Introduction
Core-binding factor (CBF) acute myeloid leukemia
(AML) associated with t(8;21; q22;q22)/runt related transcription
factor (RUNX)1-RUNX1 translocation partner 1 (T1) or inv(16;
p13q22)/t(16;16; p13;q22)/CBFβ subunit-myosin heavy chain 11 has
been reported to exhibit a favorable outcome (1–3). Jourdan
et al (4) reported that,
compared with gene mutations, minimal residual disease (MRD) levels
play a more important role in predicting early relapse in patients
with CBF-AML. Two popular approaches to detect MRD persistence are
quantitative polymerase chain reaction (qPCR) and multiparametric
flow cytometry, which possess increased submicroscopic sensitivity
compared with that possessed by conventional morphology (5–7). Although
multiple studies are attempting to stratify patients into low- or
high-risk relapse groups, the threshold to do so remains
controversial (8–11). Since it is affected by many technical
and clinical factors, a universal cut-off remains to be agreed upon
at present. Furthermore, for children with AML, PCR and
multiparametric flow cytometry have been associated with
inconsistent results (5,7).
Spatially proximal heterogeneous chromosomes are
more likely to cause chromosomal translocations compared with
spatially distant heterogeneous chromosomes, and these
translocation-prone chromosomes are frequently located towards the
center of the nucleus (12–15). To further understand the clinical
features and treatment response, our previous study followed up
with a patient with AML with maturation to delineate the spatial
organization of leukemia-specific chromosomes using
three-dimensional fluorescence in situ hybridization
(3D-FISH) as a means to detect MRD persistence (16). Disease deterioration was detected
using 3D-FISH [with 37% malignant cells (MCs); P<0.05] 4 months
prior to final relapse, whereas the PCR and 2D-FISH results were
negative with no gene fusion transcripts or chromosomal
translocations. 3D-FISH analysis in the study provided information
that may assist in predicting early relapse (16).
Studies using PCR to monitor MRD persistence in AML
have demonstrated that peripheral blood (PB) samples were as
informative as bone marrow (BM) samples in identifying patients at
high risk of relapse and may represent an alternative source of
cells for diagnosis (17–20). To further assess the similarities and
differences between BM and PB, the present study compared the
spatial organization of leukemia-specific chromosomes between BM
and PB samples from 6 patients with AML-M2 using 3D-FISH with a
confocal laser scanning microscope system.
Materials and methods
Materials
Paired BM and PB samples were collected from 6
patients (aged 24–47 years, including 5 males and 1 female) with
AML-M2, including 5 RUNX1-RUNX1T1+ patients (numbered as
1–5 in Table I) at three different
stages (initial diagnosis, post-chemotherapy and remittent) and 1
RUNX1-RUNX1T1− patient at initial diagnosis (numbered as
6 in Table I), who were treated at
Peking University Third Hospital (Beijing, China) between December
2012 and September 2015. All patients provided written informed
consent to participate in the present study. The present study was
approved by the Ethics Committee of Peking University Third
Hospital.
 | Table I.P-values for comparisons of
occurrences of three types of cells and RRP of chromosomes 8 and 21
between BM and PB samples from 6 patients with acute myeloid
leukemia-M2. |
Table I.
P-values for comparisons of
occurrences of three types of cells and RRP of chromosomes 8 and 21
between BM and PB samples from 6 patients with acute myeloid
leukemia-M2.
| Variables | Initial
diagnosis |
Post-chemotherapy | Remittent
stage | Negative |
|---|
| Patient | 1 | 2 | 3 | 4 | 5 | 6 |
| Occurrences | 0.416 | 0.420 | 0.779 | 0.115 | 0.744 | 0.431 |
| Chromosome
8-RRP | 0.054 | 0.894 | /a | /a | 0.069 | /a |
| Chromosome
21-RRP | 0.435 | 0.511 | 0.459 | 0.897 | 0.899 | 0.024b |
Reverse transcription-quantitative
poylermase chain reaction (RT-qPCR)
Mononuclear cells (MNCs) from the BM were isolated
using Ficoll-Hypaque lymphocyte separation medium (Lonza Group,
Ltd., Basel, Switzerland). Furthermore, RNA was extracted from MNCs
using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) according
to the manufacturer's guidelines. cDNA was generated using the
Maxima H Minus First Strand cDNA Synthesis kit (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). Cycling conditions were 25°C
for 10 min, followed by 50°C for 15 min and the reaction was
terminated at 85°C for 5 min. RT-qPCR was performed to monitor the
RUNX1-RUNX1T1 transcripts according to the manufacturer's standard
protocol (RUNX1-RUNX1T1 Fusion gene Quant® kit; YuanQi
Biomedical, Ltd., Shanghai, China) using the ABI PRISM 7500 DNA
Sequence Detection system (Applied Biosystems; http://home.appliedbiosystems.com). The reference
gene was Abelson (ABL). The fluorophore of TaqMan and primers
(RUNX1-RUNX1T1 fusion gene: Forward, 5′-AATCACAGTGGATGGGCCC-3′ and
reverse, 5′-GAGTCTGGCATTGTGGAGTGC-3′. ABL gene: Forward,
5′-TGGAGATAACACTCTAAGC-3′ and reverse, 5′-GATGTAGTTGCTTGGGACCCA-3′)
were all included in the kit. Cycling conditions were 42°C for 30
min, initial denaturation at 94°C for 5 min, followed by 40 cycles
of denaturation (at 94°C for 15 sec) and annealing (at 60°C for 15
sec). Clinical RT-qPCR results showed that the relative
RUNX1-RUNX1T1 transcripts, which were calculated using the
2−ΔΔCq method (7,21), for BM samples from the 6 patients were
52.00, 0.09, 0.05, 0.00, 1.80 and 0.00%, which approximately
described the disease conditions of the patients that the initial
diagnosed patient showed high RUNX1-RUNX1T1 transcripts (52.00%)
and remitted or RUNX1-RUNX1T1− patients showed low
RUNX1-RUNX1T1 transcripts (0.09, 0.05, 0.00, 1.80 and 0.00%).
3D-FISH and imaging
MNCs were obtainedd using Ficol-Hypaque density
gradient centrifugation as previously described (22), and ≥60 interphase cells from each
sample were analyzed in 3D-FISH experiments, which were performed
as previously described (22).
Suspended MNCs were adhered onto the rough surface of a microscope
slide following incubation for 150 min at 37°C with 5%
CO2. Subsequently, MNCs were fixed in 4%
paraformaldehyde for 10 min at room temperature and then underwent
certain pre-treatments (22) to
increase the likelihood of successful hybridization. Whole
chromosome 8 probes (green; Kreatech Diagnostics; Leica Biosystems,
Wetzlar, Germany) labeled with fluorescein isothiocyanate (FITC)
and whole chromosome 21 probes (red; Kreatech Diagnostics; Leica
Biosystems) labeled with tetramethylrhodamine isothiocyanate were
used to label chromosome 8 and 21, respectively, in a hybridization
oven (ThermoBrite S500; StatSpin, Inc., Westwood, MA, USA;
www.statspin.com) with denaturation for 5 min at
75°C and then hybridization for 48 h at 37°C. Following washing
with Igepal CA-630 (21), 10 µl
diamidinophenylindole (blue; 0.1 µg/ml) was used to counterstain
the nuclei at −20°C for more than 15 min.
Image stacks were acquired using a Nikon A1Rsi
confocal microscope system with a Plan Apo 100×/1.4 NA oil
immersion objective lens. Excitation spectrums for
diamidinophenylindole, FITC, and tetramethylrhodamine
isothiocyanate were 405, 488, and 561 nm, respectively. The size of
the x-y optical sections was 512×512 pixels with a pixel size of
80×80 nm, and the axial step was 500 nm.
Statistical analysis
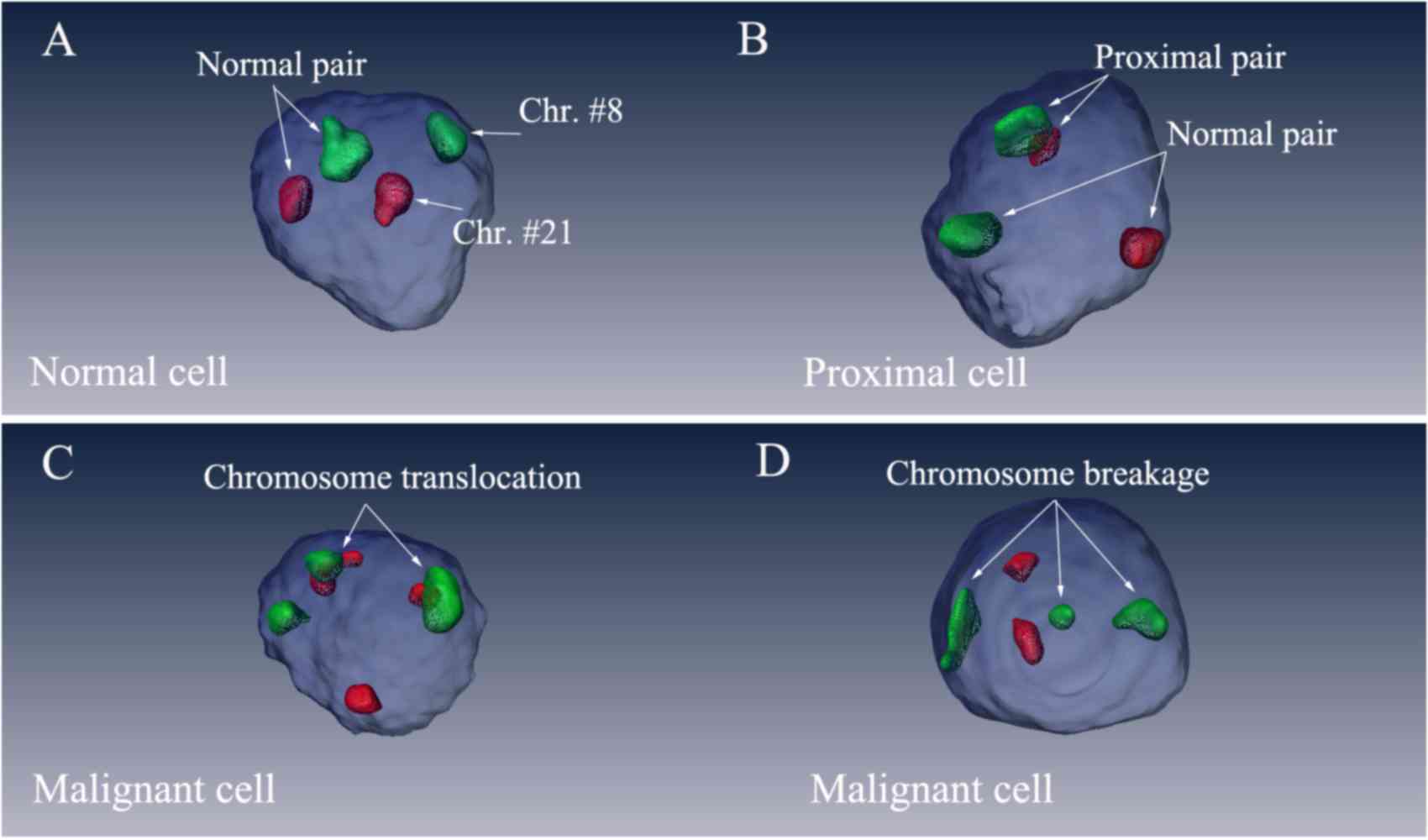
The 3D views of the nuclei were reconstructed using
Amira 5.4.3 software (FEI; Thermo Fisher Scientific, Inc.; Fig. 1). Green signals indicated chromosome 8
and red signals referred to chromosome 21. The blue spheres
represented the nuclei. Only cells that exhibited ≥2 green signals
and 2 red signals were considered in the analysis. According to the
number and spatial relative positions of the green/red signals, the
present study classified the cells into three types: Normal cells
(NCs; Fig. 1A), proximal cells (PCs;
Fig. 1B) and malignant cells (MCs;
Fig. 1C and D). Cells that exhibited
3 green or red signals (3g or 3r) were identified as MCs with
chromosomal translocations and breakages. Cells that possessed 2
red and 2 green signals (2r2g) were further categorized according
to the characterization of the physical contact between the
heterogeneous chromosomes. In cells that exhibited 2r2g signals,
heterogeneous chromosomes touching or intermingling with each other
were considered proximal and categorized as proximal pairs (PPs);
chromosomes located distantly from each other were categorized as
normal pairs (NPs). Cells that exhibited 2r2g signals were
classified as NCs when all heterogeneous chromosomes were NPs, and
were classified as PCs when there was at least one PP in the
nucleus.
The present study calculated the relative radial
positions [RRPs; the distance from the center of mass of chromosome
8 or 21 to the center of mass of the nucleus (d)/the radius
of the nucleus (R)] of chromosomes 8 and 21 in all cell
nuclei. No less than 60 cells were calculated for each sample
during the analysis. RRPs of chromosomes are presented as mean ±
standard error of the mean. SPSS 16.0 (SPSS Inc., Chicago, IL, USA)
was used to perform statistical analysis. The frequencies of
different types of cell were compared between BM and PB samples
using Pearson's χ2 test. Chromosomal RRPs were compared
between BM and PB samples using one-way analysis of variance with
the least significant difference post hoc test. RRPs were compared
between NPs and PPs using the two-sample, two-sided
Kolmogorov-Smirnov (K-S) test. P<0.05 was considered to indicate
a statistically significant difference.
Results
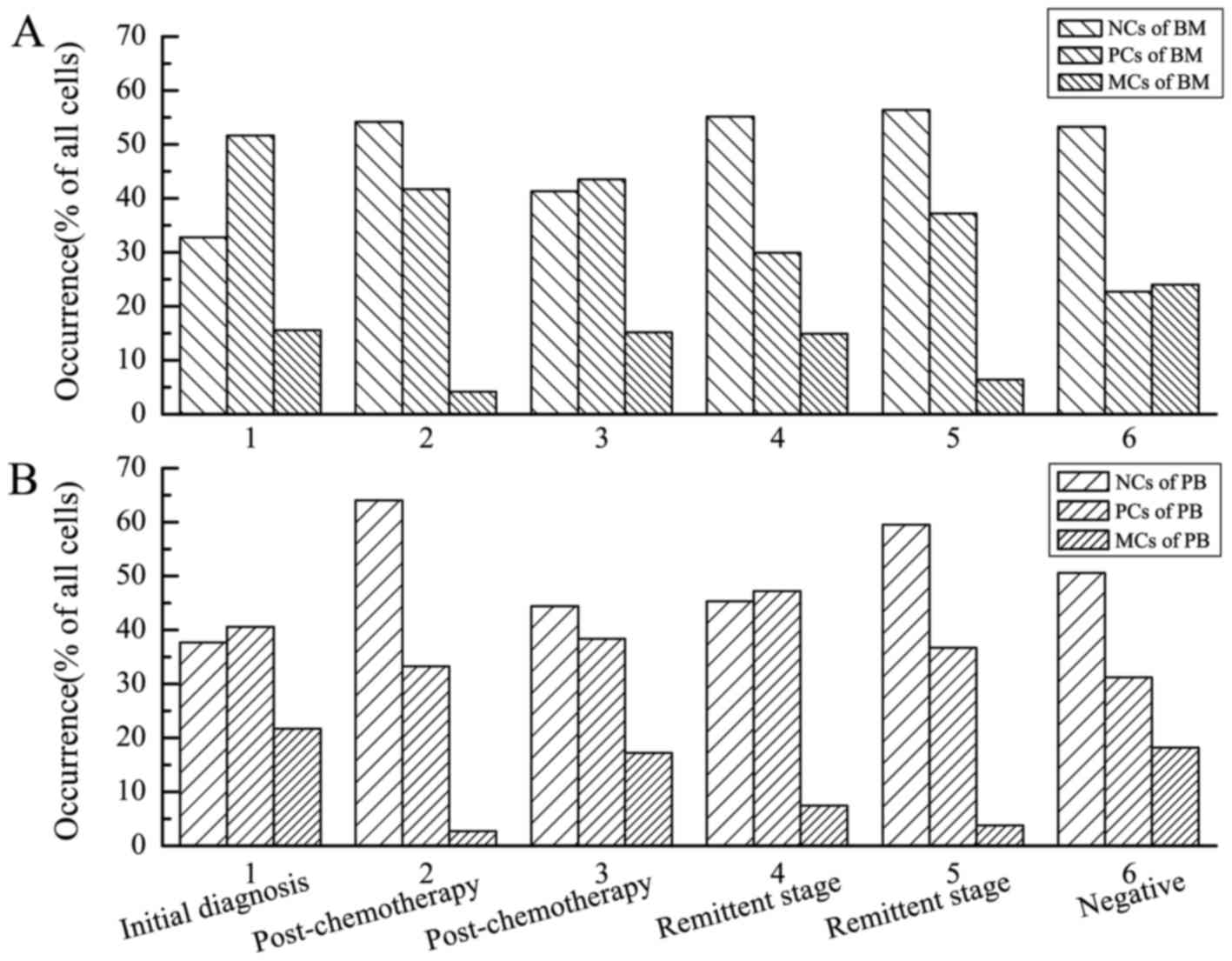
Comparing the occurrence (% of all
cells) of different types of cell in BM and PB samples
Cells were classified into three types (Fig. 1): NCs, PCs and MCs. The occurrence (%
of all cells) of different types of cell in the BM and PB samples
from the 6 patients was provided (Fig.
2). Histograms of sparse, medium and dense left-inclined
diagonals referred to the occurrence of NCs, PCs and MCs in the BM
samples, respectively (Fig. 2A).
Histograms of sparse, medium and dense right-inclined diagonals
referred to the occurrence of NCs, PCs and MCs in the PB samples,
respectively (Fig. 2B). P-values for
comparisons of the proportions between BM and PB samples, as
determined using Pearson's χ2 tests, were provided
(Table I). The comparisons revealed
no significant differences (P>0.05 for all patients), which
suggested that the occurrence (% of all cells) of the different
types of cell in the BM samples were generally consistent with
those in the PB samples.
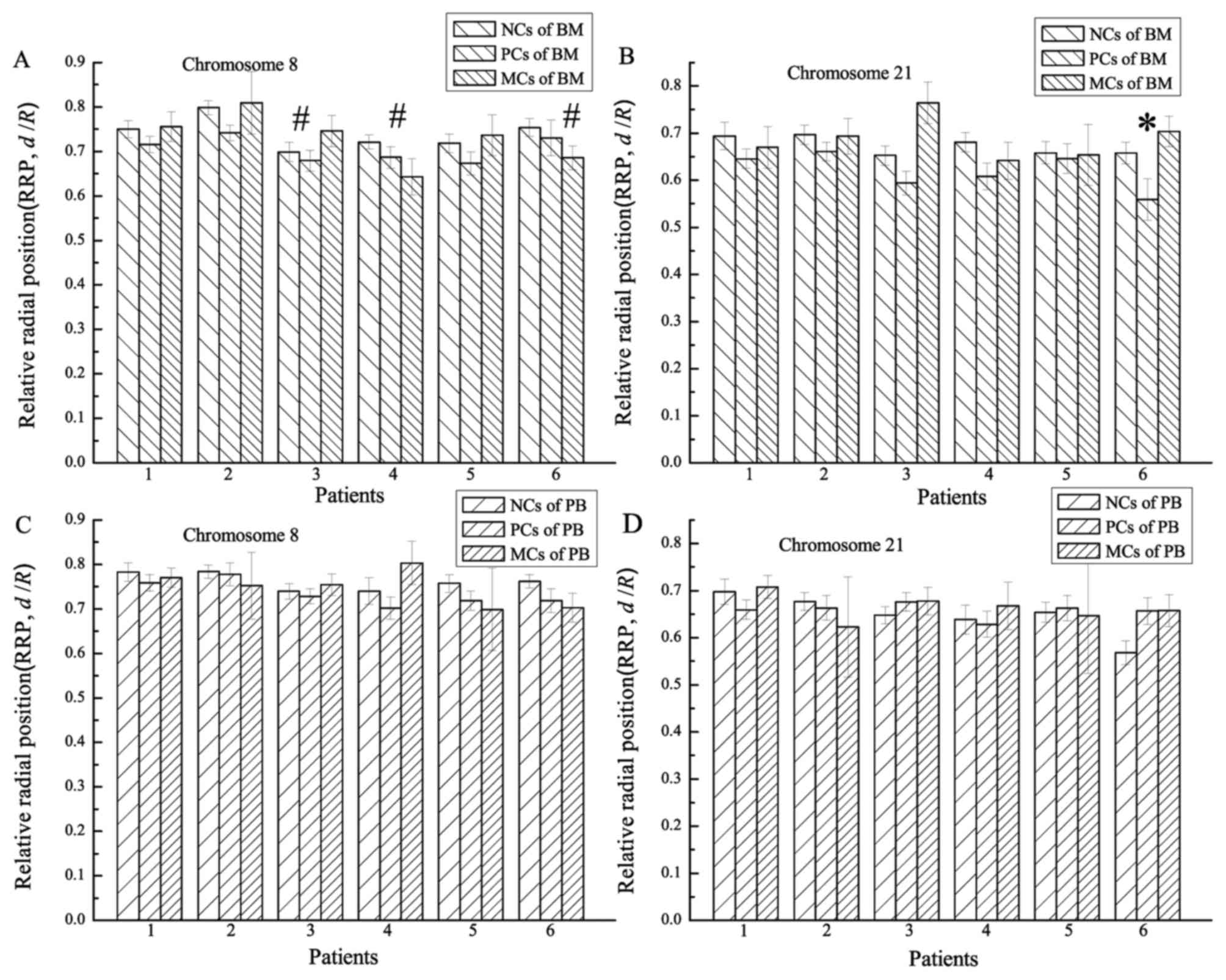
Comparing chromosomal RRPs between BM
and PB samples
RRPs (d/R) of chromosomes 8 and 21
were calculated for all cell nuclei. Chromosomal RRPs were compared
between BM and PB samples from the 6 patients (Fig. 3; Table
I). Histograms of sparse, medium and dense left-inclined
diagonals (Fig. 3A and B) referred to
the RRPs of chromosomes in NCs, PCs and MCs from the BM samples,
respectively. Histograms of sparse, medium and dense right-inclined
diagonals (Fig. 3C and D) referred to
the RRPs of chromosomes in NCs, PCs and MCs from the PB samples,
respectively. Chromosomal RRPs were compared between the BM and PB
samples using one-way analysis of variance (Table I). The comparison of RRPs of
chromosome 21 between the BM and PB samples revealed a significant
difference only for the RUNX1-RUNX1T1− patient (P=0.024;
number 6 in Fig. 3).
However, the RRPs of chromosomes 8 and 21 in the PCs
were generally more proximate to the centre of the nucleus compared
with those in the NCs. P-values for the comparison of RRPs of
chromosome 8 in NC and PC nuclei ranged from 0.089–0.508 (median,
0.378) for the 6 BM samples and from 0.010–0.964 (median, 0.583)
for the 6 PB samples. P-values for the comparison of RRPs of
chromosome 21 in NC and PC nuclei ranged from 0.024–0.980 (median,
0.334) for the 6 BM samples. For the 6 PB samples, 50% of the
chromosome 21 RRPs in PC nuclei were more interior in the nuclei
compared with those in NC nuclei (P=0.234, 0.289 and 0.607), and
50% were not (P=0.104, 0.608 and 0.014). Comparisons were made
using the two-sample, two-sided K-S test.
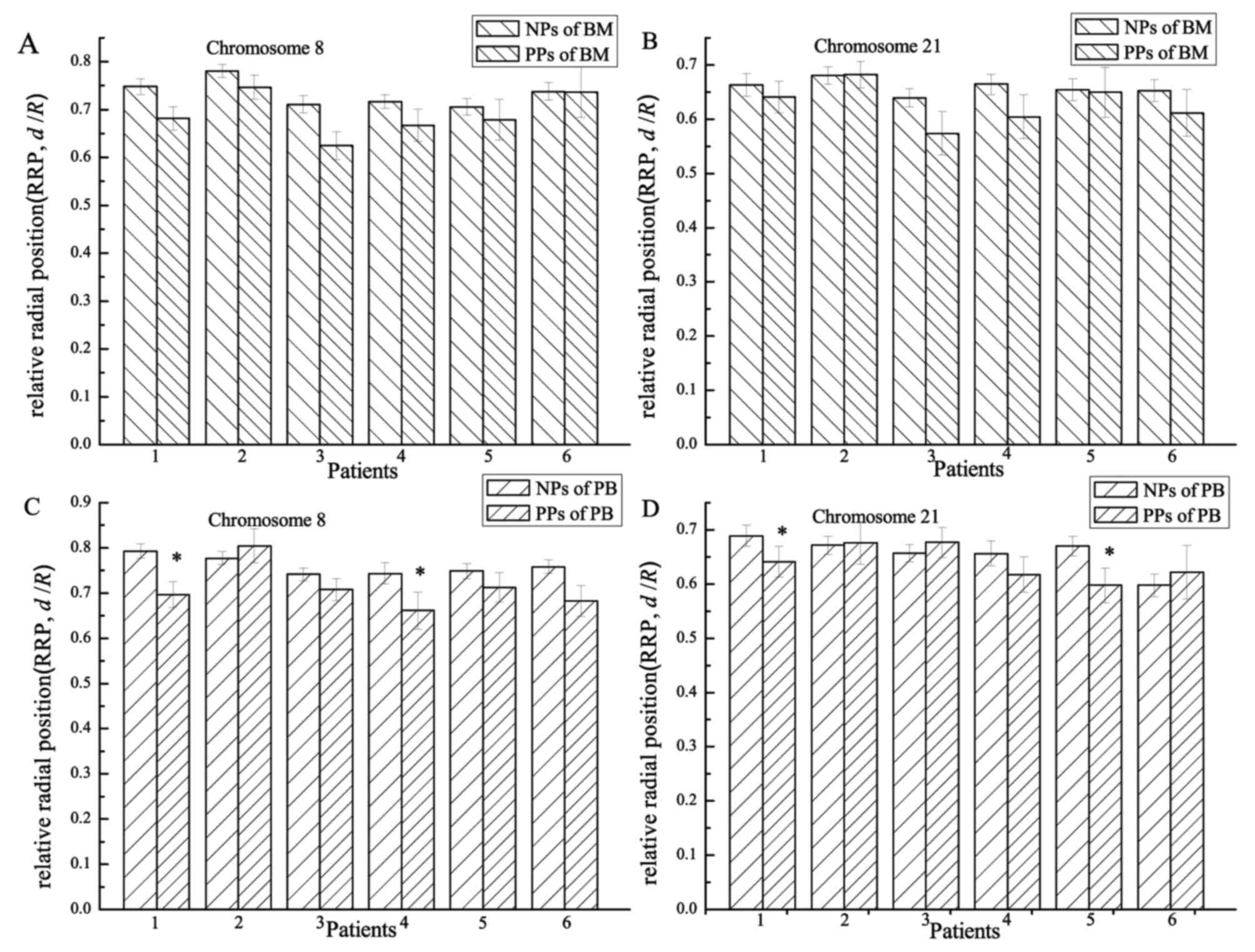
Comparing chromosomal RRPs between NPs
and PPs
Heterogeneous chromosomes were classified into NPs
and PPs according to their relative physical positions and whether
they were touching or intermingling with each other or not. RRPs of
chromosomes 8 and 21 in NPs and PPs for all BM and PB samples (12
in total) were provided (Fig. 4).
Chromosomal RRPs were compared between NPs and PPs using the
two-sample, two-sided K-S test. For 11/12 samples (6/6 for BM and
5/6 for PB), chromosome 8 was more interior (P=0.004–0.664; median,
0.232) in nuclei in PPs compared with that in NPs. For 8/12 samples
(5/6 for BM and 3/6 for PB), chromosome 21 was more interior
(P=0.029–0.923; median, 0.308) in nuclei in PPs compared with that
in NPs.
Discussion
The spatial distribution of chromosomes in
interphase is non-random, and gene-dense chromosomes are more
likely to be located proximate to the center of the nucleus
(15,23). Furthermore, chromosomes spatially
proximate to each other are more prone to translocation compared
with those that are spatially distant (12–15,24).
Therefore, studying the 3D distribution of chromosomes, even if the
patient exhibits a normal karyotype, is essential. The 3D-FISH
technique is useful since it may preserve the chromatin
architecture at the level of the 1 Mb chromatin domains (25,26). In
addition, a 3D view of interphase chromosomes may assist in
detecting complex or alternate translocations or masked runt
related transcription factor 1 (AML1)/RUNX1T1 fusions presenting as
chromosomal breakages in MCs (27–29).
A previous study revealed that MCs accounted for
<20% of all cells from healthy donors or patients in the
remittent stage (16). However, in
the present study, MCs in BM samples from the patient with
AML1/RUNX1T1− AML-M2 represented ~24% of all cells
derived from this patient. All these MCs exhibited chromosomal
breakage (3r or 3g) rather than chromosomal translocation (3r3g).
Therefore, chromosomal breakages may serve a critical function in
disease occurrence. However, 20% of MCs in BM samples does not
represent a universal cut-off for sorting patients into low- or
high-risk relapse groups. For the patient initially diagnosed in
the present study with AML1/RUNX1T1+ AML-M2, the
proportion of MCs in BM samples was only 16%. We previously also
monitored an AML-M2 AML1/RUNX1T1+ patient following
hematopoietic stem cell transplantation and demonstrated a
continuous increase in MC proportion in the patient prior to final
relapse (30). Therefore, the
proportion of MCs may be associated with patient condition, and
should be assessed for variation during follow-up rather than
tested only once.
Fritz et al (31) suggested that pairwise border distance
rather than pairwise center distance may prove preferential for
studying chromosomal interaction. Improving upon our previous study
(16), PPs were defined as
heterogeneous chromosomes touching or intermingling with each other
in the present study, thereby avoiding the complexity of
considering chromosomal size and structural orientation when
calculating the relative distance between the centers of
heterogeneous chromosomes. Previous studies have demonstrated that
PPs were positioned more interiorly in the nuclei than NPs
(12,14,15,24). The
present study compared the RRPs (d/R) of chromosomes
observed in PPs with those only observed in NPs using the
two-sample, two-sided K-S test. In general, chromosomes 8 and 21 in
PPs were more interior in the nuclei compared with those in NPs,
which is consistent with our previous study (16) and indicated that the spatial
organization of PPs in the nucleus was non-random.
Cells were classified into three types: NCs, PCs and
MCs. Occurrences (% of all cells) of the different types of cell in
BM samples were generally consistent with those in PB samples
(P>0.05), which indicated that the PB samples were as
informative as BM samples in disease detection using 3D-FISH and
may represent an alternative source of cells for studies using
3D-FISH. The results of the present study may be implicated in
cases where only PB is available for study and MRD detection due to
decreased BM availability (32).
To conclude, the 3D-FISH technique may be used to
detect the spatial organization of chromosomes in situ and
distinguish the chromosomal morphology of NPs, PPs and chromosomal
translocations or breakages. Quantitative analysis of spatial
chromosomal organization may provide significant information for
clinical diagnosis of acute myeloid leukemia. The spatial
organization of leukemia-specific chromosomes in nuclei were
generally consistent between BM and PB samples in the present
study. Therefore, PB may represent an alternative to BM in disease
follow-up or studies using 3D-FISH.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 11374180).
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NP
|
normal pair
|
|
PP
|
proximal pair
|
|
NC
|
normal cell
|
|
PC
|
proximal cell
|
|
MC
|
malignant cell
|
|
RRP
|
relative radial position
|
References
|
1
|
Grimwade D, Walker H, Oliver F, Wheatley
K, Harrison C, Harrison G, Rees J, Hann I, Stevens R, Burnett A and
Goldstone A: The importance of diagnostic cytogenetics on outcome
in AML: Analysis of 1,612 patients entered into the MRC AML 10
trial. The Medical Research Council Adult And Children's Leukaemia
Working Parties. Blood. 92:2322–2333. 1998.PubMed/NCBI
|
|
2
|
Byrd JC, Mrózek K, Dodge RK, Carroll AJ,
Edwards CG, Arthur DC, Pettenati MJ, Patil SR, Rao KW, Watson MS,
et al: Pretreatment cytogenetic abnormalities are predictive of
induction success, cumulative incidence of relapse, and overall
survival in adult patients with de novo acute myeloid leukemia:
Results from Cancer and Leukemia Group B (CALGB 8461). Blood.
100:4325–4336. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Grimwade D, Hills RK, Moorman AV, Walker
H, Chatters S, Goldstone AH, Wheatley K, Harrison CJ and Burnett
AK: National Cancer Research Institute Adult Leukaemia Working
Group: Refinement of cytogenetic classification in acute myeloid
leukemia: Determination of prognostic significance of rare
recurring chromosomal abnormalities among 5876 younger adult
patients treated in the United Kingdom Medical Research Council
trials. Blood. 116:354–365. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jourdan E, Boissel N, Chevret S, Delabesse
E, Renneville A, Cornillet P, Blanchet O, Cayuela JM, Recher C,
Raffoux E, et al: Prospective evaluation of gene mutations and
minimal residual disease in patients with core binding factor acute
myeloid leukemia. Blood. 121:2213–2223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Inaba H, Coustan-Smith E, Cao X, Pounds
SB, Shurtleff SA, Wang KY, Raimondi SC, Onciu M, Jacobsen J,
Ribeiro RC, et al: Comparative analysis of different approaches to
measure treatment response in acute myeloid leukemia. J Clin Oncol.
30:3625–32. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Coustan-Smith E and Campana D: Should
evaluation for minimal residual disease be routine in acute myeloid
leukemia? Curr Opin Hematol. 20:86–92. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
van der Velden VH, Hochhaus A, Cazzaniga
G, Szczepanski T, Gabert J and van Dongen JJ: Detection of minimal
residual disease in hematologic malignancies by real-time
quantitative PCR: Principles, approaches, and laboratory aspects.
Leukemia. 17:1013–1034. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wheatley K, Burnett AK, Goldstone AH, Gray
RG, Hann IM, Harrison CJ, Rees JK, Stevens RF and Walker H: A
simple, robust, validated and highly predictive index for the
determination of risk-directed therapy in acute myeloid leukaemia
derived from the MRC AML 10 trial. United Kingdom medical research
council's adult and childhood leukaemia working parties british. J
Haematol. 107:69–79. 1999. View Article : Google Scholar
|
|
9
|
Buccisano F, Maurillo L, Gattei V, Del
Poeta G, Del Principe MI, Cox MC, Panetta P, Consalvo MI, Mazzone
C, Neri B, et al: The kinetics of reduction of minimal residual
disease impacts on duration of response and survival of patients
with acute myeloid leukemia. Leukemia. 20:1783–1789. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Maurillo L, Buccisano F, Del Principe MI,
Del Poeta G, Spagnoli A, Panetta P, Ammatuna E, Neri B, Ottaviani
L, Sarlo C, et al: Toward optimization of postremission therapy for
residual disease-positive patients with acute myeloid leukemia. J
Clin Oncol. 26:4944–4951. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Buccisano F, Maurillo L, Del Principe MI,
Del Poeta G, Sconocchia G, Lo-Coco F, Arcese W, Amadori S and
Venditti A: Prognostic and therapeutic implications of minimal
residual disease detection in acute myeloid leukemia. Blood.
119:332–341. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Roix JJ, McQueen PG, Munson PJ, Parada LA
and Misteli T: Spatial proximity of translocation-prone gene loci
in human lymphomas. Nat Genet. 34:287–291. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Holley WR, Mian IS, Park SJ, Rydberg B and
Chatterjee A: A model for interphase chromosomes and evaluation of
radiation-induced aberrations. Radiat Res. 158:568–80. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Martin LD, Harizanova J, Righolt CH, Zhu
G, Mai S, Belch AR and Pilarski LM: Differential nuclear
organization of translocation-prone genes in nonmalignant B cells
from patients with t (14; 16) as compared with t (4; 14) or t (11;
14) myeloma. Genes Chromosomes Cancer. 52:523–537. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Parada LA and Misteli T: Chromosome
positioning in the interphase nucleus. Trends Cell Biol.
12:425–432. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tian X, Wang Y, Zhao F, Liu J, Yin J, Chen
D, Ma W and Ke X: A new classification of interphase nuclei based
on spatial organizations of chromosome 8 and 21 for t(8;21)
(q22;q22) acute myeloid leukemia by three-dimensional fluorescence
in situ hybridization. Leuk Res. 39:1414–1420. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tobal K, Newton J, Macheta M, Chang J,
Morgenstern G, Evans PA, Morgan G, Lucas GS and Yin Liu JA:
Molecular quantitation of minimal residual disease in acute myeloid
leukemia with t(8;21) can identify patients in durable remission
and predict clinical relapse. Blood. 95:815–819. 2000.PubMed/NCBI
|
|
18
|
Corbacioglu A, Scholl C, Schlenk RF, Eiwen
K, Du J, Bullinger L, Fröhling S, Reimer P, Rummel M, Derigs HG, et
al: Prognostic impact of minimal residual disease in
CBFB-MYH11-positive acute myeloid leukemia. J Clin Oncol.
28:3724–3729. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Leroy H, de Botton S, Grardel-Duflos N,
Darre S, Leleu X, Roumier C, Morschhauser F, Lai JL, Bauters F,
Fenaux P and Preudhomme C: Prognostic value of real-time
quantitative PCR (RQ-PCR) in AML with t(8;21). Leukemia.
19:367–372. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Stentoft J, Hokland P, Ostergaard M, Hasle
H and Nyvold CG: Minimal residual core binding factor AMLs by real
time quantitative PCR - Initial response to chemotherapy predicts
event free survival and close monitoring of peripheral blood
unravels the kinetics of relapse. Leuk Res. 30:389–395. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao F, Yang X, Chen D, Ma W, Zheng J and
Zhang X: A method for simultaneously delineating multiple targets
in 3D-FISH using limited channels, lasers, and fluorochromes. Eur
Biophys J. 43:53–58. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Parada LA, Sotiriou S and Misteli T:
Spatial genome organization. Exp Cell Res. 296:64–70. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Branco MR and Pombo A: Intermingling of
chromosome territories in interphase suggests role in
translocations and transcription-dependent associations. PLoS Biol.
4:e1382006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Solovei I, Cavallo A, Schermelleh L,
Jaunin F, Scasselati C, Cmarko D, Cremer C, Fakan S and Cremer T:
Spatial preservation of nuclear chromatin architecture during
three-dimensional fluorescence in situ hybridization (3D-FISH). Exp
Cell Res. 276:10–23. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Manvelyan M, Hunstig F, Mrasek K, Bhatt S,
Pellestor F, Weise A and Liehr T: Position of chromosomes 18, 19,
21 and 22 in 3D-preserved interphase nuclei of human and gorilla
and white hand gibbon. Mol Cytogenet. 1:92008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu S, Li Q, Pang W, Bo L, Qin S, Liu X,
Teng Q, Qian L and Wang J: A new complex variant t(4;15;17) in
acute promyelocytic leukemia: Fluorescence in situ hybridization
confirmation and literature review. Cancer Genet Cytogenet.
130:33–37. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hiorns LR, Min T, Swansbury GJ, Zelent A,
Dyer MJS and Catovsky D: Interstitial insertion of retinoic acid
receptor-alpha gene in acute promyelocytic leukemia with normal
chromosomes 15 and 17. Blood. 83:2946–29451. 1994.PubMed/NCBI
|
|
29
|
Brockman SR, Paternoster SF, Ketterling RP
and Dewald GW: New highly sensitive fluorescence in situ
hybridization method to detect PML/RARA fusion in acute
promyelocytic leukemia. Cancer Genet Cytogenet. 145:144–151. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tian X, Wang Y, Chen D, Ke X and Ma W:
Significance of spatial organization of chromosomes in the
progression of acute myeloid leukemia. Chin J Cancer. 36:402017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fritz AJ, Stojkovic B, Ding H, Xu J,
Bhattacharya S, Gaile D and Berezney R: Wide-scale alterations in
interchromosomal organization in breast cancer cells: Defining a
network of interacting chromosomes. Hum Mol Genet. 23:5133–5146.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Satake N, Maseki N, Kozu T, Sakashita A,
Kobayashi H, Sakurai M, Ohki M and Kaneko Y: Disappearance of
AML1-MTG8(ETO) fusion transcript in acute myloid-leukemia patients
with t(8–21) in long-term remission. Br J Haematol. 91:892–898.
1995. View Article : Google Scholar : PubMed/NCBI
|