Cancers originate from normal cells that gain the
ability to aberrantly proliferate and eventually turn malignant.
These cancerous cells then grow clonally into tumors and eventually
acquire the potential to metastasize (1). Alteration of cellular processes is a
central component in cancer development, including changes in
cancer cell growth, apoptosis, migration and invasion (2–6).
Inhibition of the abnormal growth of cancer cells and the promotion
of cancer cell apoptosis are widely recognized as crucial goals for
intervention of cancer progression. In addition, dysregulations of
oncogenes and cancer suppressors tightly correlate with cancer
occurrence (7–9). However, the lack of useful cancer
biomarkers and targets is a major contributor to the high mortality
rate and prevalence of cancer.
SAD1/UNC84 domain protein-2 (SUN2), a member of the
SUN domain protein family, is a key component of linker of
nucleoskeleton and cytoskeleton (LINC) complex. The nuclear
architecture functionally provides a framework for organizing and
regulating diverse processes within cells. Notably, cancer cells
generally exhibit variety of features indicative of atypical nuclei
(10), although the molecular
mechanism of these phenomena remains to be elucidated. A number of
studies have shown that loss of LINC complexes reduces nuclear and
cellular rigidity, increasing tissue fluidity, promoting invasive
activity, and inducing cancer progression (10–12).
Interestingly, studies have uncovered a fundamental role of SUN2 in
nuclear structure determination function (13). Therefore, we hypothesize that the
effects of SUN2 on regulating nuclear architecture may affect
biological function in cancer cells. Indeed, abnormal expression of
SUN2 and LINC complexes is associated with the occurrence of many
human diseases, especially cancers (10).
Several studies have also linked SUN2 function with
various cancers. SUN2 plays SUN2 also plays a cancer suppressor
role in miR-221/222-mediated malignant embryonal tumors of the
central nervous system (14). Another
study confirmed that expression of SUN2 was reduced in breast
cancer (10). Moreover, SUN2
exhibited suppression of lung cancer cell proliferation and
migration and promotion of lung cancer cell apoptosis; SUN2 also
enhanced the chemotherapy sensitivity of lung cancer cells exposed
to cisplatin, and higher SUN2 level predicts a better overall
survival in lung cancer progression (12). Together these studies indicate that
dysregulation of SUN2 may be involved in cancer development.
Failure to detect and repair DNA damage leads to
genomic instability, which is one of the hallmarks that drive
cancer occurrence (15). Recent
reports also suggest that SUN2 exhibits resistance to DNA damage
and maintaining the genome integrity (16). Lei et al confirmed that SUN2 is
required for attenuating excessive DNA damage in mouse embryonic
fibroblasts (MEFs) from SUN1−/−SUN2−/− double
knockout mice (16). Whether SUN2
participates in maintaining genomic stability in other types of
cancer needed further validation.
Following recent advances, this review presents
recent information regarding the functions of SUN2 in the
progression of cancer and discusses the emerging signal pathways
regulated by SUN2 in cancer.
The LINC complex is a nuclear envelope protein
complex that mainly consists of SUN and nesprin proteins,
connecting nuclear lamina and cytoskeletal filaments (10), LINC complex characteristics of
architecture framework helps to regulate the size and shape of the
cell nucleus. Several SUN proteins have been identified in several
organisms, including Schizosaccharomyces pombe Sad1,
Caenorhabditis elegans UNC-84 and SUN1, and five human SUN
proteins (17). Human SUN proteins
can be grouped into two subfamilies based on their intracellular
localization: SUN1 and SUN2 are integral membrane components of the
inner nuclear membrane (INM) (18–20); SUN3
and the sperm-associated antigen 4 localize to endoplasmic
reticulum and outer nuclear membrane (ONM) (21,22).
SUN proteins are conserved among all eukaryotes and
characterized by a C-terminal 200 amino acid SUN domain (18,19,23–25).
SUN proteins form a trimer through the SUN domain and exhibits a
perfect three-fold symmetry, resembling a cloverleaf (65 Å
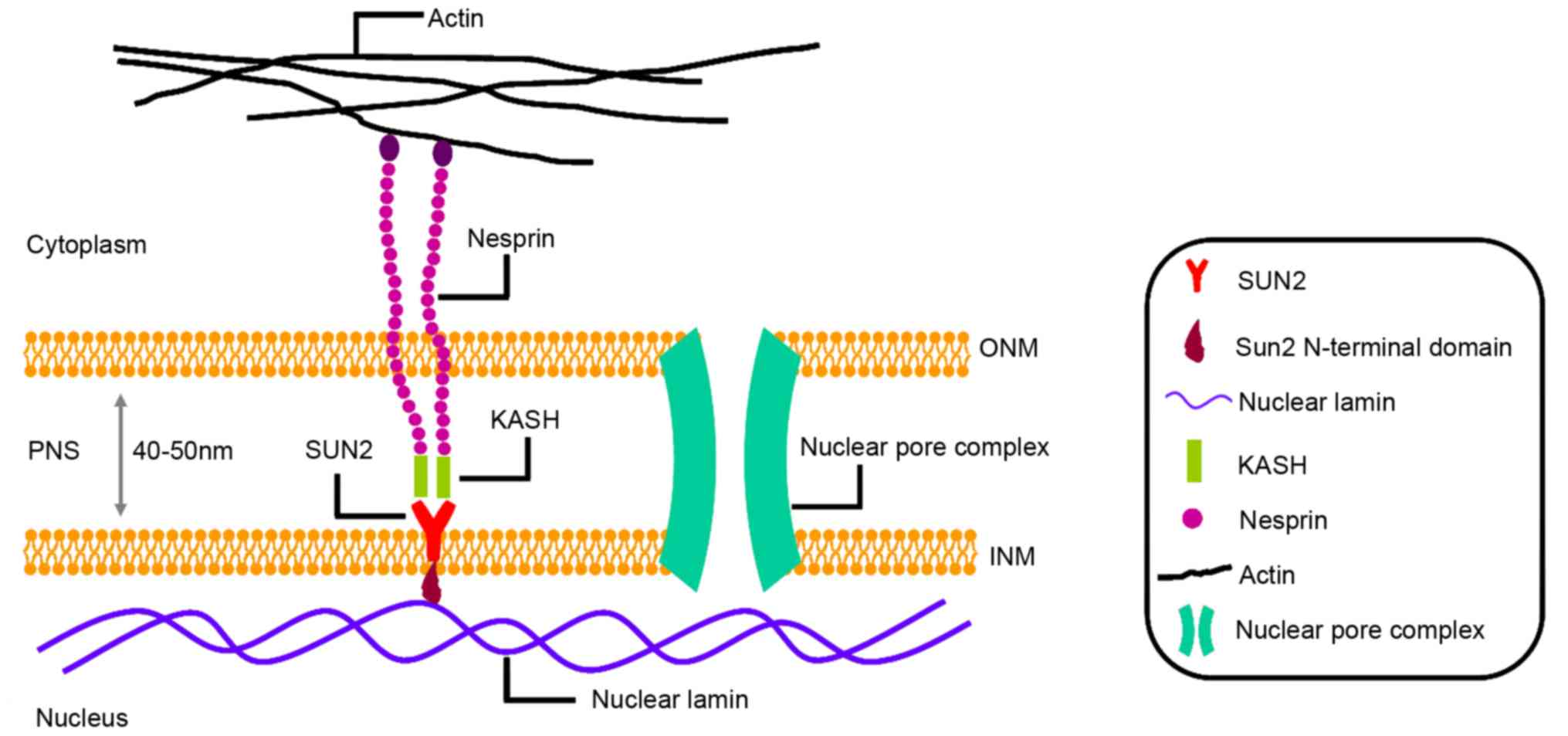
diameter) sitting on a short stem (30 Å of length) (26). As shown in Fig. 1, SUN2 extends into the perinuclear
space by its C-terminal SUN domain and interacts with nuclear
lamina via its nucleoplasmic N-terminal domain (11,27,28).
Additionally, SUN2 connects with klarsicht/ANC-1/syne-1 homology
(KASH) domain, providing mechanical transduction between the
cytoskeleton and nuclear interior, directly (19,22,28–30).
Recent reports indicate that the SUN domain is at the center of a
nucleocytoplasmic bridge that is essential for nuclear motility in
cells (31). These observations
suggest that the structural characteristics of SUN2 are crucial for
nuclear anchoring, migration, and positioning (19,22,29,30,32,33),
centromere localization (34) and
regulating the tethering of meiotic telomere (26). Thus, SUN2 may possess anti-cancer by
regulating atypical nuclei structures in cancer cells.
AT/RT frequently occur in children. However, the
pathogenesis of AT/RT remains to be uncovered. Several studies have
indicated that the miR-221/222 gene cluster serves as an oncogenic
miRNA in several types of human cancer (35,36).
Recently, miRNome and transcriptome traits in AT/RT were evaluated
using small RNA sequencing and gene expression microarray analyses.
Hsieh et al showed that miR-221/222-encoded miRNAs are
abundantly expressed in AT/RT and substantially contribute to the
malignancy of embryonal tumors (10).
In AT/RT cells, overexpression of miR-221/222 leads to faster cell
growth, and this observation is supported by previous reports that
miR-221/222 promotes AT/RT malignancy and tumor growth in nude mice
(14). AT/RT tissue microarray
demonstrated that SUN2 is markedly decreased in AT/RT specimens.
miRNAs generally execute their cellular functions through
regulating target gene expression. Notably, miR-221/222 promotes
cancer cell proliferation and tumor malignancy by targeting SUN2
mRNA in AT/RT, directly. Adherent cell growth of human
medulloblastoma Daoy and human ATRT CHLA-02-ATRT cells was
significantly increased upon transfection of SUN2 short
hairpin-producing plasmids, parallelly, while overexpression of
SUN2 reduced the proliferation rate (14). Together these studies show that SUN2
plays a critical role in miR-221/222-mediated AT/RT malignancy,
indicating that SUN2 may be a promising target of AT/RT. For the
first time, SUN2 was demonstrated closely relate to cancer
initiation and progression.
Several studies have provided evidence that
abnormalities of the LINC are associated with complex alteration of
biological processes and cancer occurrence. Reduced expression of
lamin A/C was detected in colon cancer (37), small cell lung cancer (38), leukemias and lymphomas (39,40).
However, lamin A/C is overexpressed in colorectal cancer (41), prostate cancer (42), and skin cancer (43,44).
Therefore, the precise relationship between LINC complex components
and the clinical significance of cancer still has not yet been well
elucidated. In the present study, evaluation of four LINC complex
and nuclear lamina components, SUN1, SUN2, nesprin-2, and lamin
A/C, in breast cancer was performed. Matsumoto et al
collected 73 breast cancer samples and found lower expression
levels of LINC components in tumor regions compared with
cancer-associated noncancerous regions (11). Furthermore, decreased expression of
SUN2 was detected in several breast cancer cell lines compared with
noncancerous mammary gland cells in vitro. Together this
demonstrates that the expression of SUN2 is attenuated in human
breast cancer clinical specimens, indicating that SUN2 may have
fundamental pathological functions in human breast cancer
progression.
Previous studies have demonstrated that SUN2
exhibits anti-cancer functions in lung cancer progression. Higher
SUN2 expression predicts a better overall survival (OS) in lung
cancer (12). Lv et al
confirmed that expression level of SUN2 was significantly reduced
in lung cancer tissues compared with paired normal tissues using
Oncomine Database (12). According to
the Protein Atlas Database, the expression of SUN2 is reduced in
75% (9 out of 12) of lung cancer tissue samples (12). Furthermore, in a previous study
evaluating the relationship between SUN2 and lung cancer, lung
cancer samples were subdivided into two groups and OS was analyzed.
Individuals with lower SUN2 expression levels exhibited shorter OS
than those with high SUN2 expression level (12). Together this indicates that
downregulation of SUN2 in lung cancer progression and higher
expression of SUN2 may predict a good outcome in human lung cancer
occurrence. Additionally, ectopic expression of SUN2 inhibited lung
cancer cell proliferation and colony formation abilities, and
chemotherapy sensitivity to cisplatin treatment was increased when
SUN2 was overexpressed in lung cancer cells. Further, knockdown of
SUN2 promoted lung cancer cell proliferation and migration
(12). Together these observations
suggest that SUN2 is a key player in lung cancer development.
Failure of the DNA damage response (DDR) leads to
genomic instability, which is one of enabling hallmarks that drive
cancer occurrence (46).
Phosphorylation of ataxia telangiectasia mutated (ATM) and H2A.X
are among the earliest events in response to DNA damage (47–49).
Recent studies showed that the expression level of γ-H2A.X is
significantly reduced in MEFs isolated from
SUN1−/−SUN2−/− double knockout mice compared
with wild-type mice. Although ATM is activated by 0.1 mM of
hydroxyurea (HU) in wild-type MEFs, ATM is not activated by HU in
SUN1−/−SUN2−/− MEFs (16). There was no significant difference in
tail moment between wild-type and
SUN1−/−SUN2−/− MEFs in the absence of methyl
methane-sulfonate (16), which
induces DNA damage (50).
Interestingly, after treatment of
SUN1−/−SUN2−/− MEFs with methyl
methane-sulfonate, a substantial increase occurred in the number of
cells with prominent comet tails, indicative of DNA fragmentation
(16). These observations reveal that
DNA damage may accumulate rapidly in
SUN1−/−SUN2−/− MEFs.
Phosphorylated checkpoint kinase-1 (Chk1), a
cell-cycle checkpoint factor downstream of the DDR pathway, is
reduced in SUN1−/−SUN2−/− MEFs compared with
wild-type mice (16). In addition,
perinuclear heterochromatin is decreased in
SUN1−/−SUN2−/− MEFs, indicating that SUN1 and
SUN2 participate in maintaining genomic stability, possibly by
affecting DDR or DNA repair. Furthermore, MEFs from
SUN1−/−SUN2−/− mice exhibit a premature
proliferative arrest at the S phase of cell cycle and increase in
cell apoptosis (16,51,52),
leading to the death of SUN1−/−SUN2−/− mice
shortly after birth. These reports suggest that SUN2 may have
crucial effects on evading cancer occurrence by its involvement in
the DDR, and eliminating DNA lesions, maintaining genome stability
and integrity.
SUN2 also interacts with DNA-PKcs that are
potentially involved in the DDR, especially in DNA repair (16). Of note, cancer cells exhibit a high
rate of proliferation and metabolic activities and DNA-PKcs plays
an active part in regulation of cell proliferation (53). Of further interest, a previous study
demonstrated that SUN2 exhibits suppression of cancer cell
proliferation activity. In summary, uncovering the function of
DNA-PKcs/SUN2 in regulation of cancer cells may offer potential
avenues for cancers treatment.
miRNAs mainly function by regulating the expression
of target genes at the post-transcriptional level. Up-regulation of
miR-221/222 is associated with initiation and progression of breast
cancer (54–59), liver cancer (60–63),
pancreatic cancer (64–68), gastric cancer (69–73),
colorectal cancer (74–78), glioma (79–85),
multiple myeloma (86–89), and malignant melanoma (90,91).
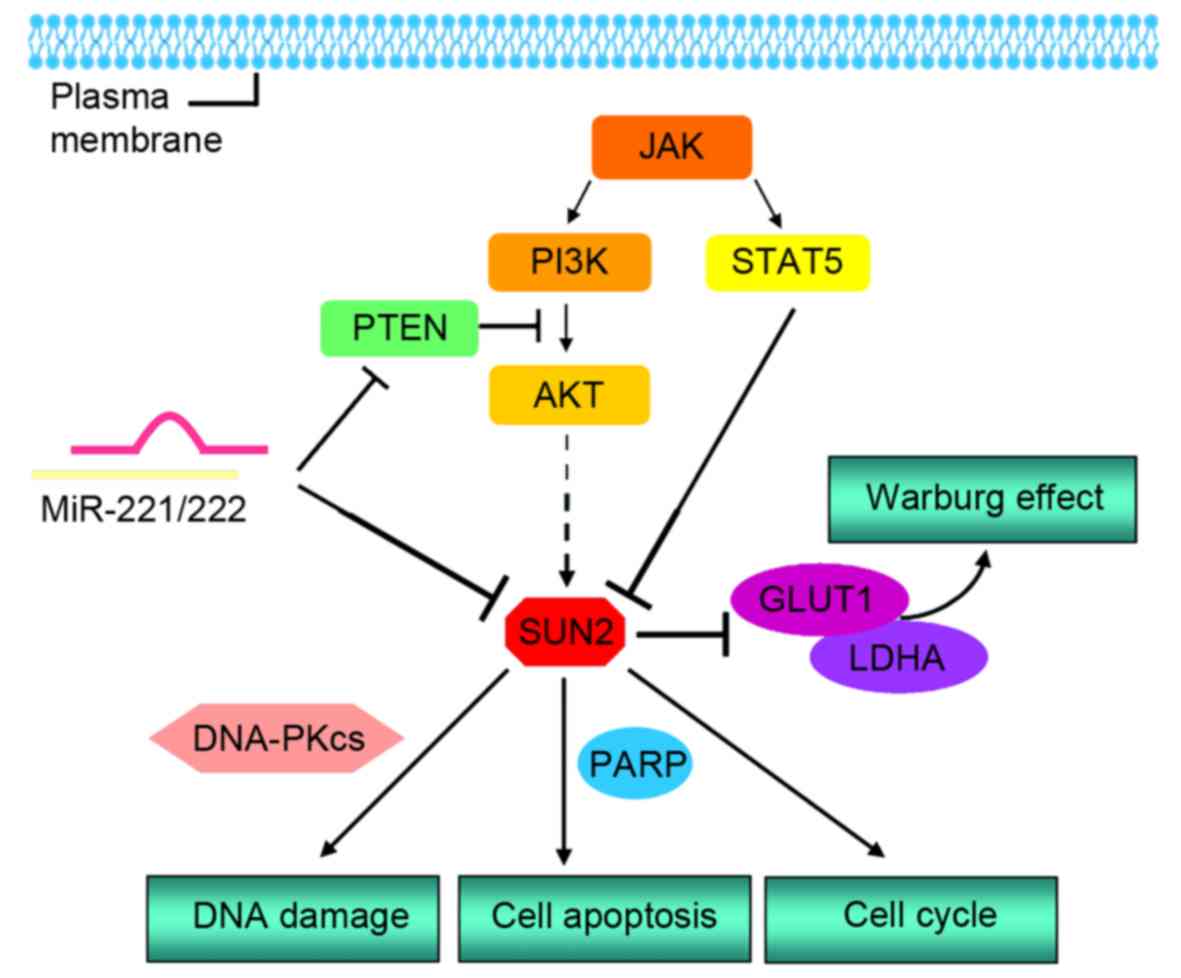
TargetScan indicated a potentially favorable interaction between
miR-221-3p/miR-222-3p and an 8-mer site at the position 255–262 in
the SUN2 3′-untranslated region (3′UTR). Luciferase assays
demonstrated that both miR-221-3p and miR-222-3p directly bind to
the recognition element and reduce activity of Luc fused to
full-length 3′UTR of SUN2 (14).
Moreover, correlation coefficients (Pearson's r) between
SUN2 and miR-221-3p as well as SUN2 and miR-222-3p are −0.777 and
−0.802, respectively, indicating negative correlation between SUN2
and miR-221/222 in AT/RT and medulloblastoma (MB) (14). A previous report also showed that the
transcript and protein level of SUN2 was reduced after ectopic
expression of miR-221/222, further supporting SUN2 as a direct
target of miR-221/222. Increasing numbers of studies illustrate
that expression of miR-221/222 induces cancer cell proliferation
and invasion by inhibiting cancer suppressors and apoptotic genes
(92). Over-expression of miR-221/222
significantly increases cell proliferation, while over-expression
of both miR-221/222 and the complete coding sequence (CDS) of SUN2,
which possesses no miR-221/222 recognition elements, counteracted
the pro-proliferative effects (14).
Together this suggests that one of the crucial pathways of
miR-221/222 increasing cancer cell proliferation may occur by
down-regulating SUN2 expression (Fig.
2).
Silent information regulator-5 (SIRT5) is a key
component of the sirtuin family. SIRT5 expression has been
associated with cancer prognosis and survival (93) via stimulating cancer cell
proliferation and tumor growth, attenuating the tumor-type
metabolism (94). Of note, the
expression of SIRT5 is decreased in squamous cell carcinoma
(95) and endometrial carcinoma
(96). Thus, inhibition of SIRT5 may
become a potential strategy to suppress the progression of cancers
(97). Nevertheless, SIRT5 has also
been found to have negative implications in certain types of
malignancies (98). For instance,
SIRT5 is highly expressed in human non-small cell lung cancer
(NSCLC) and facilitates tumor growth and drug resistance (99). SIRT5 is also downregulated with
histone deacetylase (HDACs) inhibitor treatment (100), while SUN2 expression dramatically
increased in response to nicotinamide, an inhibitors of HDAC
(101,102), indicating that SUN2 may be regulated
by SIRT5. Additionally, ectopic expression of SIRT5 significantly
reduced SUN2 expression, while knockdown of SIRT5 dramatically
increased SUN2 expression. Expression of SIRT5 inversely correlates
with SUN2, indicating that SIRT5 acts as a negative regulator of
SUN2, at least in part (12). The
precise role of SIRT5/SUN2 as a novel axis in the regulation of
different types of cancers is currently unclear, and the
relationship between SUN2 with cancer occurrence related to SIRT5
requires further exploration.
Poly (ADP-ribose) polymerase (PARP) is verified as
tightly correlated with cellular functions, such as DNA repair and
transcriptional and posttranscriptional modulation of oncogenic
gene expression, ultimately modulating carcinogenesis (110). Several studies suggested that PARP
interacts with breast cancer (111),
ovarian cancer (112), prostate
cancer, lung cancer, gastric cancer and hepatocellular carcinoma
(113,114). Cleavage of PARP is a well-known
marker of cell apoptosis, and interestingly, higher expression
level of SUN2 increases PARP cleavage events (12). Together this suggests that SUN2
expression may inhibit cancer progression by regulating
PARP-mediated cell apoptosis.
Overall, the complete underlying molecular
mechanisms of cancers are still poorly elucidated. Many studies
have established that dysregulations of oncogenes and cancer
suppressor genes distinctly correlate with the initiation and
progression of cancers. As outlined in this review, we discussed
recent insights into the function of SUN2 in cancer progression.
SUN2, as an anti-cancer member, participates in AT/RT, breast
cancer and lung cancer by regulating biological processes in cancer
cells, including cell cycle, apoptosis and migration. In addition,
deficiency of SUN2 distinctly induces DNA damage, which is
critically involved in cancer initiation.
Of note, SUN2 is widely expressed in different
organs and tissues, such as the heart, brain, spleen, lung, liver,
skeletal muscle, testis and embryos (17). Previous studies have shown an
involvement of SUN2 in human cancers, such as cervical carcinoma,
colorectal cancer, esophageal carcinoma and oral cavity squamous
cell carcinoma. Furthermore, recent findings in fission yeast
suggest that SUN2 may serve as a predictor and prognostic
biomarkers in cancer. Therefore, we hypothesize that SUN2 may act
as a potential biomarker in multiple cancer cell types. However,
current findings suggest that SUN2 may present different functions
in various cancers, and thus, we cannot definitively conclude that
SUN2 solely functions as a cancer suppressor in all types of
cancers. Undoubtedly, the precise functions and potential signaling
pathways of SUN2 in cancer progression remain to be elucidated, and
further studies and validations are urgently needed.
Not applicable.
This work is supported by the National Science
Foundation of China (grant no. 81473268); the Anhui Provincial
Higher Education Natural Science Foundation (grant no. KJ2016A364)
and Anhui Provincial Natural Science Foundation (grant no.
21408085MKL31).
Not applicable.
In this review, study concept and design: JL, XC,
YC. Draft of the manuscript: XC, YC. Analysis of data: HMH, HDL,
CH, XMM. Critical revision of the manuscript for important
intellectual content: FTB, XYP, YY, WXL, XFL. All authors agreed
the final version.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Lobo NA, Shimono Y, Qian D and Clarke MF:
The biology of cancer stem cells. Annu Rev Cell Dev Biol.
23:675–699. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Peng XH, Huang HR, Lu J, Liu X, Zhao FP,
Zhang B, Lin SX, Wang L, Chen HH, Xu X, et al: MiR-124 suppresses
tumor growth and metastasis by targeting Foxq1 in nasopharyngeal
carcinoma. Mol Cancer. 13:1862014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu Y, Li Z, Wu L, Wang Z, Wang X, Yu Y,
Zhao Q and Luo F: MiRNA-125a-5p: A regulator and predictor of
gefitinib's effect on nasopharyngeal carcinoma. Cancer Cell Int.
14:242014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu X, Lv XB, Wang XP, Sang Y, Xu S, Hu K,
Wu M, Liang Y, Liu P, Tang J, et al: MiR-138 suppressed
nasopharyngeal carcinoma growth and tumorigenesis by targeting the
CCND1 oncogene. Cell Cycle. 11:2495–2506. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qi X, Li J, Zhou C, Lv C and Tian M:
MicroRNA-320a inhibits cell proliferation, migration and invasion
by targeting BMI-1 in nasopharyngeal carcinoma. FEBS Lett.
588:3732–3738. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cheung CC, Chung GT, Lun SW, To KF, Choy
KW, Lau KM, Siu SP, Guan XY, Ngan RK, Yip TT, et al: miR-31 is
consistently inactivated in EBV-associated nasopharyngeal carcinoma
and contributes to its tumorigenesis. Mol Cancer. 13:1842014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu X, Yu X, Xie J, Zhan M, Yu Z, Xie L,
Zeng H, Zhang F, Chen G, Yi X and Zheng J: ANGPTL2/LILRB2 signaling
promotes the propagation of lung cancer cells. Oncotarget.
6:21004–21015. 2015.PubMed/NCBI
|
|
8
|
Wang J, Tian X, Han R, Zhang X, Wang X,
Shen H, Xue L, Liu Y, Yan X, Shen J, et al: Downregulation of
miR-486-5p contributes to tumor progression and metastasis by
targeting protumorigenic ARHGAP5 in lung cancer. Oncogene.
33:1181–1189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hecht I, Natan S, Zaritsky A, Levine H,
Tsarfaty I and Ben-Jacob E: The motility-proliferation-metabolism
interplay during metastatic invasion. Sci Rep. 5:135382015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hsieh TH, Chien CL, Lee YH, Lin CI, Hsieh
JY, Chao ME, Liu DJ, Chu SS, Chen W, Lin SC, et al: Downregulation
of SUN2, a novel tumor suppressor, mediates miR-221/222-induced
malignancy in central nervous system embryonal tumors.
Carcinogenesis. 35:2164–2174. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Matsumoto A, Hieda M, Yokoyama Y, Nishioka
Y, Yoshidome K, Tsujimoto M and Matsuura N: Global loss of a
nuclear lamina component, lamin A/C, and LINC complex components
SUN1, SUN2, and nesprin-2 in breast cancer. Cancer Med.
4:1547–1557. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lv XB, Liu L, Cheng C, Yu B, Xiong L, Hu
K, Tang J, Zeng L and Sang Y: SUN2 exerts tumor suppressor
functions by suppressing the Warburg effect in lung cancer. Sci
Rep. 5:179402015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meinke P, Nguyen TD and Wehnert MS: The
LINC complex and human disease. Biochem Soc Trans. 39:1693–1697.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Khatau SB, Hale CM, Stewart-Hutchinson PJ,
Patel MS, Stewart CL, Searson PC, Hodzic D and Wirtz D: A
perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci
USA. 106:19017–19022. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Z, Zhu WG and Xu X: Ubiquitin-like
modifications in the DNA damage response. Mutat Res. 803–805.
56–75. 2017.
|
|
16
|
Lei K, Zhu X, Xu R, Shao C, Xu T, Zhuang Y
and Han M: Inner nuclear envelope proteins SUN1 and SUN2 play a
prominent role in the DNA damage response. Curr Biol. 22:1609–1615.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Q, Du X, Cai Z and Greene MI:
Characterization of the structures involved in localization of the
SUN proteins to the nuclear envelope and the centrosome. DNA Cell
Biol. 25:554–562. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hodzic DM, Yeater DB, Bengtsson L, Otto H
and Stahl PD: Sun2 is a novel mammalian inner nuclear membrane
protein. J Biol Chem. 279:25805–25812. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Padmakumar VC, Libotte T, Lu W, Zaim H,
Abraham S, Noegel AA, Gotzmann J, Foisner R and Karakesisoglou I:
The inner nuclear membrane protein Sun1 mediates the anchorage of
Nesprin-2 to the nuclear envelope. J Cell Scie. 118:3419–3430.
2005. View Article : Google Scholar
|
|
20
|
Dreger M, Bengtsson L, Schöneberg T, Otto
H and Hucho F: Nuclear envelope proteomics: Novel integral membrane
proteins of the inner nuclear membrane. Proc Natl Acad Sci USA.
98:11943–11948. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kennedy C, Sebire K, de Kretser DM and
O'Bryan MK: Human sperm associated antigen 4 (SPAG4) is a potential
cancer marker. Cell Tissue Res. 315:279–283. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tzur YB, Wilson KL and Gruenbaum Y:
SUN-domain proteins: ‘Velcro’ that links the nucleoskeleton to the
cytoskeleton. Nat Rev Mol Cell Biol. 7:782–788. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Starr DA and Fischer JA: KASH'n Karry: The
KASH domain family of cargo-specific cytoskeletal adaptor proteins.
Bioessays. 27:1136–1146. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hagan I and Yanagida M: The product of the
spindle formation gene sad1+ associates with the fission yeast
spindle pole body and is essential for viability. J Cell Biol.
129:1033–1047. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Malone CJ, Fixsen WD, Horvitz HR and Han
M: UNC-84 localizes to the nuclear envelope and is required for
nuclear migration and anchoring during C. elegans development.
Development. 126:3171–3181. 1999.PubMed/NCBI
|
|
26
|
Zhou Z, Du X, Cai Z, Song X, Zhang H,
Mizuno T, Suzuki E, Yee MR, Berezov A, Murali R, et al: Structure
of Sad1-UNC84 homology (SUN) domain defines features of molecular
bridge in nuclear envelope. J Biol Chem. 287:5317–5326. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Starr DA and Fridolfsson HN: Interactions
between nuclei and the cytoskeleton are mediated by SUN-KASH
nuclear-envelope bridges. Annu Rev Cell Dev Biol. 26:421–444. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Stewart-Hutchinson PJ, Hale CM, Wirtz D
and Hodzic D: Structural requirements for the assembly of LINC
complexes and their function in cellular mechanical stiffness. Exp
Cell Res. 314:1892–1905. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Crisp M, Liu Q, Roux K, Rattner JB,
Shanahan C, Burke B, Stahl PD and Hodzic D: Coupling of the nucleus
and cytoplasm: Role of the LINC complex. J Cell Biol. 172:41–53.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Haque F, Lloyd DJ, Smallwood DT, Dent CL,
Shanahan CM, Fry AM, Trembath RC and Shackleton S: SUN1 interacts
with nuclear lamin A and cytoplasmic nesprins to provide a physical
connection between the nuclear lamina and the cytoskeleton. Mol
Cell Biol. 26:3738–3751. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schmitt J, Benavente R, Hodzic D, Höög C,
Stewart CL and Alsheimer M: Transmembrane protein Sun2 is involved
in tethering mammalian meiotic telomeres to the nuclear envelope.
Proc Natl Acad Sci USA. 104:7426–7431. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee KK, Starr D, Cohen M, Liu J, Han M,
Wilson KL and Gruenbaum Y: Lamin-dependent localization of UNC-84,
a protein required for nuclear migration in Caenorhabditis
elegans. Mol Biol Cell. 13:892–901. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Starr DA and Han M: ANChors away: An actin
based mechanism of nuclear positioning. J Cell Scie. 116:211–216.
2003. View Article : Google Scholar
|
|
34
|
Ostlund C, Folker ES, Choi JC, Gomes ER,
Gundersen GG and Worman HJ: Dynamics and molecular interactions of
linker of nucleoskeleton and cytoskeleton (LINC) complex proteins.
J Cell Sci. 122:4099–4108. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chistiakov DA, Sobenin IA, Orekhov AN and
Bobryshev YV: Human miR-221/222 in physiological and
atherosclerotic vascular remodeling. Biomed Res Int.
2015:3545172015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Song J, Ouyang Y, Che J, Li X, Zhao Y,
Yang K, Zhao X, Chen Y, Fan C and Yuan W: Potential value of
miR-221/222 as diagnostic, prognostic and therapeutic biomarkers
for diseases. Front Immunol. 8:562017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu Z, Wu L, Weng D, Xu D, Geng J and Zhao
F: Reduced expression of lamin A/C correlates with poor
histological differentiation and prognosis in primary gastric
carcinoma. J Exp Clin Cancer Res. 28:82009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Broers JL, Raymond Y, Rot MK, Kuijpers H,
Wagenaar SS and Ramaekers FC: Nuclear A-type lamins are
differentially expressed in human lung cancer subtypes. Am J
Pathol. 143:211–220. 1993.PubMed/NCBI
|
|
39
|
Stadelmann B, Khandjian E, Hirt A, Lüthy
A, Weil R and Wagner HP: Repression of nuclear lamin A and C gene
expression in human acute lymphoblastic leukemia and non-Hodgkin's
lymphoma cells. Leuk Res. 14:815–821. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Agrelo R, Setien F, Espada J, Artiga MJ,
Rodriguez M, Pérez-Rosado A, Sanchez-Aguilera A, Fraga MF, Piris MA
and Esteller M: Inactivation of the lamin A/C gene by CpG island
promoter hypermethylation in hematologic malignancies, and its
association with poor survival in nodal diffuse large B-cell
lymphoma. J Clin Oncol. 23:3940–3947. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Willis ND, Cox TR, Rahman-Casañs SF, Smits
K, Przyborski SA, van den Brandt P, van Engeland M, Weijenberg M,
Wilson RG, de Bruïne A and Hutchison CJ: Lamin A/C is a risk
biomarker in colorectal cancer. PLoS One. 3:e29882008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kong L, Schäfer G, Bu H, Zhang Y and
Klocker H: Lamin A/C protein is overexpressed in tissue-invading
prostate cancer and promotes prostate cancer cell growth, migration
and invasion through the PI3K/AKT/PTEN pathway. Carcinogenesis.
33:751–759. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tilli CM, Ramaekers FC, Broers JL,
Hutchison CJ and Neumann HA: Lamin expression in normal human skin,
actinic keratosis, squamous cell carcinoma and basal cell
carcinoma. Br J Dermatol. 148:102–109. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Venables RS, McLean S, Luny D, Moteleb E,
Morley S, Quinlan RA, Lane EB and Hutchison CJ: Expression of
individual lamins in basal cell carcinomas of the skin. Br J
Cancer. 84:512–519. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ellenbroek SI and van Rheenen J: Imaging
hallmarks of cancer in living mice. Nat Rev Cancer. 14:406–418.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ciccia A and Elledge SJ: The DNA damage
response: Making it safe to play with knives. Mol Cell. 40:179–204.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Paull TT, Rogakou EP, Yamazaki V,
Kirchgessner CU, Gellert M and Bonner WM: A critical role for
histone H2AX in recruitment of repair factors to nuclear foci after
DNA damage. Curr Biol. 10:886–895. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Harper JW and Elledge SJ: The DNA damage
response: Ten years after. Mol Cell. 28:739–745. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sobol RW, Horton JK, Kühn R, Gu H, Singhal
RK, Prasad R, Rajewsky K and Wilson SH: Requirement of mammalian
DNA polymerase-beta in base-excision repair. Nature. 379:183–186.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Majidinia M and Yousefi B: DNA repair and
damage pathways in breast cancer development and therapy. DNA
Repair (Amst). 54:22–29. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu
T, Xu R and Han M: SUN1/2 and Syne/Nesprin-1/2 complexes connect
centrosome to the nucleus during neurogenesis and neuronal
migration in mice. Neuron. 64:173–187. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Davidson D, Amrein L, Panasci L and Aloyz
R: Small molecules, inhibitors of DNA-PK, targeting DNA repair, and
beyond. Front Pharmacol. 4:52013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Stinson S, Lackner MR, Adai AT, Yu N, Kim
HJ, O'Brien C, Spoerke J, Jhunjhunwala S, Boyd Z, Januario T, et
al: miR-221/222 targeting of trichorhinophalangeal 1 (TRPS1)
promotes epithelial-to-mesenchymal transition in breast cancer. Sci
Signal. 4:pt52011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hwang MS, Yu N, Stinson SY, Yue P, Newman
RJ, Allan BB and Dornan D: miR-221/222 targets adiponectin receptor
1 to promote the epithelial-to-mesenchymal transition in breast
cancer. PLoS One. 8:e665022013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Li Y, Liang C, Ma H, Zhao Q, Lu Y, Xiang
Z, Li L, Qin J, Chen Y, Cho WC, et al: miR-221/222 promotes S-phase
entry and cellular migration in control of basal-like breast cance.
Molecules. 19:7122–7137. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gan R, Yang Y, Yang X, Zhao L, Lu J and
Meng QH: Downregulation of miR-221/222 enhances sensitivity of
breast cancer cells to tamoxifen through upregulation of TIMP3.
Cancer Gene Ther. 21:290–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Pichiorri F, Palmieri D, De Luca L,
Consiglio J, You J, Rocci A, Talabere T, Piovan C, Lagana A,
Cascione L, et al: In vivo NCL targeting affects breast cancer
aggressiveness through miRNA regulation. J Exp Med. 210:951–968.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Falkenberg N, Anastasov N, Rappl K,
Braselmann H, Auer G, Walch A, Huber M, Höfig I, Schmitt M, Höfler
H, et al: MiR-221/-222 differentiate prognostic groups in advanced
breast cancers and influence cell invasion. Br J Cancer.
109:2714–2723. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Gramantieri L, Fornari F, Ferracin M,
Veronese A, Sabbioni S, Calin GA, Grazi GL, Croce CM, Bolondi L and
Negrini M: MicroRNA-221 targets Bmf in hepatocellular carcinoma and
correlates with tumor multifocality. Clin Cancer Res. 15:5073–5081.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Bae HJ, Jung KH, Eun JW, Shen Q, Kim HS,
Park SJ, Shin WC, Yang HD, Park WS, Lee JY and Nam SW: MicroRNA-221
governs tumor suppressor HDAC6 to potentiate malignant progression
of liver cancer. J Hepatol. 63:408–419. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Callegari E, Elamin BK, Giannone F,
Milazzo M, Altavilla G, Fornari F, Giacomelli L, D'Abundo L,
Ferracin M, Bassi C, et al: Liver tumorigenicity promoted by
microRNA-221 in a mouse transgenic model. Hepatology. 56:1025–1033.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Li J, Wang Y, Yu W, Chen J and Luo J:
Expression of serum miR-221 in human hepatocellular carcinoma and
its prognostic significance. Biochem Biophys Res Commun. 406:70–73.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Duan M, Yao H, Hu G, Chen X, Lund AK and
Buch S: HIV Tat induces expression of ICAM-1 in HUVECs:
implications for miR-221/-222 in HIV-associated cardiomyopathy.
PLoS One. 8:e601702013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Sarkar S, Dubaybo H, Ali S, Goncalves P,
Kollepara SL, Sethi S, Philip PA and Li Y: Down-regulation of
miR-221 inhibits proliferation of pancreatic cancer cells through
up-regulation of PTEN, p27(kip1), p57(kip2), and PUMA. Am J Cancer
Res. 3:465–477. 2013.PubMed/NCBI
|
|
66
|
Passadouro M, Pedroso de Lima MC and
Faneca H: MicroRNA modulation combined with sunitinib as a novel
therapeutic strategy for pancreatic cancer. Int J Nanomedicine.
9:3203–3217. 2014.PubMed/NCBI
|
|
67
|
Tanaka R, Tomosugi M, Horinaka M, Sowa Y
and Sakai T: Metformin causes G1-phase arrest via down-regulation
of MiR-221 and enhances TRAIL sensitivity through DR5 Up-regulation
in pancreatic cancer cells. PLoS One. 10:e01257792015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Lee C, He H, Jiang Y, Di Y, Yang F, Li J,
Jin C and Fu D: Elevated expression of tumor miR-222 in pancreatic
cancer is associated with Ki67 and poor prognosis. Med Oncol.
30:7002013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kim YK, Yu J, Han TS, Park SY, Namkoong B,
Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK and Kim VN: Functional links
between clustered microRNAs: suppression of cell-cycle inhibitors
by microRNA clusters in gastric cancer. Nucleic Acids Res.
37:1672–1681. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Liu W, Song N, Yao H, Zhao L, Liu H and Li
G: miR-221 and miR-222 simultaneously target RECK and regulate
growth and invasion of gastric cancer cells. Med Sci Monit.
21:2718–2725. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F,
Xiao Y, Guang-Xiu W, Zhi-Fan J, Pei-Yu P, Qing-Yu Z and Chun-Sheng
K: MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell
proliferation and radioresistance by targeting PTEN. BMC Cancer.
10:3672010. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Song MY, Pan KF, Su HJ, Zhang L, Ma JL, Li
JY, Yuasa Y, Kang D, Kim YS and You WC: Identification of serum
microRNAs as novel non-invasive biomarkers for early detection of
gastric cancer. PLoS One. 7:e336082012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Fu Z, Qian F, Yang X, Jiang H, Chen Y and
Liu S: Circulating miR-222 in plasma and its potential diagnostic
and prognostic value in gastric cancer. Med Oncol. 31:1642014.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Sun K, Wang W, Zeng JJ, Wu CT, Lei ST and
Li GX: MicroRNA-221 inhibits CDKN1C/p57 expression in human
colorectal carcinoma. Acta Pharmacol Sin. 32:375–384. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Qin J and Luo M: MicroRNA-221 promotes
colorectal cancer cell invasion and metastasis by targeting RECK.
FEBS Lett. 588:99–104. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Liu S, Sun X, Wang M, Hou Y, Zhan Y, Jiang
Y, Liu Z, Cao X, Chen P, Liu Z, et al: A microRNA 221- and
222-mediated feedback loop maintains constitutive activation of
NFκB and STAT3 in colorectal cancer cells. Gastroenterology.
147:847–859, e811. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Xue Q, Sun K, Deng HJ, Lei ST, Dong JQ and
Li GX: Anti-miRNA-221 sensitizes human colorectal carcinoma cells
to radiation by upregulating PTEN. World J Gastroenterol.
19:9307–9317. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Pu XX, Huang GL, Guo HQ, Guo CC, Li H, Ye
S, Ling S, Jiang L, Tian Y and Lin TY: Circulating miR-221 directly
amplified from plasma is a potential diagnostic and prognostic
marker of colorectal cancer and is correlated with p53 expression.
J Gastroenterol Hepatol. 25:1674–1680. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhang C, Zhang J, Hao J, Shi Z, Wang Y,
Han L, Yu S, You Y, Jiang T, Wang J, et al: High level of
miR-221/222 confers increased cell invasion and poor prognosis in
glioma. J Transl Med. 10:1192012. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Medina R, Zaidi SK, Liu CG, Stein JL, van
Wijnen AJ, Croce CM and Stein GS: MicroRNAs 221 and 22χ2 bypass
quiescence and compromise cell survival. Cancer Res. 68:2773–2780.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhang C, Kang C, You Y, Pu P, Yang W, Zhao
P, Wang G, Zhang A, Jia Z, Han L and Jiang H: Co-suppression of
miR-221/222 cluster suppresses human glioma cell growth by
targeting p27kip1 in vitro and in vivo. Int J Oncol. 34:1653–1660.
2009.PubMed/NCBI
|
|
82
|
Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han
L, Jia ZF, Yang WD, Wang GX, Jiang T, You YP, et al: MiR-221 and
miR-222 target PUMA to induce cell survival in glioblastoma. Mol
Cancer. 9:2292010. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Quintavalle C, Garofalo M, Zanca C, Romano
G, Iaboni M, del Basso De Caro M, Martinez-Montero JC, Incoronato
M, Nuovo G, Croce CM and Condorelli G: miR-221/222 overexpession in
human glioblastoma increases invasiveness by targeting the protein
phosphate PTPµ. Oncogene. 31:858–868. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Chen L, Zhang J, Han L, Zhang A, Zhang C,
Zheng Y, Jiang T, Pu P, Jiang C and Kang C: Downregulation of
miR-221/222 sensitizes glioma cells to temozolomide by regulating
apoptosis independently of p53 status. Oncol Rep. 27:854–860.
2012.PubMed/NCBI
|
|
85
|
Li W, Guo F, Wang P, Hong S and Zhang C:
miR-221/222 confers radioresistance in glioblastoma cells through
activating Akt independent of PTEN status. Curr Mol Med.
14:185–195. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Di Martino MT, Gullà A, Cantafio ME,
Lionetti M, Leone E, Amodio N, Guzzi PH, Foresta U, Conforti F,
Cannataro M, et al: In vitro and in vivo anti-tumor activity of
miR-221/222 inhibitors in multiple myeloma. Oncotarget. 4:242–255.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Di Martino MT, Gullà A, Gallo Cantafio ME,
Altomare E, Amodio N, Leone E, Morelli E, Lio SG, Caracciolo D,
Rossi M, et al: In vitro and in vivo activity of a novel locked
nucleic acid (LNA)-inhibitor-miR-221 against multiple myeloma
cells. PLoS One. 9:e896592014. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Gullà A, Di Martino MT, Gallo Cantafio ME,
Morelli E, Amodio N, Botta C, Pitari MR, Lio SG, Britti D, Stamato
MA, et al: A 13 mer LNA-i-miR-221 inhibitor restores drug
sensitivity in melphalan-refractory multiple myeloma cells. Clin
Cancer Res. 22:1222–1233. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Huang JJ, Yu J, Li JY, Liu YT and Zhong
RQ: Circulating microRNA expression is associated with genetic
subtype and survival of multiple myeloma. Med Oncol. 29:2402–2408.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Kanemaru H, Fukushima S, Yamashita J,
Honda N, Oyama R, Kakimoto A, Masuguchi S, Ishihara T, Inoue Y,
Jinnin M and Ihn H: The circulating microRNA-221 level in patients
with malignant melanoma as a new tumor marker. J Dermatol Sci.
61:187–193. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Felicetti F, De Feo A, Coscia C, Puglisi
R, Pedini F, Pasquini L, Bellenghi M, Errico MC, Pagani E and Carè
A: Exosome-mediated transfer of miR-222 is sufficient to increase
tumor malignancy in melanoma. J Transl Med. 14:562016. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Alamolhodaei NS, Behravan J, Mosaffa F and
Karimi G: MiR 221/222 as new players in tamoxifen resistance. Curr
Pharm Des. 22:6946–6955. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Linher-Melville K and Singh G: The complex
roles of STAT3 and STAT5 in maintaining redox balance: Lessons from
STAT-mediated xCT expression in cancer cells. Mol Cell Endocrinol.
451:40–52. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Li F, He X, Ye D, Lin Y, Yu H, Yao C,
Huang L, Zhang J, Wang F, Xu S, et al: NADP(+)-IDH mutations
promote hypersuccinylation that impairs mitochondria respiration
and induces apoptosis resistance. Mol Cell. 60:661–675. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Lai CC, Lin PM, Lin SF, Hsu CH, Lin HC, Hu
ML, Hsu CM and Yang MY: Altered expression of SIRT gene family in
head and neck squamous cell carcinoma. Tumour Biol. 34:1847–1854.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Kim DH, Kwak Y, Kim ND and Sim T:
Antitumor effects and molecular mechanisms of ponatinib on
endometrial cancer cells harboring activating FGFR2 mutations.
Cancer Biol Ther. 17:65–78. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Xiangyun Y, Xiaomin N, Linping G, Yunhua
X, Ziming L, Yongfeng Y, Zhiwei C and Shun L: Desuccinylation of
pyruvate kinase M2 by SIRT5 contributes to antioxidant response and
tumor growth. Oncotarget. 8:6984–6993. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Osborne B, Bentley NL, Montgomery MK and
Turner N: The role of mitochondrial sirtuins in health and disease.
Free Radic Biol Med. 100:164–174. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Lu W, Zuo Y, Feng Y and Zhang M: SIRT5
facilitates cancer cell growth and drug resistance in non-small
cell lung cancer. Tumour Biol. 35:10699–10705. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Kyrylenko S, Kyrylenko O, Suuronen T and
Salminen A: Differential regulation of the Sir2 histone deacetylase
gene family by inhibitors of class I and II histone deacetylases.
Cell Mol Life Sci. 60:1990–1997. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Ding S, Khoury-Hanold W, Iwasaki A and
Robek MD: Epigenetic reprogramming of the type III interferon
response potentiates antiviral activity and suppresses tumor
growth. PLoS Biol. 12:e10017582014. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Vanhaecke T, Papeleu P, Elaut G and
Rogiers V: Trichostatin A-like hydroxamate histone deacetylase
inhibitors as therapeutic agents: toxicological point of view. Curr
Med Chem. 11:1629–1643. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Zhou W, Liotta LA and Petricoin EF: The
Warburg effect and mass spectrometry-based proteomic analysis.
Cancer Genomics Proteomics. 14:211–218. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Sica A, Strauss L, Consonni FM, Travelli
C, Genazzani A and Porta C: Metabolic regulation of suppressive
myeloid cells in cancer. Cytokine Growth Factor Rev. 35:27–35.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Cairns RA: Drivers of the Warburg
phenotype. Cancer J. 21:56–61. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
He X, Li C, Ke R, Luo L and Huang D:
Down-regulation of adenosine monophosphate-activated protein kinase
activity: A driver of cancer. Tumour Biol. 39:10104283176975762017.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo
F, Lyssiotis CA, Aldape K, Cantley LC and Lu Z: ERK1/2-dependent
phosphorylation and nuclear translocation of PKM2 promotes the
Warburg effect. Nat Cell Biol. 14:1295–1304. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Liu J, Zhang C, Wu R, Lin M, Liang Y, Liu
J, Wang X, Yang B and Feng Z: RRAD inhibits the Warburg effect
through negative regulation of the NF-κB signaling. Oncotarget.
6:14982–14992. 2015.PubMed/NCBI
|
|
109
|
Dueregger A, Schöpf B, Eder T, Höfer J,
Gnaiger E, Aufinger A, Kenner L, Perktold B, Ramoner R, Klocker H
and Eder IE: Differential utilization of dietary fatty acids in
benign and malignant cells of the prostate. PLoS One.
10:e01357042015. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
He JX, Yang CH and Miao ZH:
Poly(ADP-ribose) polymerase inhibitors as promising cancer
therapeutics. Acta Pharmacol Sin. 31:1172–1180. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Livraghi L and Garber JE: PARP inhibitors
in the management of breast cancer: current data and future
prospects. BMC Med. 13:1882015. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Evans T and Matulonis U: PARP inhibitors
in ovarian cancer: Evidence, experience and clinical potential.
Ther Adv Med Oncol. 9:253–267. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Rajawat J, Shukla N and Mishra DP:
Therapeutic targeting of poly(ADP-Ribose) polymerase-1 (PARP1) in
cancer: Current developments, therapeutic strategies, and future
opportunities. Med Res Rev. 37:1461–1491. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Vici P, Mariani L, Pizzuti L, Sergi D, Di
Lauro L, Vizza E, Tomao F, Tomao S, Mancini E, Vincenzoni C, et al:
Emerging biological treatments for uterine cervical carcinoma. J
Cancer. 5:86–97. 2014. View Article : Google Scholar : PubMed/NCBI
|