Introduction
Cancer is the second leading cause of death globally
(1). In China, the numbers of newly
diagnosed cases and deaths were approximately 3.0 million and 1.9
million, respectively, in 2010 (2).
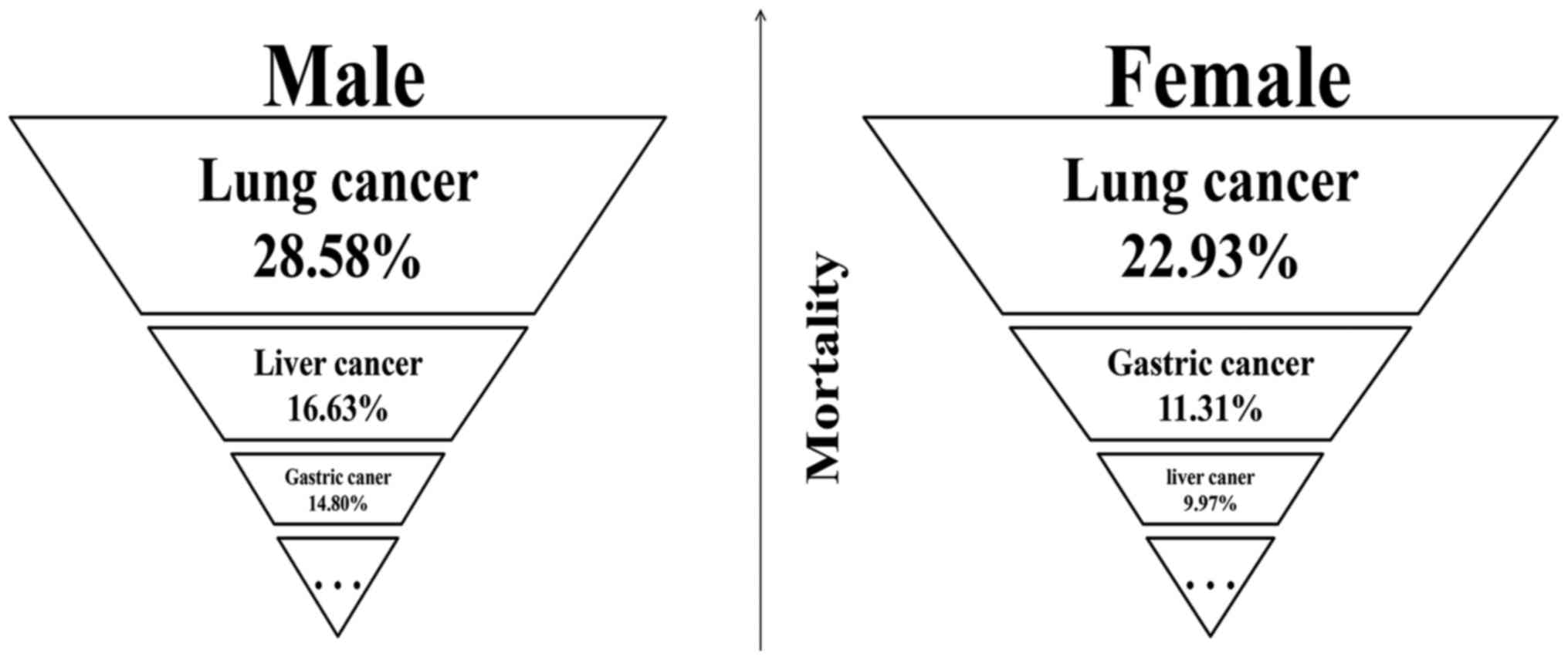
According to 2013 data, lung cancer, liver cancer and gastric
cancer are the top three leading causes of mortality in males in
China, whereas lung cancer, gastric cancer and liver cancer are the
top three leading causes of mortality in females (3) (Fig.
1).
A growing number of studies have focused on the
biology, function and clinical implications of exosomes in cancers
(4,5),
and it has been demonstrated that exosomal miRNAs and proteins can
act as tumor biomarkers for clinical diagnosis or prognosis and
that exosomes shuttle between cells to exchange genetic material,
which promotes tumor progression, metastasis and prognosis;
regulates the immune response; and affects the sensitivity of tumor
cells to chemotherapy drugs (6–8).
Therefore, exosomal miRNAs and proteins potentially play critical
roles in cancers with high mortality rates.
Exosome composition
Exosomes are extracellular vesicles (EVs) that are
produced and released by many different cells; and these vesicles
range in size from 30 to 100 nm in diameter and contain a lipid
bilayer (9,10). Proteins, DNA, mRNAs, miRNAs and lipids
are enriched in exosomes (11).
Exosomes transfer nucleic acids and proteins between different
cells, leading to both the transportation of materials and
cell-cell communication (6,12,13).
A set of distinct proteins are contained in exosomes
(14), including heat-shock proteins
(Hsp70, Hsp90), tetraspanins (CD9, CD81), ESCRT-related proteins
(Alix, Tsg101), cytoskeletal proteins (actin, Tubulin) and GTPases
(EEF1A1, EEF2) (15,16). These proteins are known to be involved
in biogenesis, the sorting and secretion of exosomes (17), antigen presentation, the organization
of membrane microdomains, the cytoskeleton, and the endosomal
system (18,19). Typically, exosomes contain both
cell-type specific proteins and proteins that are expressed in
various cell types (20).
In addition to proteins, exosomes contain a
significant amount of nucleic acids, including DNA, mRNAs, miRNAs,
circular RNAs (circRNAs) and long noncoding RNAs (lncRNAs)
(21). Of these, miRNAs are a class
of well-known regulatory molecules that control posttranscriptional
gene regulation (22). Increasing
evidence has shown that exosomal miRNAs are associated with many
diseases, such as cancers, diabetes and obesity (23–26).
Interestingly, the miRNA content of exosomes is similar to that of
the original tumor; thus, a series of studies has focused on
exosomal miRNA profiles for cancer diagnostics (16). In particular, the shuttling of miRNAs
may act as a tumor promoter or a tumor suppressor during
tumorigenesis (27). Previous studies
have uncovered exosomal miRNAs that are closely associated with
tumorigenesis, metastasis and drug resistance in various kinds of
cancers (28,29). All of these findings suggest that
exosomal miRNAs play a pivotal role in the diagnosis, treatment and
prognosis of cancers (30,31).
Additionally, cholesterols, diglycerides,
phospholipids, glycerophospholipids, sphingomyelins and ceramides
are enriched in exosomes (32). These
lipids participate in exosome biogenesis, function and release. For
example, the cellular trafficking of the tetraspanin CD82 to
endosomes is regulated by the cholesterol content of the membrane,
and ceramides can protect miRNAs from degradation by circulating
RNases and govern the cellular distribution of the tetraspanin
CD81. In addition, bioactive lipids such as prostaglandins,
leukotrienes, fatty acids and lipid-related enzymes such as
phospholipases A2 have been detected in exosomes (33).
Exosome isolation
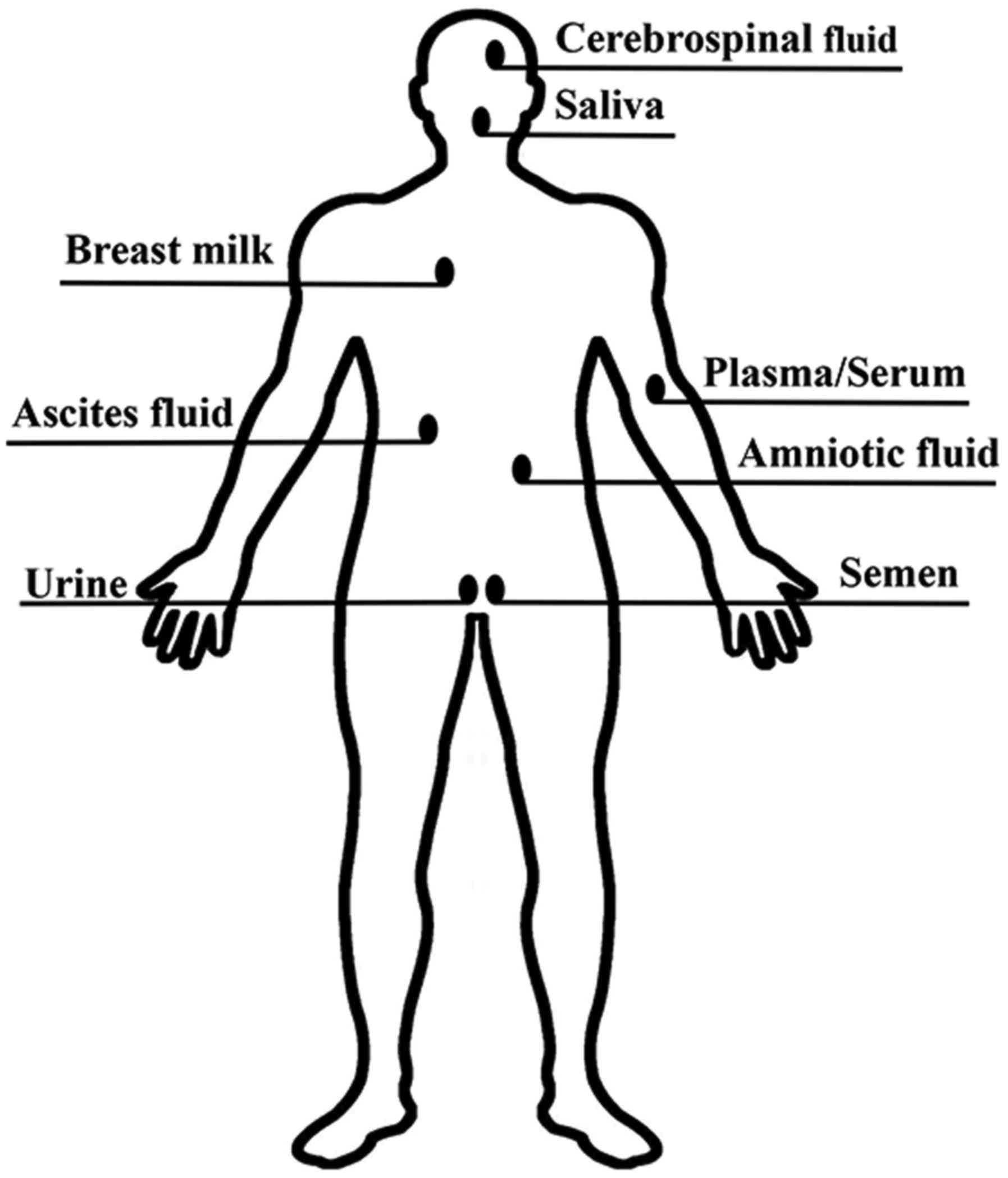
Exosomes secreted by various types of living cells
have been detected in a diverse range of bodily fluids, including
peripheral blood, saliva, cerebrospinal fluid, ascites fluid,
amniotic fluid, urine, breast milk and semen (31,34)
(Fig. 2). It is clear that the
utility of exosomes goes beyond basic research and extends to
clinical practice. For this reason, an efficient and accurate
method for exosome isolation is crucial.
Here, we compare the common methods for exosome
isolation (Table I), including
ultracentrifugation (UC), ultrafiltration (UF), immunomagnetic
beads, size exclusion chromatography (SEC) and ExoQuick™
(35,36). UC is a common and simple method
(37); however, recent studies
indicated that more contaminants were found in exosomes isolated by
UC compared to other methods mainly due to the presence of
albumins. Furthermore, the high-velocity ultracentrifugation
process could cause some exosomes to rupture, resulting in exosome
loss (38). Recently, the challenges
of UC approach have been again discussed, the conventional
biophysical UC cannot distinguish exosomes from lipoproteins and
oncosomes, other types of small EVs with sedimentation velocities
and gradient densities similar to those of exosomes (39). UF does not require special equipment,
although it leads to a reduction in the membranes' lifespan and a
low isolation efficiency (35,40). The
use of immunomagnetic beads is an alternative method with high
specificity and purity, but it is limited to exosomes with a known
antigen and has a high reagent cost (35). Although SEC does not lead to
significant albumin contamination, the efficiency is low (35,37,41).
ExoQuick™ produces excellent reproducibility and
sensitivity. However, the proprietary reagents exhibit
contamination from unknown sources, and the polymer leads to
protein aggregation (35,36,42,43).
Moreover, the ExoQuick™ kit does not specifically
precipitate exosomes, which means that other types of nanovesicles
with similar sizes (30–100 nm) might also be coprecipitated
(39). Recently, a new technique
developed by the microfluidics community has been used to approach
some of the problems with exosome isolation mentioned above. The
most important feature of this method is exosome enrichment during
isolation, which is beneficial for the detection of early-stage
cancers. This microfluidics approach showed a superior recovery of
60–80% compared to the conventional techniques of UC (6%) and
ExoQuick™ (30%) based on nanoparticle tracking analysis
(NTA) (43).
 | Table I.Comparison of exosome isolation
methods. |
Table I.
Comparison of exosome isolation
methods.
| Author, year | Method | Principle | Advantages | Disadvantages | (Refs.) |
|---|
| Baranyai et
al, 2015; Peterson et al, 2015 | UC | Separating the
exosomes through differential mass, density and shape | • Available
technology | • The high velocity
ultracentrifugation process could cause some exosomes rupture that
results in some exosomes loss | (37,38) |
|
|
|
| • Simple
operation |
|
|
|
|
|
|
| • Contaminated with
albumin and IgG |
|
|
|
|
|
| • Time consuming
(16–20 h) |
|
| Li et al,
2017; Zeringer et al, 2015 | UF | Depending on
exosomal size or molecular weight | • No need of
special equipment | • Clogging and
vesicle trapping lead to reduce the membranes' lifetime and low
isolation efficiency | (35,40) |
|
|
|
| • Good
portability |
|
|
| Li et al,
2017 | Immunom-agnetic
beads | Specific exosomal
antigens (receptors) can be captured by magnetic beads
(ligands) | • High specificity
and purity | • High reagent
cost | (35) |
|
|
|
|
| • Low yield |
|
|
|
|
| • No damage on the
integrity of the exosomes' morphology and structure |
|
|
| Li et al,
2017; Baranyai et al, 2015; Taylor and Shah, 2015 | SEC | A porous stationary
phase is utilized to sort exosomes out according to the size | • Obtaining
high-purity exosomes without significant albumin contamination | • Require dedicated
equipment | (35,37,41) |
|
|
|
|
| • Low
efficiency |
|
|
|
|
| • Excellent
reproducibility and sensitivity |
|
|
| Li et al,
2017; Caradec et al, 2014; Ban et al, 2015 | ExoQuick™ | By the
precipitation approach | • Efficient (around
100%) and reproducible | • Isolation
procedure should be under acidic conditions (pH=4) | (35,36,42) |
|
|
|
| • Decreasing
albumin contamination | • Polymer
precipitates protein aggregation |
|
|
|
|
| • Fast (within 30
min) |
|
|
Indeed, the high quantity and purity of exosomes are
extremely important for exosomal biology studies. Thus, western
blotting should be used to determine whether exosomal protein
markers (Alix, Tsg101, Hsp70 or others) are present in exosome
isolations (44). Simultaneously,
transmission electron microscopy (TEM) is often utilized to observe
exosome morphology, NTA is used to measure particle size, and the
bicinchoninic acid assay (BCA) is performed to examine the protein
concentration of exosomes (45).
Additionally, to ensure the sensitivity of isolations and achieve a
robust result, pre-analytical factors should be taken into
consideration (Table II) (46,47).
 | Table II.Pre-analytical considerations. |
Table II.
Pre-analytical considerations.
| Author, year | Pre-analytical
considerations | (Refs.) |
|---|
| Muller et
al, 2014; Witwer et al, 2013 | Venous blood from
patients or healthy volunteers is collected into tubes without
heparin-based anticoagulants, EDTA may be more appropriate. | (46,47) |
| Witwer et
al, 2013 | Blood should be
processed quickly at room temperature. | (47) |
| Witwer et
al, 2013 | Collected blood
should be handled gently and tubes should be vertically positioned
prior to centrifugation. | (47) |
| Witwer et
al, 2013 | Both plasma and
serum can be used, but most studies indicate the isolation of
exosomes prefers to plasma. | (47) |
| Muller et
al, 2014 | Harvested plasma or
serum should be immediately used or stored at −80°C. | (46) |
Exosomal miRNAs and proteins in lung
cancer
The latest report showed that lung cancer caused
approximately 597,000 deaths in China in 2013 (3). Of lung cancer cases, approximately 95%
are non-small-cell lung cancer (NSCLC) and small-cell lung cancer
(SCLC) (48), which together
represent the most common cause of cancer-related death globally
(49,50).
Serving as biomarkers
Exosomes and exosomal miRNAs differed between
patients with lung cancer and controls (51). By comparing 12 specific tumor- and
exosome-derived miRNAs (miR-17-3p, miR-21, miR-106a, miR-146,
miR-155, miR-191, miR-192, miR-203, miR-205, miR-210, miR-212, and
miR-214) in lung cancer, previous studies revealed that there was
no significant difference between circulating miRNAs and tumor
miRNAs, demonstrating that exosome-derived miRNAs can serve as
diagnostic biomarkers for lung cancer (51). In a nude mouse model of subcutaneous
primary and recurrent lung cancer xenografts in vivo, miR-21
and miR-155 were found to be up-regulated in serum exosomes derived
from recurrent tumor-bearing nude mice compared to nontumor- or
primary tumor-bearing nude mice (52), suggesting that these two miRNAs might
be potential prognostic biomarkers for noninvasive diagnosis of
recurrent lung cancer. In addition, Liu et al (53) first reported that elevation of plasma
exosomal miR-23b-3p, miR-10b-5p and miR-21-5p predicted a
significantly poor survival, implying that these three exosomal
miRNAs could serve as independent prognostic biomarkers for
NSCLC.
Exosomal membrane-bound proteins, for example, the
epidermal growth factor receptor (EGFR), NY-ESO-1 and CD91, are
also promising diagnostic or prognostic biomarker candidates for
lung cancer. Yamashita et al (54) demonstrated that the measurement of
plasma exosomal proteins might be helpful for in vitro
diagnosis, and exosomal EGFR was a potential diagnostic biomarker
for the characterization of lung cancer. In NSCLC patients,
exosomal NY-ESO-1 was a strong prognostic biomarker of poorer
survival (55). CD91 expression was
significantly increased in serum exosomes derived from patients
with lung adenocarcinoma (ADC), and its detection power for
early-stage patients was higher than that of carcinoembryonic
antigen (CEA) (56).
Stimulating angiogenesis and inducing
metastasis
Angiogenesis is essential for tumor growth,
progression and metastasis (57). Liu
et al (58) found that
exosomal miR-21 derived from cigarette smoke extract
(CSE)-transformed human bronchial epithelial (HBE) cells was
elevated, and this increased exosomal miR-21 led to STAT3
activation and altered the vascular endothelial growth factor
(VEGF) expression of recipient cells, promoting CSE-induced
angiogenesis and the malignant transformation of HBE cells. These
results provided a novel intervention strategy to prevent
carcinogenesis of lung cancer. In addition, hypoxic lung cancer
cell (hypoxic CL1-5)-derived exosomal miR-23a enhanced
neovascularization and tumor growth, and serum exosomal miR-23a was
also elevated in patients with lung cancer. These findings provided
strong evidence that an increase in exosomal miR-23a contributes to
angiogenesis, intravasation and extravasation in lung cancer
(59).
Exosomes play a fundamental role in the
premetastatic niche and metastasis (4). Results from Fabbri et al
(60) indicated that miRNAs
(miR-21/29a) derived from lung cancer cell line (A549 and SK-MES)
exosomes activate members of the Toll-like receptor (TLR) family
(murine TLR7 and human TLR8) in immune cells, leading to a
TLR-mediated prometastatic inflammatory response that might
ultimately trigger tumor growth and metastasis.
Mediating cisplatin (DDP)
resistance
Lung cancer cell-derived exosomes could confer DDP
resistance to other cancer cells. Qin et al (61) established A549 cells that were
resistant to DDP (A549/DDP). Compared with A549 exosomes,
miR-100-5p was downregulated by 75% in A549/DDP cell exosomes.
Lower expression of miR-100-5p induced DDP resistance in recipient
cells (other lung cancer cell lines). miR-100-5p negatively
regulated mTOR, the mammalian target of rapamycin, to alter the
recipient lung cancer cells' resistance to DDP. Additionally, the
chemosensitivity of NSCLC to DDP could be regulated by serum
exosomal miR-146a-5p. The overexpression of miR-146a-5p reversed
the resistance of A549/DDP cells by targeting Atg12 to inhibit
autophagy (62). Furthermore, in a
human bronchial epithelial cell (HBEC) model, exosomes derived from
chemoresistant mesenchymal NSCLC cells were able to transfer
chemoresistance and mesenchymal phenotypes to recipient cells,
thereby enhancing resistance to gemcitabine and
cisplatin/gemcitabine combination therapy (63).
Exosomal miRNAs and proteins in liver
cancer
Liver cancer is a common malignancy with a high
mortality rate both in China and around the world (64,65). Liver
cancer includes primary liver cancer (PLC) and secondary liver
cancer. Hepatocellular carcinoma (HCC) and intrahepatic
cholangiocarcinoma (ICC) are two different histologic types of PLC,
which is the second most common cause of cancer-related deaths
worldwide (66).
Serving as biomarkers
Differential expression of exosomal miRNAs in serum
could serve as a diagnostic biomarker for HCC. Sohn et al
(67) reported that the levels of
serum exosomal miR-18a, miR-221, miR-222 and miR-224 were
remarkably higher in HCC patients compared with patients with liver
cirrhosis (LC) or chronic hepatitis B (CHB); however, the levels of
serum exosomal miR-101, miR-106b, miR-122 and miR-195 were lower in
HCC patients than in CHB patients. In addition, other studies have
shown that expression of exosomal miR-21 and miR-125b was
upregulated in HCC patients compared with CHB patients or healthy
controls. More importantly, the levels of miR-21 and miR-125b were
higher in exosomes than in serum samples (68,69).
Promoting proliferation, invasion and
metastasis
Exosomal miRNAs could affect cellular gene
expression and cellular behaviors in target cells (70). Wei et al (71) showed that exosomes derived from HCC
cells (SMMC-7721, Hep3B, and Huh-7) could functionally deliver
miRNAs to target cells and that Vps4A regulated the secretion and
uptake of these miRNAs in hepatoma cells by utilizing exosomes as
mediators. Vps4A-associated miRNAs are believed to regulate the
PI3K/AKT signaling pathway and promote the proliferation, invasion
and metastasis of HCC cells. It has been suggested that a large
number of protumorigenic RNAs and proteins, such as the MET
proto-oncogene, caveolins (CAV1, CAV2) and an S100 family member
(S100A4), are enriched in metastatic HCC-derived exosomes (72–74).
Moreover, He et al (75)
showed that uptake of these shuttling molecules in exosomes derived
from motile HCC cell lines (HKCI-C3, HKCI-8 and MHCC97 L) markedly
enhanced the invasive and migratory abilities of nonmotile
immortalized hepatocyte (MIHA) cell lines by activating the
PI3K/AKT and MAPK signaling pathways and increasing the secretion
of matrix metalloproteinases (MMP)-2 and MMP-9, which induced cell
invasion.
Mediating sensitivity to
sorafenib
Sorafenib is predominantly used for the treatment of
liver cancer and can improve the overall survival of patients with
advanced HCC (76). Exosomes may
mediate sorafenib resistance in HCC cells. Guo et al
(77) revealed that miR-122 contained
in adipose tissue mesenchymal stem cell (AMSC) exosomes enhanced
HCC cell sensitivity to chemotherapeutic agents. Compared with the
control groups, the inhibitory effect of 5-fluorouracil (5-FU) or
sorafenib on HCC cells (HepG2 and Huh7) treated with AMSC-derived
exosomes (122-Exo) was significantly enhanced, thereby providing a
new strategy for HCC therapy. An important mechanism of sorafenib
resistance is the overexpression of c-Met, a proto-oncogene that
serves as a receptor for hepatocyte growth factor (HGF) in tumor
cells (78). Further investigations
confirmed that HGF upregulation and c-Met/AKT pathway activation
triggered sorafenib resistance induced by exosomes derived from HCC
cells (MHCC-97L and MHCC-97H), indicating that HGF/c-Met might be a
possible target for decreasing sorafenib resistance of HCC cells
(79).
Exosomal miRNAs and proteins in gastric
cancer
Gastric cancer (GC), a malignant tumor of the
digestive system, is the second leading cause of cancer-related
death and the fourth most common cancer worldwide (80). Although its incidence and mortality
have appreciably decreased globally over recent decades, the
mortality of GC is still relatively high in Asia (81).
Serving as biomarkers
Recent research suggested that serum exosomal
miR-19b-3p and miR-106a-5 could be potential biomarkers for the
early diagnosis of GC (82).
Additionally, Tokuhisa et al (83) assessed exosomal miRNA profiles in
peritoneal fluid and found that miR-21 and miR-1225-5p might be
prognostic biomarkers for peritoneal recurrence after curative GC
resection. miR-10b-5p, miR-195-5p, miR-20a-3p and miR-296-5p were
significantly upregulated in serum exosomes derived from patients
with GC and were able to discriminate GC patients from healthy
controls (84).
Promoting metastasis
miR-214, miR-221 and miR-222 are commonly
upregulated in gastric cancer tissue-derived mesenchymal stem cells
(GC-MSCs) and tumor tissues; moreover, GC-MSC-derived exosomes
deliver miR-221 to HGC-27 cells and promote the proliferation and
migration (85). The serum exosomes
of GC patients transport EGFR to liver cells, and EGFR activates
HGF by suppressing miR-26a/b, stimulating the development of a
liver-like microenvironment that promotes gastric cancer liver
metastasis (86). In later studies,
proliferation and Matrigel invasion of gastric cancer cells in the
presence of exosomes derived from gastric cancer cells (SGC-7901)
with either high (SGC/wt) or low (SGC/kd) CD97 expression were
investigated, and the results indicated that CD97 promoted gastric
cancer cell proliferation and invasion through exosome-mediated
activation of the MAPK signaling pathway (87,88).
Regulating the immune response
Compared with exosomes derived from the untreated
malignant ascites of GC patients, exosomes derived from
heat-treated malignant ascites contained higher concentrations of
the heat shock proteins Hsp70 and Hsp60, which might play an
important role in inducing a tumor-specific cytotoxic T lymphocyte
(CTL) response in vitro and are involved in the promotion of
dendritic cell (DC) maturation (89).
Additionally, HSPs have been identified as damage-associated
molecular patterns (DAMPs), a class of self-danger signals released
by stressed cells that elicited immune responses. Mechanistically,
HSPs respond to the innate immune system both directly with
inflammation and indirectly by recruiting reinforcements (90). However, there is some evidence showing
that HSPs have a dampening effect on the immune system under
physiological conditions, indicating that HSPs are actually
DAMPERs, a class of molecules that reduces the activity of the
innate immune system (91).
Mediating DDP resistance
The level of miR-21 in exosomes derived from
tumor-associated macrophages (M2 macrophages) has been shown to be
increased, and exosomal miR-21 can be directly transferred from
tumor-associated macrophages to gastric cancer cells, conferring
DDP resistance to gastric cancer cells by downregulating PTEN and
activating signaling through the PI3K/AKT pathway (92). However, exosome-delivered anti-miR-214
was able to reverse the resistance of gastric cancer cells to DDP,
leading to suppressed migration in vitro, inhibited tumor
growth in vivo, and increased cellular apoptosis (93). Additionally, MSC-derived exosomes
significantly induced gastric cancer cell resistance to 5-FU both
in vivo and ex vivo by activating the
calmodulin-dependent protein kinase (CaM-K)/Raf/MEK/ERK pathway
(94).
Conclusion and future studies
Exosomes have established a role in cancer biology,
immunology, drug sensitivity and clinical diagnosis. In particular,
exosomal miRNAs and proteins play important roles in cancers with
high mortality rates (lung cancer, liver cancer and gastric cancer)
(Tables III and IV).
 | Table III.Exosomal miRNAs in the top three
mortality cancer types. |
Table III.
Exosomal miRNAs in the top three
mortality cancer types.
| A, Lung cancer |
|---|
|
|---|
| Author, year | miRNAs | Study design | Sample | Clinical
significance | Approach | Performance | (Refs.) |
|---|
| Rabinowits et
al, 2009 |
miR-17-p/21/106a/146/155/191/192/203/205/210/212/214 | Case-control | Human plasma | Diagnostic
biomarkers for NSCLC | Microarray | Increase | (51) |
| Munagala et
al, 2016 | miR-21/155 | Animal model Cell
model | Athymic nude mice
H1299, Beas-2b | Possible prognostic
markers for lung cancer recurrence | Microarray,
qPCR | Increase | (52) |
| Liu et al,
2017 |
miR-23b-3p/10b-5p/21-5p | Case-control | Human plasma | Independent
non-invasive prognostic markers for NSCLC | qPCR | Increase | (53) |
| Liu et al,
2016 | miR-21 | Patients Cell
model | Human serum
CSE-transformed HBE cells | Promoting
CSE-induced angiogenesis and malignant transformation of HBE
cells | qPCR | Increase | (58) |
| Hsu et al,
2017 | miR-23a | Patients Cell
model | Human serum Hypoxic
CL1-5 | Stimulating the
angiogenesis, intrava-sation and extravasation in lung cancer | qPCR | Increase | (59) |
| Fabbri et
al, 2012 | miR-21/29a | Cell model, Animal
model | A549, SK-MES WT B6
mice B6 TLR7−/−mice | Triggering tumour
growth and metastasis | qPCR | Increase | (60) |
| Qin et al,
2017 | miR-100-5p | Cell model | A549/DDP | Altering the
recipient lung cancer cells' resistance to DDP | Microarray,
qPCR | Decrease | (61) |
| Yuwen et al,
2017 | miR-146a-5p | Patients Cell
model | Human serum
A549/DDP | Reversing the
resistance of A549/DDP | qPCR | Increase | (62) |
|
| B, Liver
cancer |
|
| Author,
year | miRNAs | Study
design | Sample | Clinical
significance |
Approach |
Performance | (Refs.) |
|
| Sohn et al,
2015 |
miR-18a/221/222/224 | Case-control | Human serum | Discriminating HCC
from LC or CHB | qPCR | Increase | (67) |
| Sohn et al,
2015 |
miR-101/106b/122/195 | Case-control | Human serum | Discriminating HCC
from CHB | qPCR | Decrease | (67) |
| Wang et al,
2014; Liu et al, 2017 | miR-21/125b | Case-control | Human serum | Discriminating HCC
from CHB or healthy controls | qPCR | Increase | (68,69) |
| Wei et al,
2015 | Vps4A-related
miRNAs | Cell model | SMMC-7721, Hep3B,
Huh-7 | Regulating PI3K/AKT
signaling pathway and promoting proliferation, invasion and
metastasis of HCC cells | RNA sequencing | Increase | (71) |
| Lou et al,
2015 | miR-122 | Cell model | AMSC | Enhancing the
effect 5-FU or sorafenib on HCC cells | qPCR | Increase | (77) |
|
| C, Gastric
cancer |
|
| Author,
year | miRNAs | Study
design | Sample | Clinical
significance |
Approach |
Performance | (Refs.) |
|
| Wang et al,
2017 |
miR-19b-3p/106a-5 | Case-control | Human serum | Potential
biomarkers for the early diagnosis of GC | qPCR | Increase | (82) |
| Tokuhisa, et
al, 2015 | miR-21/1225-5p | Patients Cell
model | Peritoneum lavage
fluid, OCUM-2M OCUM-2MD3 | Prognostic
biomarkers for peritoneal recurrence after curative GC
resection | Microarray,
qPCR | Increase | (83) |
| Huang et al,
2017 |
miR-10b-5p/miR-195-5p/miR-20a-3p/miR-296-5p | Case-control | Human serum | Discriminating GC
patients from healthy controls | qPCR | Increase | (84) |
| Wang et al,
2014 | miR-221 | Patients Cell model
Animal model | Human tissue
GC-MSCs BALB/cnu/nu nude mice | Promoting HGC-27
cells proliferation and migration | Microarray,
qPCR | Increase | (85) |
| Zheng et al,
2017 | miR-21 | Cell model Animal
model | M2 macrophages
athymic C57-nudemice | Conferring DDP
resistance in GC cells | Microarray,
qPCR | Increase | (92) |
| Wang et al,
2018 | Anti-miR-214 | Cell model Animal
model | SGC7901,
SGC7901/DDP BALB/c-nude mice | Reversing the
resistance of GC cells to DDP | qPCR | Increase | (93) |
 | Table IV.Exosomal proteins in the top three
mortality cancer types. |
Table IV.
Exosomal proteins in the top three
mortality cancer types.
| A, Lung cancer |
|---|
|
|---|
| Author, year | Protein | Study design | Sample | Clinical
significance | Approach | Performance | (Refs.) |
|---|
| Yamashita et
al, 2017 | EGFR | Case-control | Human plasma | Potential
diagnostic biomarker for characterization of lung cancer | ELISA | Increase | (54) |
| Sandfeld-Paulsen
et al, 2016 | NY-ESO-1 | Case-control | Human plasma | A strongly
prognostic markers for poor survival of NSCLC | Microarray | Increase | (55) |
| Ueda et al,
2014 | CD91 | Case-control | Human serum | Diagnostic markers
for ADC | ELISA Mass
spectrometry | Increase | (56) |
|
| B, Liver
cancer |
|
| Author,
year | Protein | Study
design | Sample | Clinical
significance |
Approach |
Performance | (Refs.) |
|
| He et al,
2015 |
CAV1/CAV2/S100A4 | Cell model | HKCI-C3, HKCI-8
MHCC97L | Enhancing the
invasive and migratory abilities of non-motile MIHA cells | Western blot Mass
spectrometry | Increase | (75) |
| Qu et al,
2016 | HGF | Cell model Animal
model | MHCC-97L, MHCC-97H
BALB/c nu/nu mice | Improving sorafenib
resistance of HCC cells | ELISA Western
blot | Increase | (79) |
|
| C, Gastric
cancer |
|
| Author,
year | Protein | Study
design | Sample | Clinical
significance |
Approach |
Performance | (Refs.) |
|
| Zhang et al,
2017 | EGFR | Patients Animal
model Cell model | Human serum/tissue
BALB/c-nu nude mice SGC7901 | Promoting GC liver
metastasis | ELISA Western
blot | Increase | (86) |
| Li et al,
2015; Liu et al, 2016 | CD97 | Cell model | SGC-7901 | Promoting GC cells
proliferation and invasion | Western blot | Increase | (87,88) |
| Zhong et al,
2011 | Hsp70, Hsp60 | Patients | Heat-treated
malignant ascites | Inducing a CTL
response in vitro and involving in the promotion of DC
maturation | Western blot | Increase | (89) |
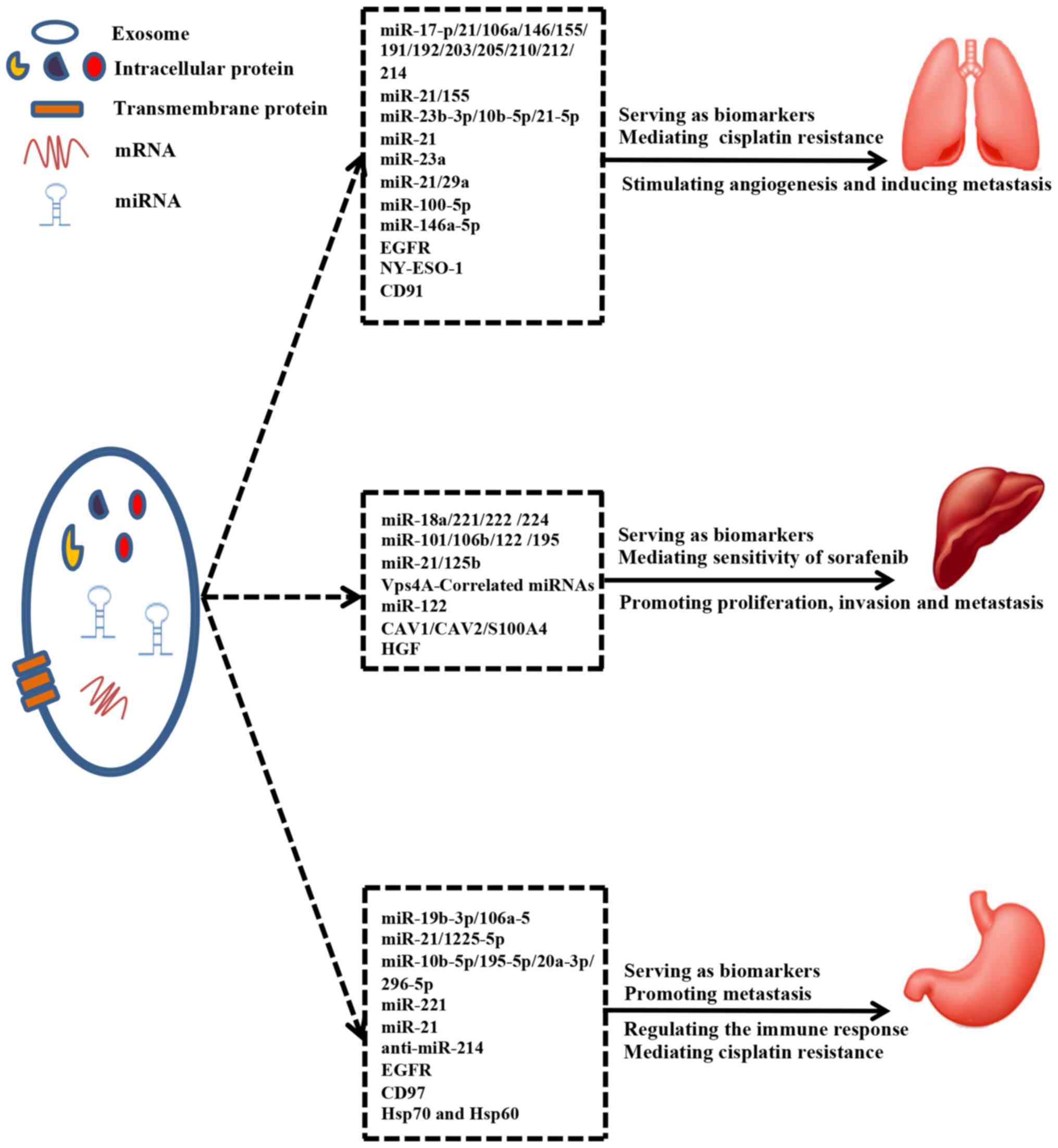
On one hand, existing data indicate that the
packaging of miRNAs into exosomes is a selective process and that
the levels of specific exosomal miRNAs and proteins are changed in
exosomes upon tumorigenesis. For these reasons, exosomal miRNAs and
proteins can be served as a class of novel biomarkers for clinical
applications in high-mortality cancers. Moreover, the specificity,
sensitivity and diagnostic value of exosomal miRNAs and proteins
may be superior to that of traditional tumor markers. On the other
hand, exosomal miRNAs and proteins are delivered between tumor
cells to transmit information and modulate signaling pathways.
Taken together, exosomal miRNAs and proteins perform the essential
function of promoting tumor progression and metastasis as well as
mediating the immune response and sensitivity of tumor cells to
chemotherapy drugs (Fig. 3).
In the future, more robust techniques, such as
RNA-Seq and mass spectrometry, can be used for the detection,
characterization and discovery of exosomal miRNAs and proteins.
Moreover, exosomes could efficiently deliver chemotherapeutic
agents to cells and tissues. Therefore, these bioengineered,
drug-loaded exosomes can serve as promising exosome mimetics for
effective chemotherapeutic agent delivery, which will be applied
for the target treatment of malignant tumors. Currently, the
majority of research on chemotherapy resistance and exosomal
microRNAs focuses on cisplatin, and little is known about other
drugs. To identify more sensitive and specific exosomal miRNAs and
proteins to guide personal chemotherapy selection, future studies
should further elucidate the role and underlying mechanism of
exosomal miRNAs and proteins in more diverse cancers with more
chemotherapy drugs.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 81772276) and the
disciplines group construction project of Pudong Health Bureau of
Shanghai (grant no. PWZxq2017-15).
Availability of data and materials
The datasets used or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LML was a major contributor in writing the
manuscript. HL and XHL were responsible for the collection of the
relevant literatures. HBH and SML revised the manuscript critically
for important intellectual content. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
References
|
1
|
Global Burden of Disease Cancer
Collaboration, . Fitzmaurice C, Allen C, Barber RM, Barregard L,
Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, et al:
Global, regional, and National cancer incidence, mortality, years
of life lost, years lived with disability, and disability-adjusted
life-years for 32 cancer groups, 1990 to 2015: A systematic
analysis for the global burden of disease study. JAMA Oncol.
3:524–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Zhang S, Zhao P, Zeng H
and Zou X: Report of cancer incidence and mortality in China, 2010.
Ann Transl Med. 2:612014.PubMed/NCBI
|
|
3
|
Zheng R, Zeng H, Zhang S and Chen W:
Estimates of cancer incidence and mortality in China, 2013. Chin J
Cancer. 36:662017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou L, Lv T, Zhang Q, Zhu Q, Zhan P, Zhu
S, Zhang J and Song Y: The biology, function and clinical
implications of exosomes in lung cancer. Cancer Lett. 407:84–92.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qi H, Liu C, Long L, Ren Y, Zhang S, Chang
X, Qian X, Jia H, Zhao J, Sun J, et al: Blood exosomes endowed with
magnetic and targeting properties for cancer therapy. ACS Nano.
10:3323–3333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Greening DW, Gopal SK, Xu R, Simpson RJ
and Chen W: Exosomes and their roles in immune regulation and
cancer. Semin Cell Dev Biol. 40:72–81. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Taylor DD and Gercel-Taylor C:
Exosomes/microvesicles: Mediators of cancer-associated
immunosuppressive microenvironments. Semin Immunopathol.
33:441–454. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao L, Liu W, Xiao J and Cao B: The role
of exosomes and ‘exosomal shuttle microRNA’ in tumorigenesis and
drug resistance. Cancer Lett. 356:339–346. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kalluri R: The biology and function of
exosomes in cancer. J Clin Invest. 126:1208–1215. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Frydrychowicz M, Kolecka-Bednarczyk A,
Madejczyk M, Yasar S and Dworacki G: Exosomes-structure, biogenesis
and biological role in non-small-cell lung cancer. Scand J Immunol.
81:2–10. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Aqil F, Munagala R, Jeyabalan J, Agrawal
AK and Gupta R: Exosomes for the enhanced tissue bioavailability
and efficacy of curcumin. AAPS J. 19:1691–1702. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Simons M and Raposo G: Exosomes-vesicular
carriers for intercellular communication. Curr Opin Cell Biol.
21:575–581. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schneider A and Simons M: Exosomes:
Vesicular carriers for intercellular communication in
neurodegenerative disorders. Cell Tissue Res. 352:33–47. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mathivanan S, Ji H and Simpson RJ:
Exosomes: Extracellular organelles important in intercellular
communication. J Proteomics. 73:1907–1920. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Villarroya-Beltri C, Baixauli F,
Gutiérrez-Vázquez C, Sánchez-Madrid F and Mittelbrunn M: Sorting it
out: Regulation of exosome loading. Semin Cancer Biol. 28:3–13.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hannafon BN and Ding WQ: Intercellular
communication by exosome-derived microRNAs in cancer. Int J Mol
Sci. 14:14240–14269. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Keerthikumar S, Chisanga D, Ariyaratne D,
Al Saffar H, Anand S, Zhao K, Samuel M, Pathan M, Jois M,
Chilamkurti N, et al: ExoCarta: A web-based compendium of exosomal
cargo. J Mol Biol. 428:688–692. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ludwig AK and Giebel B: Exosomes: Small
vesicles participating in intercellular communication. Int J
Biochem Cell Biol. 44:11–15. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Théry C, Zitvogel L and Amigorena S:
Exosomes: Composition, biogenesis and function. Nat Rev Immunol.
2:569–579. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Iraci N, Leonardi T, Gessler F, Vega B and
Pluchino S: Focus on extracellular vesicles: Physiological role and
signalling properties of extracellular membrane vesicles. Int J Mol
Sci. 17:1712016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li W, Li C, Zhou T, Liu X, Liu X, Li X and
Chen D: Role of exosomal proteins in cancer diagnosis. Mol Cancer.
16:1452017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hannafon BN, Carpenter KJ, Berry WL,
Janknecht R, Dooley WC and Ding WQ: Exosome-mediated microRNA
signaling from breast cancer cells is altered by the
anti-angiogenesis agent docosahexaenoic acid (DHA). Mol Cancer.
14:1332015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kapetanakis NI, Baloche V and Busson P:
Tumor exosomal microRNAs thwarting anti-tumor immune responses in
nasopharyngeal carcinomas. Ann Transl Med. 5:1642017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Thomou T, Mori MA, Dreyfuss JM, Konishi M,
Sakaguchi M, Wolfrum C, Rao TN, Winnay JN, Garcia-Martin R,
Grinspoon SK, et al: Adipose-derived circulating miRNAs regulate
gene expression in other tissues. Nature. 542:450–455. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ying W, Riopel M, Bandyopadhyay G, Dong Y,
Birmingham A, Seo JB, Ofrecio JM, Wollam J, Hernandez-Carretero A,
Fu W, et al: Adipose tissue macrophage-derived exosomal miRNAs can
modulate in vivo and in vitro insulin sensitivity. Cell.
171:372–384.e12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shi R, Zhao L, Cai W, Wei M, Zhou X, Yang
G and Yuan L: Maternal exosomes in diabetes contribute to the
cardiac development deficiency. Biochem Biophys Res Commun.
483:602–608. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Milane L, Singh A, Mattheolabakis G,
Suresh M and Amiji MM: Exosome mediated communication within the
tumor microenvironment. J Control Release. 219:278–294. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ge Q, Zhou Y, Lu J, Bai Y, Xie X and Lu Z:
miRNA in plasma exosome is stable under different storage
conditions. Molecules. 19:1568–1575. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pfeffer SR, Grossmann KF, Cassidy PB, Yang
CH, Fan M, Kopelovich L, Leachman SA and Pfeffer LM: Detection of
exosomal miRNAs in the plasma of melanoma patients. J Clin Med.
4:2012–2027. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Watahiki A, Macfarlane RJ, Gleave ME, Crea
F, Wang Y, Helgason CD and Chi KN: Plasma miRNAs as biomarkers to
identify patients with castration-resistant metastatic prostate
cancer. Int J Mol Sci. 14:7757–7770. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu R, Greening DW, Rai A, Ji H and Simpson
RJ: Highly-purified exosomes and shed microvesicles isolated from
the human colon cancer cell line LIM1863 by sequential centrifugal
ultrafiltration are biochemically and functionally distinct.
Methods. 87:11–25. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Subra C, Laulagnier K, Perret B and Record
M: Exosome lipidomics unravels lipid sorting at the level of
multivesicular bodies. Biochimie. 89:205–212. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Record M, Carayon K, Poirot M and
Silvente-Poirot S: Exosomes as new vesicular lipid transporters
involved in cell-cell communication and various pathophysiologies.
Biochim Biophys Acta. 1841:108–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Raposo G and Stoorvogel W: Extracellular
vesicles: Exosomes, microvesicles, and friends. J Cell Biol.
200:373–383. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li P, Kaslan M, Lee SH, Yao J and Gao Z:
Progress in exosome isolation techniques. Theranostics. 7:789–804.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Caradec J, Kharmate G, Hosseini-Beheshti
E, Adomat H, Gleave M and Guns E: Reproducibility and efficiency of
serum-derived exosome extraction methods. Clin Biochem.
47:1286–1292. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Baranyai T, Herczeg K, Onódi Z, Voszka I,
Módos K, Marton N, Nagy G, Mäger I, Wood MJ, El Andaloussi S, et
al: Isolation of exosomes from blood plasma: Qualitative and
quantitative comparison of ultracentrifugation and size exclusion
chromatography methods. PLoS One. 10:e01456862015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Peterson MF, Otoc N, Sethi JK, Gupta A and
Antes TJ: Integrated systems for exosome investigation. Methods.
87:31–45. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lu L and Risch HA: Exosomes: Potential for
early detection in pancreatic cancer. Future Oncol. 12:1081–1090.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zeringer E, Barta T, Li M and Vlassov AV:
Strategies for isolation of exosomes. Cold Spring Harb Protoc.
2015:319–323. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Taylor DD and Shah S: Methods of isolating
extracellular vesicles impact down-stream analyses of their
cargoes. Methods. 87:3–10. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ban JJ, Lee M, Im W and Kim M: Low pH
increases the yield of exosome isolation. Biochem Biophys Res
Commun. 461:76–79. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Marczak S, Richards K, Ramshani Z, Smith
E, Senapati S, Hill R, Go DB and Chang HC: Simultaneous isolation
and preconcentration of exosomes by ion concentration polarization.
Electrophoresis. Feb 27–2018.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tauro BJ, Greening DW, Mathias RA, Ji H,
Mathivanan S, Scott AM and Simpson RJ: Comparison of
ultracentrifugation, density gradient separation, and
immunoaffinity capture methods for isolating human colon cancer
cell line LIM1863-derived exosomes. Methods. 56:293–304. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Vaswani K, Koh YQ, Almughlliq FB, Peiris
HN and Mitchell MD: A method for the isolation and enrichment of
purified bovine milk exosomes. Reprod Biol. 17:341–348. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Muller L, Hong CS, Stolz DB, Watkins SC
and Whiteside TL: Isolation of biologically-active exosomes from
human plasma. J Immunol Methods. 411:55–65. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Witwer KW, Buzás EI, Bemis LT, Bora A,
Lässer C, Lötvall J, Nolte-'t Hoen EN, Piper MG, Sivaraman S, Skog
J, et al: Standardization of sample collection, isolation and
analysis methods in extracellular vesicle research. J Extracell
Vesicles. 2:2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kadota T, Yoshioka Y, Fujita Y, Kuwano K
and Ochiya T: Extracellular vesicles in lung cancer-From bench to
bedside. Semin Cell Dev Biol. 67:39–47. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Nanavaty P, Alvarez MS and Alberts WM:
Lung cancer screening: Advantages, controversies, and applications.
Cancer Control. 21:9–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Pletnikoff PP, Laukkanen JA, Tuomainen TP,
Kauhanen J, Rauramaa R, Ronkainen K and Kurl S: Cardiorespiratory
fitness, C-reactive protein and lung cancer risk: A prospective
population-based cohort study. Eur J Cancer. 51:1365–1370. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Rabinowits G, Gercel-Taylor C, Day JM,
Taylor DD and Kloecker GH: Exosomal microRNA: A diagnostic marker
for lung cancer. Clin Lung Cancer. 10:42–46. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Munagala R, Aqil F and Gupta RC: Exosomal
miRNAs as biomarkers of recurrent lung cancer. Tumour Biol.
37:10703–10714. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu Q, Yu Z, Yuan S, Xie W, Li C, Hu Z,
Xiang Y, Wu N, Wu L, Bai L and Li Y: Circulating exosomal microRNAs
as prognostic biomarkers for non-small-cell lung cancer.
Oncotarget. 8:13048–13058. 2017.PubMed/NCBI
|
|
54
|
Yamashita T, Kamada H, Kanasaki S, Maeda
Y, Nagano K, Abe Y, Inoue M, Yoshioka Y, Tsutsumi Y, Katayama S, et
al: Epidermal growth factor receptor localized to exosome membranes
as a possible biomarker for lung cancer diagnosis. Pharmazie.
68:969–973. 2013.PubMed/NCBI
|
|
55
|
Sandfeld-Paulsen B, Aggerholm-Pedersen N,
Bæk R, Jakobsen KR, Meldgaard P, Folkersen BH, Rasmussen TR,
Varming K, Jørgensen MM and Sorensen BS: Exosomal proteins as
prognostic biomarkers in non-small cell lung cancer. Mol Oncol.
10:1595–1602. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ueda K, Ishikawa N, Tatsuguchi A, Saichi
N, Fujii R and Nakagawa H: Antibody-coupled monolithic silica
microtips for highthroughput molecular profiling of circulating
exosomes. Sci Rep. 4:62322014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ostrowski K and Kinsner A: Inhibition of
angiogenesis in the treatment of tumors. Arch Immunol Ther Exp
(Warsz). 49:27–31. 2001.PubMed/NCBI
|
|
58
|
Liu Y, Luo F, Wang B, Li H, Xu Y, Liu X,
Shi L, Lu X, Xu W, Lu L, et al: STAT3-regulated exosomal miR-21
promotes angiogenesis and is involved in neoplastic processes of
transformed human bronchial epithelial cells. Cancer Lett.
370:125–135. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hsu YL, Hung JY, Chang WA, Lin YS, Pan YC,
Tsai PH, Wu CY and Kuo PL: Hypoxic lung cancer-secreted exosomal
miR-23a increased angiogenesis and vascular permeability by
targeting prolyl hydroxylase and tight junction protein ZO-1.
Oncogene. 36:4929–4942. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Fabbri M, Paone A, Calore F, Galli R,
Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, et al:
MicroRNAs bind to Toll-like receptors to induce prometastatic
inflammatory response. Proc Natl Acad Sci USA. 109:E2110–E2116.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Qin X, Yu S, Zhou L, Shi M, Hu Y, Xu X,
Shen B, Liu S, Yan D and Feng J: Cisplatin-resistant lung cancer
cell-derived exosomes increase cisplatin resistance of recipient
cells in exosomal miR-100-5p-dependent manner. Int J Nanomedicine.
12:3721–3733. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yuwen DL, Sheng BB, Liu J, Wenyu W and Shu
YQ: MiR-146a-5p level in serum exosomes predicts therapeutic effect
of cisplatin in non-small cell lung cancer. Eur Rev Med Pharmacol
Sci. 21:2650–2658. 2017.PubMed/NCBI
|
|
63
|
Lobb RJ, van Amerongen R, Wiegmans A, Ham
S, Larsen JE and Möller A: Exosomes derived from mesenchymal
non-small cell lung cancer cells promote chemoresistance. Int J
Cancer. 141:614–620. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang FS, Fan JG, Zhang Z, Gao B and Wang
HY: The global burden of liver disease: The major impact of China.
Hepatology. 60:2099–2108. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li X and Xu WF: China's efforts to shed
its title of ‘Leader in liver disease’. Drug Discov Ther. 1:84–85.
2007.PubMed/NCBI
|
|
66
|
Wong MC, Jiang JY, Goggins WB, Liang M,
Fang Y, Fung FD, Leung C, Wang HH, Wong GL, Wong VW and Chan HL:
International incidence and mortality trends of liver cancer: A
global profile. Sci Rep. 7:458462017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Sohn W, Kim J, Kang SH, Yang SR, Cho JY,
Cho HC, Shim SG and Paik YH: Serum exosomal microRNAs as novel
biomarkers for hepatocellular carcinoma. Exp Mol Med. 47:e1842015.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wang H, Hou L, Li A, Duan Y, Gao H and
Song X: Expression of serum exosomal microRNA-21 in human
hepatocellular carcinoma. Biomed Res Int.
2014:8648942014.PubMed/NCBI
|
|
69
|
Liu W, Hu J, Zhou K, Chen F, Wang Z, Liao
B, Dai Z, Cao Y, Fan J and Zhou J: Serum exosomal miR-125b is a
novel prognostic marker for hepatocellular carcinoma. Onco Targets
Ther. 10:3843–3851. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Kogure T, Lin WL, Yan IK, Braconi C and
Patel T: Intercellular nanovesicle-mediated microRNA transfer: A
mechanism of environmental modulation of hepatocellular cancer cell
growth. Hepatology. 54:1237–1248. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wei JX, Lv LH, Wan YL, Cao Y, Li GL, Lin
HM, Zhou R, Shang CZ, Cao J, He H, et al: Vps4A functions as a
tumor suppressor by regulating the secretion and uptake of exosomal
microRNAs in human hepatoma cells. Hepatology. 61:1284–1294. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Mishra SK, Siddique HR and Saleem M:
S100A4 calcium-binding protein is key player in tumor progression
and metastasis: Preclinical and clinical evidence. Cancer
Metastasis Rev. 31:163–172. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Tse EY, Ko FC, Tung EK, Chan LK, Lee TK,
Ngan ES, Man K, Wong AS, Ng IO and Yam JW: Caveolin-1
overexpression is associated with hepatocellular carcinoma
tumourigenesis and metastasis. J Pathol. 226:645–653. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Cokakli M, Erdal E, Nart D, Yilmaz F,
Sagol O, Kilic M, Karademir S and Atabey N: Differential expression
of Caveolin-1 in hepatocellular carcinoma: Correlation with
differentiation state, motility and invasion. BMC Cancer. 9:652009.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
He M, Qin H, Poon TC, Sze SC, Ding X, Co
NN, Ngai SM, Chan TF and Wong N: Hepatocellular carcinoma-derived
exosomes promote motility of immortalized hepatocyte through
transfer of oncogenic proteins and RNAs. Carcinogenesis.
36:1008–1018. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S,
Kim JS, Luo R, Feng J, Ye S, Yang TS, et al: Efficacy and safety of
sorafenib in patients in the Asia-Pacific region with advanced
hepatocellular carcinoma: A phase III randomised, double-blind,
placebo-controlled trial. Lancet Oncol. 10:25–34. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Lou G, Song X, Yang F, Wu S, Wang J, Chen
Z and Liu Y: Exosomes derived from miR-122-modified adipose
tissue-derived MSCs increase chemosensitivity of hepatocellular
carcinoma. J Hematol Oncol. 8:1222015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
You H, Ding W, Dang H, Jiang Y and
Rountree CB: c-Met represents a potential therapeutic target for
personalized treatment in hepatocellular carcinoma. Hepatology.
54:879–889. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Qu Z, Wu J, Wu J, Luo D, Jiang C and Ding
Y: Exosomes derived from HCC cells induce sorafenib resistance in
hepatocellular carcinoma both in vivo and in vitro. J Exp Clin
Cancer Res. 35:1592016. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Ferro A, Peleteiro B, Malvezzi M, Bosetti
C, Bertuccio P, Levi F, Negri E, La Vecchia C and Lunet N:
Worldwide trends in gastric cancer mortality (1980–2011), with
predictions to 2015, and incidence by subtype. Eur J Cancer.
50:1330–1344. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Peleteiro B, Severo M, La Vecchia C and
Lunet N: Model-based patterns in stomach cancer mortality
worldwide. Eur J Cancer Prev. 23:524–531. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wang N, Wang L, Yang Y, Gong L, Xiao B and
Liu X: A serum exosomal microRNA panel as a potential biomarker
test for gastric cancer. Biochem Biophys Res Commun. 493:1322–1328.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Tokuhisa M, Ichikawa Y, Kosaka N, Ochiya
T, Yashiro M, Hirakawa K, Kosaka T, Makino H, Akiyama H, Kunisaki C
and Endo I: Exosomal miRNAs from peritoneum lavage fluid as
potential prognostic biomarkers of peritoneal metastasis in gastric
cancer. PLoS One. 10:e01304722015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Huang Z, Zhu D, Wu L, He M, Zhou X, Zhang
L, Zhang H, Wang W, Zhu J, Cheng W, et al: Six serum-based miRNAs
as potential diagnostic biomarkers for gastric cancer. Cancer
Epidemiol Biomarkers Prev. 26:188–196. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Wang M, Zhao C, Shi H, Zhang B, Zhang L,
Zhang X, Wang S, Wu X, Yang T, Huang F, et al: Deregulated
microRNAs in gastric cancer tissue-derived mesenchymal stem cells:
Novel biomarkers and a mechanism for gastric cancer. Br J Cancer.
110:1199–1210. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhang H, Deng T, Liu R, Bai M, Zhou L,
Wang X, Li S, Wang X, Yang H, Li J, et al: Exosome-delivered EGFR
regulates liver microenvironment to promote gastric cancer liver
metastasis. Nat Commun. 8:150162017. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Li C, Liu DR, Li GG, Wang HH, Li XW, Zhang
W, Wu YL and Chen L: CD97 promotes gastric cancer cell
proliferation and invasion through exosome-mediated MAPK signaling
pathway. World J Gastroenterol. 21:6215–6228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Liu D, Li C, Trojanowicz B, Li X, Shi D,
Zhan C, Wang Z and Chen L: CD97 promotion of gastric carcinoma
lymphatic metastasis is exosome dependent. Gastric Cancer.
19:754–766. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Zhong H, Yang Y, Ma S, Xiu F, Cai Z, Zhao
H and Du L: Induction of a tumour-specific CTL response by exosomes
isolated from heat-treated malignant ascites of gastric cancer
patients. Int J Hyperthermia. 27:604–611. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Broere F, van der Zee R and van Eden W:
Heat shock proteins are no DAMPs, rather ‘DAMPERs’. Nat Rev
Immunol. 11:5652011. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
van Eden W, Spiering R, Broere F and van
der Zee R: A case of mistaken identity: HSPs are no DAMPs but
DAMPERs. Cell Stress Chaperones. 17:281–292. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Zheng P, Chen L, Yuan X, Luo Q, Liu Y, Xie
G, Ma Y and Shen L: Exosomal transfer of tumor-associated
macrophage-derived miR-21 confers cisplatin resistance in gastric
cancer cells. J Exp Clin Cancer Res. 36:532017. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Wang X, Zhang H, Bai M, Ning T, Ge S, Deng
T, Liu R, Zhang L, Ying G and Ba Y: Exosomes serve as nanoparticles
to deliver anti-miR-214 to reverse chemoresistance to cisplatin in
gastric cancer. Mol Ther. 26:774–783. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Ji R, Zhang B, Zhang X, Xue J, Yuan X, Yan
Y, Wang M, Zhu W, Qian H and Xu W: Exosomes derived from human
mesenchymal stem cells confer drug resistance in gastric cancer.
Cell Cycle. 14:2473–2483. 2015. View Article : Google Scholar : PubMed/NCBI
|