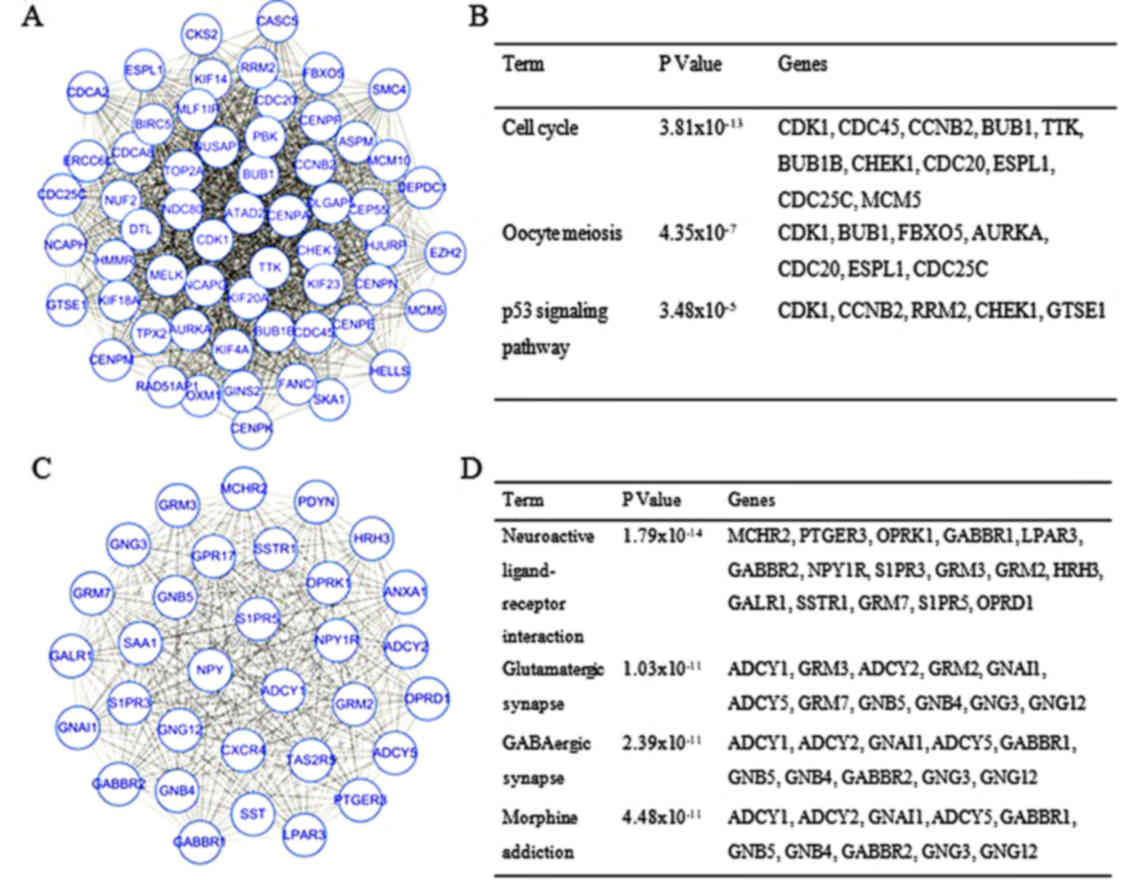
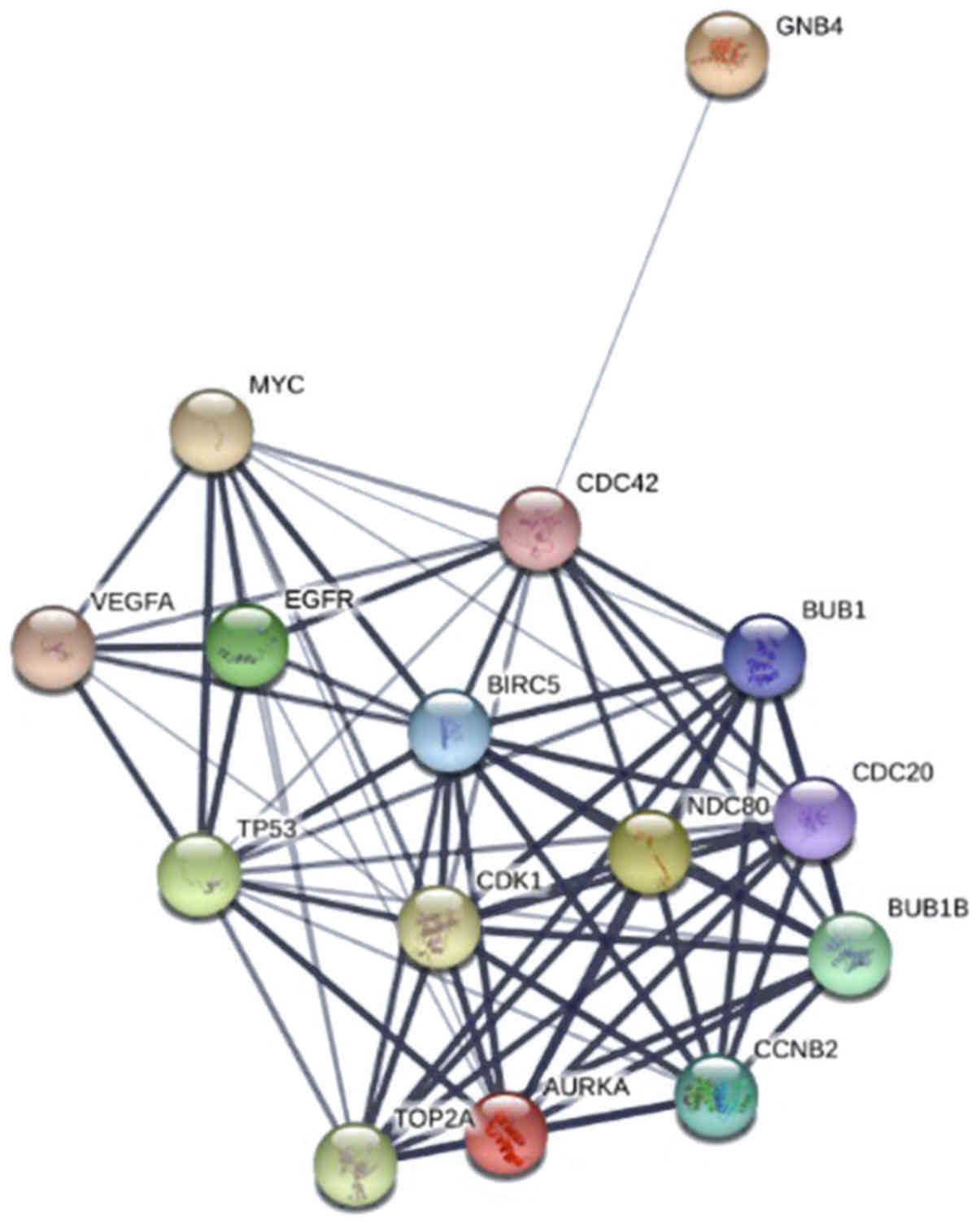
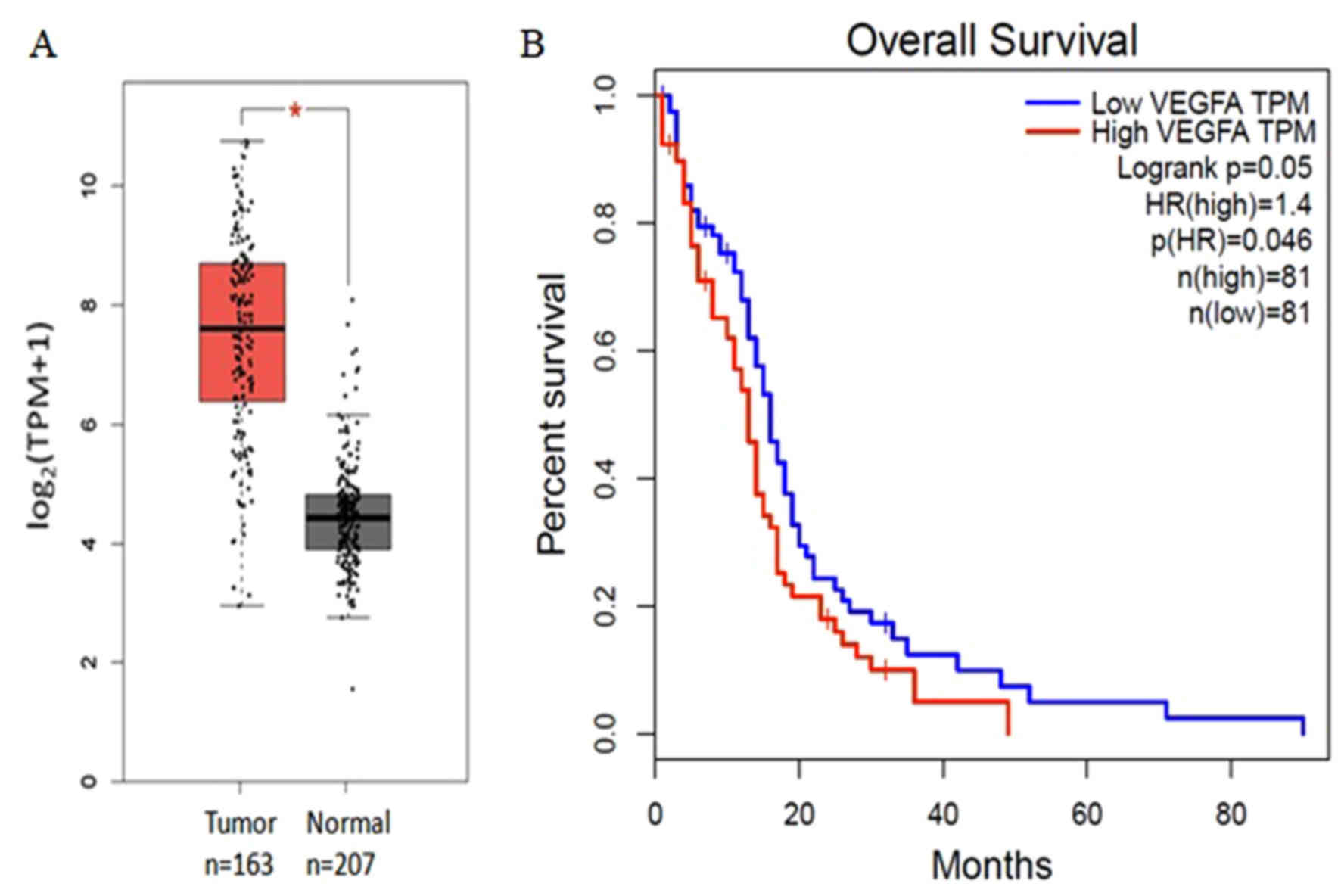
|
1
|
Dolecek TA, Propp JM, Stroup NE and
Kruchko C: CBTRUS statistical report: Primary brain and central
nervous system tumors diagnosed in the United States in 2005-2009.
Neuro Oncol. 14 Suppl 5:v1–v49. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yeom SY, Nam DH and Park C: RRAD promotes
EGFR-mediated STAT3 activation and induces temozolomide resistance
of malignant glioblastoma. Mol Cancer Ther. 13:3049–3061. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Z, Lu J, Guo G, Yang Y, Dong S, Liu
Y, Nan Y, Zhong Y, Yu K and Huang Q: IKBKE promotes glioblastoma
progression by establishing the regulatory feedback loop of
IKBKE/YAP1/miR-Let-7b/i. Tumour Biol. 39:10104283177055752017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kaynar MY, Sanus GZ, Hnimoglu H, Kacira T,
Kemerdere R, Atukeren P, Gumustas K, Canbaz B and Tanriverdi T:
Expression of hypoxia inducible factor-1alpha in tumors of patients
with glioblastoma multiforme and transitional meningioma. J Clin
Neurosci. 15:1036–1042. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Qiu Z, Yuan W, Chen T, Zhou C, Liu C,
Huang Y, Han D and Huang Q: HMGCR positively regulated the growth
and migration of glioblastoma cells. Gene. 576:22–27. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gao X, Mi Y, Ma Y and Jin W: LEF1
regulates glioblastoma cell proliferation, migration, invasion, and
cancer stem-like cell self-renewal. Tumour Biol. 35:11505–11511.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun L, Hui AM, Su Q, Vortmeyer A,
Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey
R, et al: Neuronal and glioma-derived stem cell factor induces
angiogenesis within the brain. Cancer Cell. 9:287–300. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Davis S and Meltzer PS: GEOquery: A bridge
between the gene expression omnibus (GEO) and BioConductor.
Bioinformatics. 23:1846–1847. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Deng L, Xiong P, Luo Y, Bu X, Qian S and
Zhong W: Bioinformatics analysis of the molecular mechanism of
diffuse intrinsic pontine glioma. Oncol Lett. 12:2524–2530. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma K, Cheng Z, Sun L and Li H:
Identification of potential therapeutic targets for gliomas by
bioinformatics analysis. Oncol Lett. 14:5203–5210. 2017.PubMed/NCBI
|
|
15
|
Sun C, Yuan Q, Wu D, Meng X and Wang B:
Identification of core genes and outcome in gastric cancer using
bioinformatics analysis. Oncotarget. 8:70271–70280. 2017.PubMed/NCBI
|
|
16
|
Jiao X, Sherman BT, Huang da W, Stephens
R, Baseler MW, Lane HC and Lempicki RA: DAVID-WS: A stateful web
service to facilitate gene/protein list analysis. Bioinformatics.
28:1805–1806. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:(Database Issue). D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Behin A, Hoang-Xuan K, Carpentier AF and
Delattre JY: Primary brain tumours in adults. Lancet. 361:323–331.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fueyo J, Gomez-Manzano C, Alemany R, Lee
PS, McDonnell TJ, Mitlianga P, Shi YX, Levin VA, Yung WK and
Kyritsis AP: A mutant oncolytic adenovirus targeting the Rb pathway
produces anti-glioma effect in vivo. Oncogene. 19:2–12. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fueyo J, Gomez-Manzano C, Liu TJ and Yung
WK: Delivery of cell cycle genes to block astrocytoma growth. J
Neurooncol. 51:277–287. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Alexiou GA, Vartholomatos G, Goussia A,
Batistatou A, Tsamis K, Voulgaris S and Kyritsis AP: Fast cell
cycle analysis for intraoperative characterization of brain tumor
margins and malignancy. J Clin Neurosci. 22:129–132. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu LJ, Wall BA, Wangari-Talbot J and Chen
S: Metabotropic glutamate receptors in cancer. Neuropharmacology.
115:193–202. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Y, Jiang M, Yao Y and Cai Z: WWC3
inhibits glioma cell proliferation through suppressing the
Wnt/β-catenin signaling pathway. DNA Cell Biol. 37:31–37. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ulrich TA, de Juan Pardo EM and Kumar S:
The mechanical rigidity of the extracellular matrix regulates the
structure, motility, and proliferation of glioma cells. Cancer Res.
69:4167–4174. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bellail AC, Hunter SB, Brat DJ, Tan C and
Van Meir EG: Microregional extracellular matrix heterogeneity in
brain modulates glioma cell invasion. Int J Biochem Cell Biol.
36:1046–1069. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cantrell DA: Phosphoinositide 3-kinase
signalling pathways. J Cell Sci. 114:1439–1445. 2001.PubMed/NCBI
|
|
28
|
Ashcroft M, Ludwig RL, Woods DB, Copeland
TD, Weber HO, Macrae EJ and Vousden KH: Phosphorylation of HDM2 by
Akt. Oncogene. 21:1955–1962. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vassilev LT, Vu BT, Graves B, Carvajal D,
Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et
al: In vivo activation of the p53 pathway by small-molecule
antagonists of MDM2. Science. 303:844–848. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bond GL, Hu W, Bond EE, Robins H, Lutzker
SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, et al: A
single nucleotide polymorphism in the MDM2 promoter attenuates the
p53 tumor suppressor pathway and accelerates tumor formation in
humans. Cell. 119:591–602. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kang SS, Han KS, Ku BM, Lee YK, Hong J,
Shin HY, Almonte AG, Woo DH, Brat DJ, Hwang EM, et al:
Caffeine-mediated inhibition of calcium release channel inositol
1,4,5-trisphosphate receptor subtype 3 blocks glioblastoma invasion
and extends survival. Cancer Res. 70:1173–1183. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pal J, Patil V, Kumar A, Kaur K, Sarkar C
and Somasundaram K: Loss-of-function mutations in Calcitonin
receptor (CALCR) identify highly aggressive glioblastoma with poor
outcome. Clin Cancer Res. 24:1448–1458. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wong ML, Prawira A, Kaye AH and Hovens CM:
Tumour angiogenesis: Its mechanism and therapeutic implications in
malignant gliomas. J Clin Neurosci. 16:1119–1130. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang SB, Gao KD, Jiang T, Cheng SJ and Li
WB: Bevacizumab combined with chemotherapy for glioblastoma: A
meta-analysis of randomized controlled trials. Oncotarget.
8:57337–57344. 2017.PubMed/NCBI
|
|
35
|
Paruthiyil S, Cvoro A, Tagliaferri M,
Cohen I, Shtivelman E and Leitman DC: Estrogen receptor β causes a
G2 cell cycle arrest by inhibiting CDK1 activity through the
regulation of cyclin B1, GADD45A, and BTG2. Breast Cancer Res
Treat. 129:777–784. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fan X and Chen JJ: Role of Cdk1 in DNA
damage-induced G1 checkpoint abrogation by the human papillomavirus
E7 oncogene. Cell Cycle. 13:3249–3259. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang J, Zhou F, Li Y, Li Q, Wu Z, Yu L,
Yuan F, Liu J, Tian Y, Cao Y, et al: Cdc20 overexpression is
involved in temozolomide-resistant glioma cells with
epithelial-mesenchymal transition. Cell Cycle. 16:2355–2365. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xia Z, Wei P, Zhang H, Ding Z, Yang L,
Huang Z and Zhang N: AURKA governs self-renewal capacity in
glioma-initiating cells via stabilization/activation of
β-catenin/Wnt signaling. Mol Cancer Res. 11:1101–1111. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang R, Wei J, Li Z, Tian Y and Du C:
Bioinformatical analysis of gene expression signatures of different
glioma subtypes. Oncol Lett. 15:2807–2814. 2018.PubMed/NCBI
|
|
40
|
Hu G, Wei B, Wang L, Wang L, Kong D, Jin Y
and Sun Z: Analysis of gene expression profiles associated with
glioma progression. Mol Med Rep. 12:1884–1890. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li W, Li K, Zhao L and Zou H:
Bioinformatics analysis reveals disturbance mechanism of MAPK
signaling pathway and cell cycle in Glioblastoma multiforme. Gene.
547:346–350. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wei B, Wang L, Du C, Hu G, Wang L, Jin Y
and Kong D: Identification of differentially expressed genes
regulated by transcription factors in glioblastomas by
bioinformatics analysis. Mol Med Report. 11:2548–2554. 2015.
View Article : Google Scholar
|