and Wahl AF: Cyclin G1 and cyclin G2 comprise a new
family of cyclins with contrasting tissue-specific and cell
cycle-regulated expression. J Biol Chem. 1271:6050–6061. 1996.
View Article : Google Scholar
|
|
22
|
Ye XX, Liu CB, Chen JY, Tao BH and Zhi-Yi
C: The expression of cyclin G in nasopharyngeal carcinoma and its
significance. Clin Exp Med. 12:21–24. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liang J, Bian ML, Chen QY, Liu X, Ou H, Li
M and Liu J: Relationship between cyclin G1 and human papilloma
virus infection in cervical intraepithelial neoplasia and cervical
carcinoma. Chin Med Sci J. 21:81–85. 2006.PubMed/NCBI
|
|
24
|
Wen W, Han T, Chen C, Huang L, Sun W, Wang
X, Chen SZ, Xiang DM, Tang L, Cao D, et al: Cyclin G1 expands liver
tumor-initiating cells by Sox2 induction via Akt/mTOR signaling.
Mol Cancer Ther. 12:1796–1804. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Reimer CL, Borras AM, Kurdistani SK,
Garreau JR, Chung M, Aaronson SA and Lee SW: Altered regulation of
cyclin G in human breast cancer and its specific localization at
replication foci in response to DNA damage in p53+/+
cells. J Biol Chem. 274:11022–11029. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao X, Liu M and Li D: Oleanolic acid
suppresses the proliferation of lung carcinoma cells by
miR-122/Cyclin G1/MEF2D axis. Mol Cell Biochem. 400:1–7. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu X, Ma L, Rao Q, Mao Y, Xin Y, Xu H, Li
C and Wang X: MiR-1271 inhibits ovarian cancer growth by targeting
cyclin G1. Med Sci Monit. 21:3152–3158. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yan J, Jiang JY, Meng XN, Xiu YL and Zong
ZH: miR-23b targets cyclin G1 and suppresses ovarian cancer
tumorigenesis and progression. J Exp Clin Cancer Res. 35:312016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jiang C, Chen X, Alattar M, Wei J and Liu
H: MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis
of gastric cancer. Cancer Gene Ther. 22:291–301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu Y, Li T, Chen G, Yan G, Zhang X, Wan
Y, Li Q, Zhu B and Zhuo W: Identification of a serum microRNA
expression signature for detection of lung cancer, involving
miR-23b, miR-221, miR-148b and miR-423-3p. Lung Cancer. 114:6–11.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pan Y, Ye C, Tian Q, Yan S, Zeng X, Xiao
C, Wang L and Wang H: miR-145 suppresses the proliferation,
invasion and migration of NSCLC cells by regulating the BAX/BCL-2
ratio and the caspase-3 cascade. Oncol Lett. 15:4337–4343.
2018.PubMed/NCBI
|
|
34
|
Seo HR, Lee DH, Lee HJ, Baek M, Bae S, Soh
JW, Lee SJ, Kim J and Lee YS: Cyclin G1 overcomes radiation-induced
G2 arrest and increases cell death through transcriptional
activation of cyclin B1. Cell Death Differ. 13:1475–1484. 2006.
View Article : Google Scholar : PubMed/NCBI
|