Introduction
With an estimated 21,290 novel cases of epithelial
ovarian cancer (EOC) and 14,180 cases of associated mortality in
2015, EOC is the fifth leading cause of cancer-associated mortality
in women in the USA (1). Owing to a
lack of effective biomarkers and disease-specific symptoms,
particularly for early-stage EOC, a marked proportion of patients
are not diagnosed until an advanced stage. Cytoreductive surgery
with cisplatin-based chemotherapy is the preferred treatment.
However, resistance to chemotherapy leads to a dismal prognosis
(2,3).
Therefore, an extensive understanding of the molecular mechanisms
in EOC is crucial.
Previous studies have emphasized that epigenetic
modifications, particularly DNA hypermethylation, may be among the
molecular mechanisms underlying acquired resistance to cisplatin
(4,5).
Multiple DNA methylation changes in the cancer methylome are
associated with the acquisition of drug resistance (5–7). A
significant upregulation of DNA methyltransferases (DNMTs) has been
observed in cisplatin-resistant ovarian cancer (8). Three DNMTs have been identified in
humans: DNMT1, DNMT3A and DNMT3B. DNMT1 is the most abundant DNMT
in mammalian cells, and is the key enzyme for the maintenance of
hemimethylated DNA during DNA replication and the development and
differentiation of somatic cells (9);
it serves an important function in the silencing of several tumor
suppressor genes and accumulates in the promoter regions of these
genes (10–12). Decitabine is one of the most widely
used DNMT inhibitors in research and in cancer therapy. Although it
can have a major impact in combination with other chemotherapeutic
drugs, its narrow therapeutic window and effective dosage limit its
clinical use (13).
MicroRNAs (miRNAs/miRs) are a class of short
non-coding RNAs, between 19 and 25 nucleotides in length, that
regulate gene expression by targeting mRNAs and that have functions
in multiple physiological and pathological functions (14). It has been identified that ~30% of
genes are regulated by miRNAs (15),
and >60% of protein-coding genes are computationally predicted
as being miRNA targets (16). miRNAs
may be controlled or may be used to control target genes in
aberrant DNA hypermethylation.
One particular miRNA family, the miRNA-200 family,
regulates DNA methylation in a number of types of cancer (12,17).
Ectopic overexpression of the two miRNAs increased the sensitivity
of the resistant ovarian cancer cells to cisplatin by promoting
apoptosis by directly suppressing DNMT3A and DNMT3B, and also
indirectly decreasing the expression of DNMT1 via the
downregulation of specificity protein (Sp)1, a transactivating
factor of the DNMT1 gene (12,18). This
provides attractive novel avenues for the development of
therapeutic approaches based on the molecules involved in DNA
methylation.
Materials and methods
Ethical approval
The present study was approved by the Institutional
Review Board of The Second Affiliated Hospital of South China
University (Hengyang, China). All tissues were obtained following
written informed consent from the patients.
Patient samples and cell lines
Frozen human primary ovarian tumor and corresponding
adjacent non-cancerous tissues used in the present study were
obtained from patients diagnosed between October 2007 to September
2014 who underwent radical resection at The Second Affiliated
Hospital, University of South China. The average age was 55±6.5
years. Patients who received some form of chemotherapy or
radiotherapy prior to surgery were excluded from the study. The
human ovarian cancer cell lines and human immortalized ovarian
surface epithelial (HIOSE-80 and MCC-3) cell lines used were
described previously (19). The human
ovarian cancer cell lines SKOV3, A2780CP and A2780, and human
ovarian surface epithelial cell lines were obtain from the American
Type Culture Collection (Manassas, VA, USA). OV119 cells were
purchased from the Beijing Institute for Cancer Research (Beijing,
China). HIOSE-80 cells were cultured with 199/MDCB 105 (1:1) medium
(Sigma; Merck KGaA, Darmstadt, Germany) supplemented with 5% fetal
bovine serum (FBS). All other cells were cultured in Dulbecco's
modified Eagle's medium (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% FBS and 1%
penicillin/streptomycin.
Transfections and luciferase
assay
Cells were seeded in 6-well plates at
1×105 cells/well followed by culture for 24 h and
transfection with 20 nmol/l miR-200b mimic,
5′-CAUCUUACUGGGCAGCAUUGGA-3′, miR-200c mimic,
5′-CGUCUUACCCAGCAGUGUUUGG-3′ or negative control mimics (NC) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). The NC consisted of synthetic scrambled
double-stranded oligonucleotides that do not target any mRNA. The
effect of the mimics was determined in triplicate at 24 h
post-transfection.
MTT assay
Non-transfected or transfected cells were re-seeded
in 96-well plates; 24 h later, freshly prepared cisplatin (Sigma;
Merck KGaA) at 20 µM cisplatin treatment was added, and the cells
were cultured for an additional 48 h. Cell viability was determined
using an MTT assay (Thermo Fisher Scientific, Inc.). The resulting
absorbance of each well was determined at 492 nm on a
spectrophotometer. At least three independent experiments were
performed in quadruplicate.
In situ hybridization (ISH) and
immunohistochemistry (IHC) assays
ISH procedures were carried out as described
previously (20). miR-200b and
miR-200c miRCURY locked nucleic acid custom detection probes
miR-200b mimic, 5′-CAUCUUACUGGGCAGCAUUGGA-3′, miR-200c mimic,
5′-CGUCUUACCCAGCAGUGUUUGG-3′ (Qiagen, Inc., Valencia, CA, USA) were
used for ISH. Hybridization, washing and scanning were performed
according to the manufacturer's protocol. Paraffin-embedded blocks
of tumors were sectioned into 5-µm slices, and the IHC protocol was
performed as described previously (21). Staining intensity was scored as
following: >0 and ≤1, no staining; >1 and ≤2, weak staining;
>2 and ≤3, medium staining; and >3 and ≤4, strong staining.
The proportion of positive cells was divided into four groups:
0–25, 26–50, 51–75 and 76–100%. The final score was determined by
multiplying the intensity score and the quantity score; the maximum
was 4, and the minimum was 0. All specimens were evaluated by at
least two blinded pathologists. Expression scores ≥2 were
classified as high expression, and scores <2 were classified as
low expression.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). RT of specific miRNAs (from 10 ng of total RNA) was
performed using the real-time loop primers for each type of miRNAs
and the TaqMan miRNA RT kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocols. cDNA
obtained from this step was used for quantitative TaqMan PCR using
the real-time primers provided. Reverse-transcribed cDNA was
synthesized with random primers or miRNA-specific stem-loop
primers. LightCycler Fast Start DNA Master SYBR Green Mix (Roche
Diagnostics GmbH, Mannheim, Germany) was added to each PCR reaction
along with cDNA and 1 pmol primer in a total volume of 10 µl. The
primer sequence for miR-200b was as follows: Forward,
5′-CACACTGAAATCCTGTCAGCTTC-3′ and reverse, 5-CTA ACT. The primer
sequence for miR-200b mimics was sense, 5′-UUCUCCGAACGUGUCACGUTT-3′
and anti-sense, 5′-ACGUGACACGUUCGGAGAATT-3′. The PCR thermocycling
conditions were as follows: One cycle at 95°C for 3 min, followed
by 40 cycles at 95°C for 12 sec and 62°C for 35 sec, 94°C for 5
min, 50°C for 10 min and finally 1 cycle at 62–95°C for 15 sec. The
relative expression level of each RNA was quantified using the
2−ΔΔCq method (22). CR
was performed in triplicate using a standard SYBR Green PCR kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol.
Western blot analysis
The cells were lysed in radioimmunoprecipitation
assay buffer in the presence of proteinase inhibitor cocktail
(Sigma-Aldrich; Merck KGaA) on ice. The lysates were centrifuged at
12,000 × g, for 10 min at 4°C, and SDS gel loading buffer was
added. A total of 20 µg of protein were separated by 10% SDS-PAGE
and transferred onto polyvinylidene difluoride membranes (EMD
Millipore, Billerica, MA, USA). The membranes were blocked with 10%
skimmed milk powder with PBS followed by incubation for 1 h at room
temperature with the following primary antibodies (1:500):
Anti-DNMT1 (cat. no. sc-10222; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA), anti-DNMT3A (cat. no. sc-20703; Santa Cruz
Biotechnology, Inc.), anti-DNMT3B (cat. no. sc-10236; Santa Cruz
Biotechnology, Inc.) and anti-GAPDH (cat. no., SF-126; EMD
Millipore). Following washing 3 times with 0.1% PBS, the membranes
were incubated with rabbit anti-mouse HRP (cat. no., BA1058) and
goat anti-rabbit HRP (cat. no., BA1058) secondary antibodies
purchased from Wuhan Boster Biological Technology, Ltd., (Wuhan,
China) at a dilution of 1:5,000 for 1 h at room temperature.
Protein band were visualized using enhanced chemiluminescence
western blot detection reagents (New England and Biolabs, Inc.,
Ipswich, MA, USA), according to the manufacturer's protocols. The
protein levels were normalized to the levels of GAPDH and quantifed
by a Bio Image Intelligent Quantifier 1-D 2.2.1 (Nikon Corporation,
Tokyo, Japan), according to the manufacturer's protocols.
Flow cytometry-based apoptosis
Cells were cultured in cisplatin-containing medium
and incubated for 48 h at room temperature. Following incubation,
the cells were harvested and stained with annexin V-fluorescein
isothiocyanate and propidium iodide. The mixture was incubated at
room temperature in the dark for 15 min and analyzed by
fluorescence-activated cell sorting (FACS), using BD FACSCanto I.
Flowjo 7.6 software (BD Biosciences, Franklin Lakes, NJ, USA).
Nude mouse model
A total of ~107 cells were injected
intraperitoneally into nude mice. Cells were transfected with 20
nmol/l miR-200b mimic, miR-200c mimic or NC using
Lipofectamine® 2000 respectively 48 h before being
injected into mice. After 1 week, cisplatin therapy was initiated
at a dose of 5 mg/kg twice weekly. Tumor size was calculated every
4 days according to the following formula: Tumor size=(π/6) ×
larger diameter × (smaller diameter)2. After 4
consecutive weeks of therapy, the mice were sacrificed, and the wet
weights of the tumors were determined.
Luciferase reporter assay
Cells were co-transfected with 500 ng
PGL3-DNMT1/DNMT3A/DNMT3B-WT or PGL3-DNMT1/DNMT3A/DNMT3B-Mut
constructs (both from GeneCopoeia, Inc., Rockville, MD, USA) with
miR-200b and miR-200c mimic or NC. Each sample was co-transfected
with pRL-TK plasmid (GeneCopoeia, Inc.) to determine the
transfection efficiency. Luciferase activity was examined 48 h
after transfection using a dual-luciferase reporter assay system
(Promega Corporation, Madison, WI, USA), according to the
manufacturer's protocol.
Statistical analysis
All statistical analyses were performed using SPSS
(version 17.0; (SPSS, Inc., Chicago, IL, USA). Differences between
samples were analyzed using two-tailed Student's t-test, and the
comparisons of multiple groups were performed by one-way analysis
of variance and Bonferroni's post hoc test. Spearman's correlation
analysis was used to evaluate the association between miR-200b/c
and DNMT1/3A/3B. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-200b and miR-200c are
downregulated in cisplatin-resistant ovarian cancer
The expression levels of miR-200b and miR-200c were
determined in 93 ovarian tumors and 32 normal ovarian tissues using
ISH, and it was identified that miR-200b and miR-200c were
downregulated in ovarian tumors compared with normal tissues. Among
the 93 patients with primary ovarian tumors, 35 had recurrent
(chemoresistant) ovarian cancer. ISH analysis revealed that the
levels of miR-200b and miR-200c were low or undetectable in these
recurrent ovarian cancer tissues (Table
I). These results indicated that miR-200b and miR-200c were
significantly downregulated in chemoresistant ovarian tumors
compared with normal tissues, and implied that miR-200b and
miR-200c may be involved in cisplatin resistance in patients with
ovarian cancer.
 | Table I.miRNA expression levels in different
tissues. |
Table I.
miRNA expression levels in different
tissues.
|
|
| miR-200b
expression | miR-200c
expression |
|---|
|
|
|
|
|
|---|
| Tissue
characteristic | No. of
patients | High | Low | P-value | High | Low | P-value |
|---|
| Normal | 32 | 27 | 5 | <0.001 | 24 | 8 | <0.005 |
| Tumor (total) | 93 | 36 | 57 |
| 41 | 52 |
|
| Tumor
(sensitive) | 58 | 34 | 24 | <0.001 | 36 | 22 | <0.001 |
| Tumor
(resistant) | 35 | 2 | 33 |
| 5 | 30 |
|
To further verify the biological function of
miR-200b and miR-200c in human ovarian cancer, their expression was
confirmed in ovarian cancer cell lines using RT-qPCR analysis. The
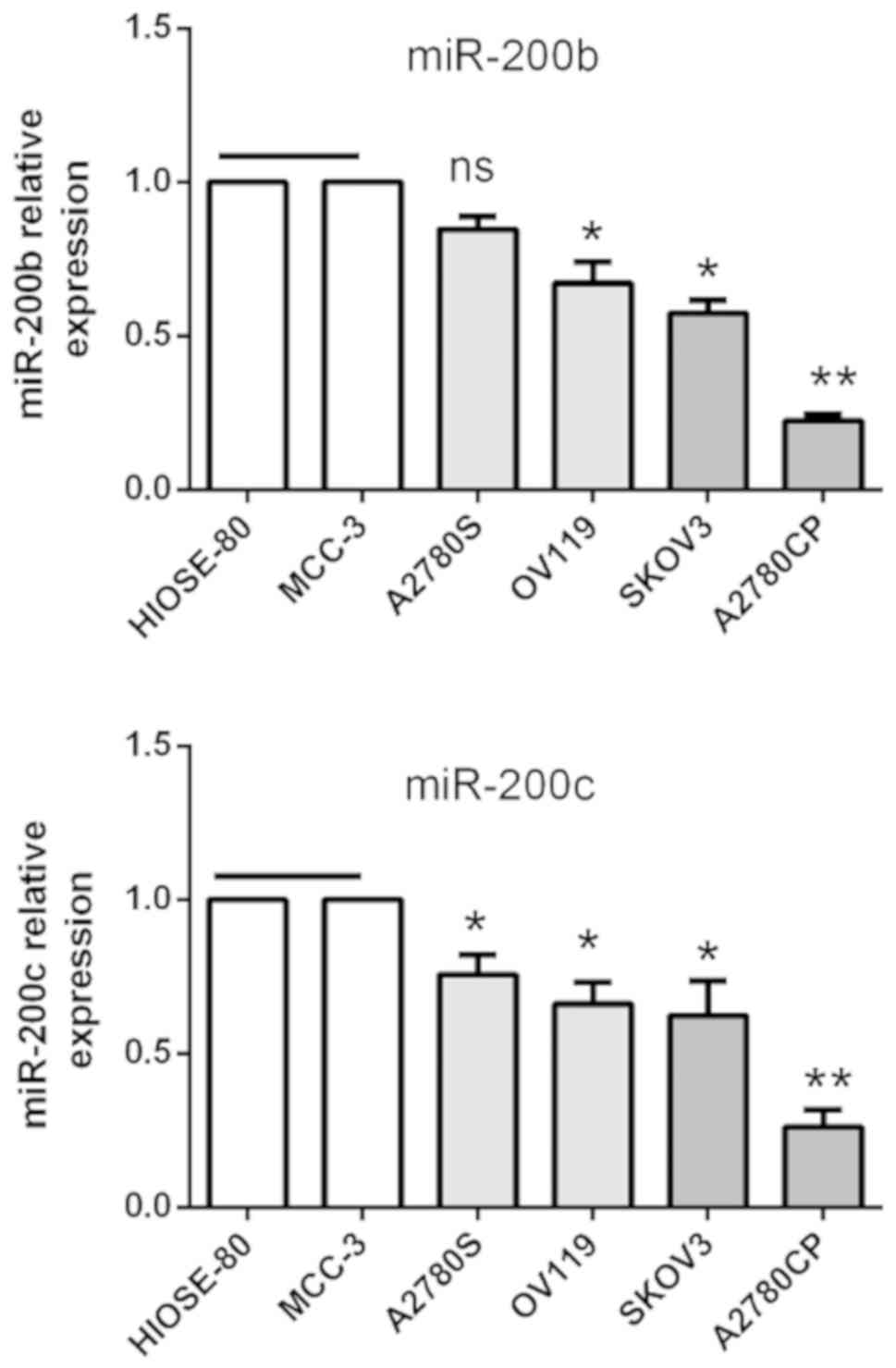
expression of miR-200b and miR-200c was downregulated in cancer
cell lines compared with immortalized human surface epithelial cell
lines HIOSE-80 and MCC-3, and A2780CP cells had the lowest
expression compared with the other ovarian cancer cell lines
investigated (Fig. 1). These results
suggested that the expression of miR-200b and miR-200c is
significantly altered in EOC, and that the low expression is
associated with poor prognosis of patients with EOC.
Overexpression of miR-200b and
miR-200c increases the cisplatin sensitivity of ovarian cancer
cells
It has been identified that A2780CP cells are
markedly resistant to cisplatin treatment, therefore it was
investigated whether miR-200b and miR-200c served any function in
the sensitivity of cisplatin treatment in EOC. miR-200b mimic,
miR-200c mimic and NC were transfected into the ovarian cancer cell
line A2780CP. Total RNA was extracted, and the transfection
efficiency was evaluated at 48 h after transfection (Fig. 2A). No significant alteration in cell
proliferation among the control, miR-200b and miR-200c groups was
identified. This was consistent with the results of a previous
study in hepatocellular carcinoma (23). The transfected cells were then exposed
to various concentrations of cisplatin for 48 h and viability was
determined using an MTT assay. It was identified that the
half-maximal inhibitory concentration (IC50) of
cisplatin was lower in cells that had been transfected with
miR-200b and miR-200c compared with the cells that had been
transfected with the NC (Fig. 2B).
These results indicated that miR-200b and miR-200c are involved in
cisplatin sensitivity in EOC cells, and the overexpression of
miR-200b and miR-200c markedly reversed the cisplatin sensitivity
of A2780CP cells. To test whether the effect of miR-200b and
miR-200c on cisplatin sensitivity was associated with the duration
of treatment, a cell survival assay was performed at 24, 48 and 72
h after transfection. The results indicated a stable effect of the
two miRNAs following transfection (Fig.
2C). Cell death was determined using annexin V staining and
FACS analysis.
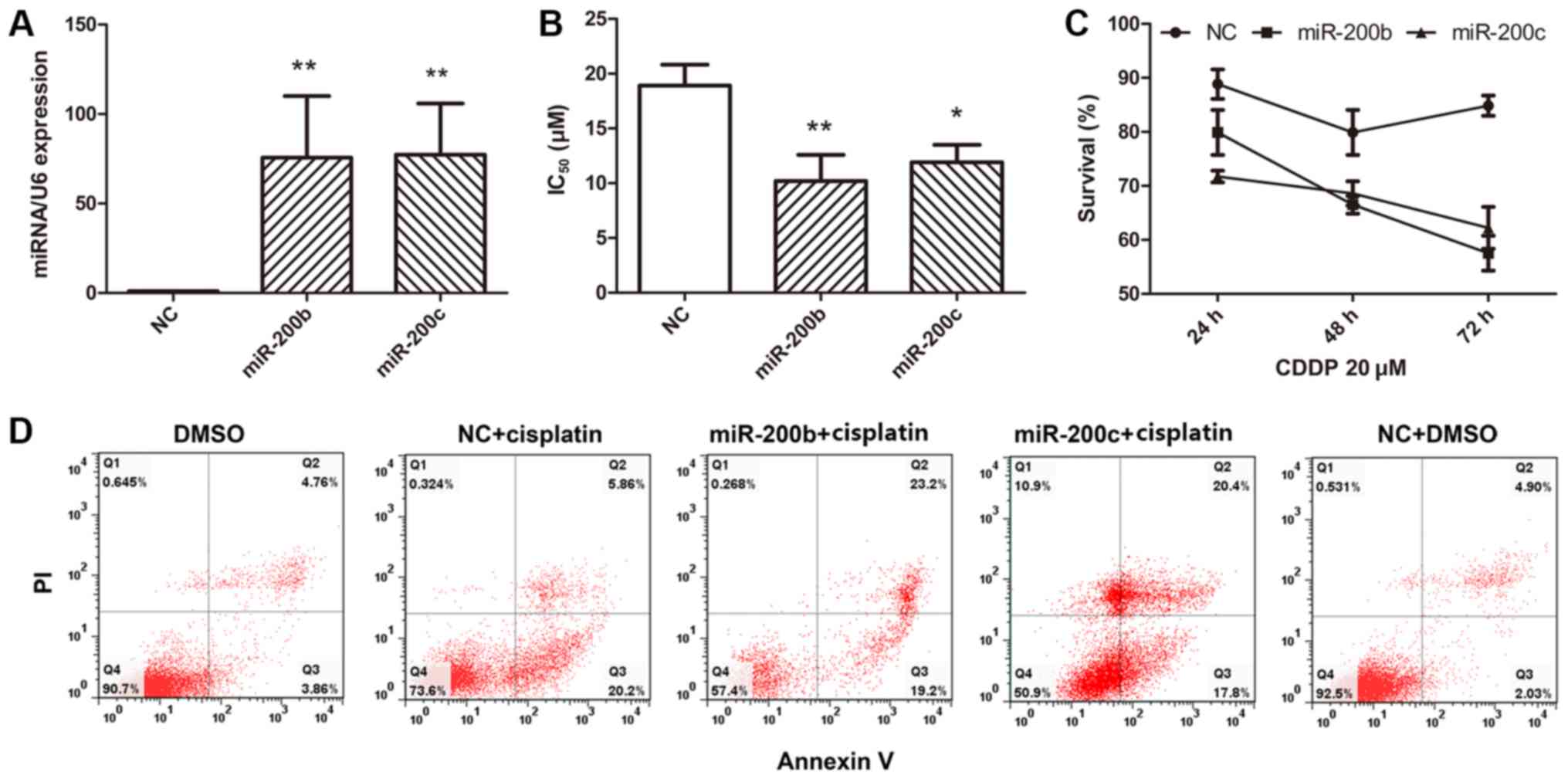 | Figure 2.Ectopic expression of miR-200b and
miR-200c reverses cisplatin resistance and induces apoptosis. (A)
A2780CP cells transfected with miR-200b mimic, miR-200c mimic or
NC. (B) Various concentrations of cisplatin were added, and the
viability of the A2780CP cells that were transfected with miR-200b
mimic, miR-200c mimic or NC was assessed using an MTT assay. The
IC50 of cisplatin is presented as the mean ± standard
deviation. (C) Cell survival was assayed at 24, 48 and 72 h after
transfection with 20 µM cisplatin treatment for another 48 h. (D)
NC- and miR-200b/miR-200c-transfected cells were treated with
cisplatin or DMSO, and after 48 h of treatment, cell apoptosis was
determined using flow cytometry. Results are presented as the mean
± standard error of the mean. *P<0.05, **P<0.01 vs. NC. NC,
negative control mimics; IC50, half-maximal inhibitory
concentration; DMSO, dimethylsulfoxide. |
Overexpression of miR-200b and miR-200c did not
affect cell death, and the overexpression of the two miRNAs in
these cells led to marked cell death when the cells were treated
with cisplatin (Fig. 2D). These
results indicated that miR-200b and miR-200c are involved in
cisplatin sensitivity in EOC cells.
Overexpression of miR-200b and
miR-200c reverses cisplatin resistance in vivo
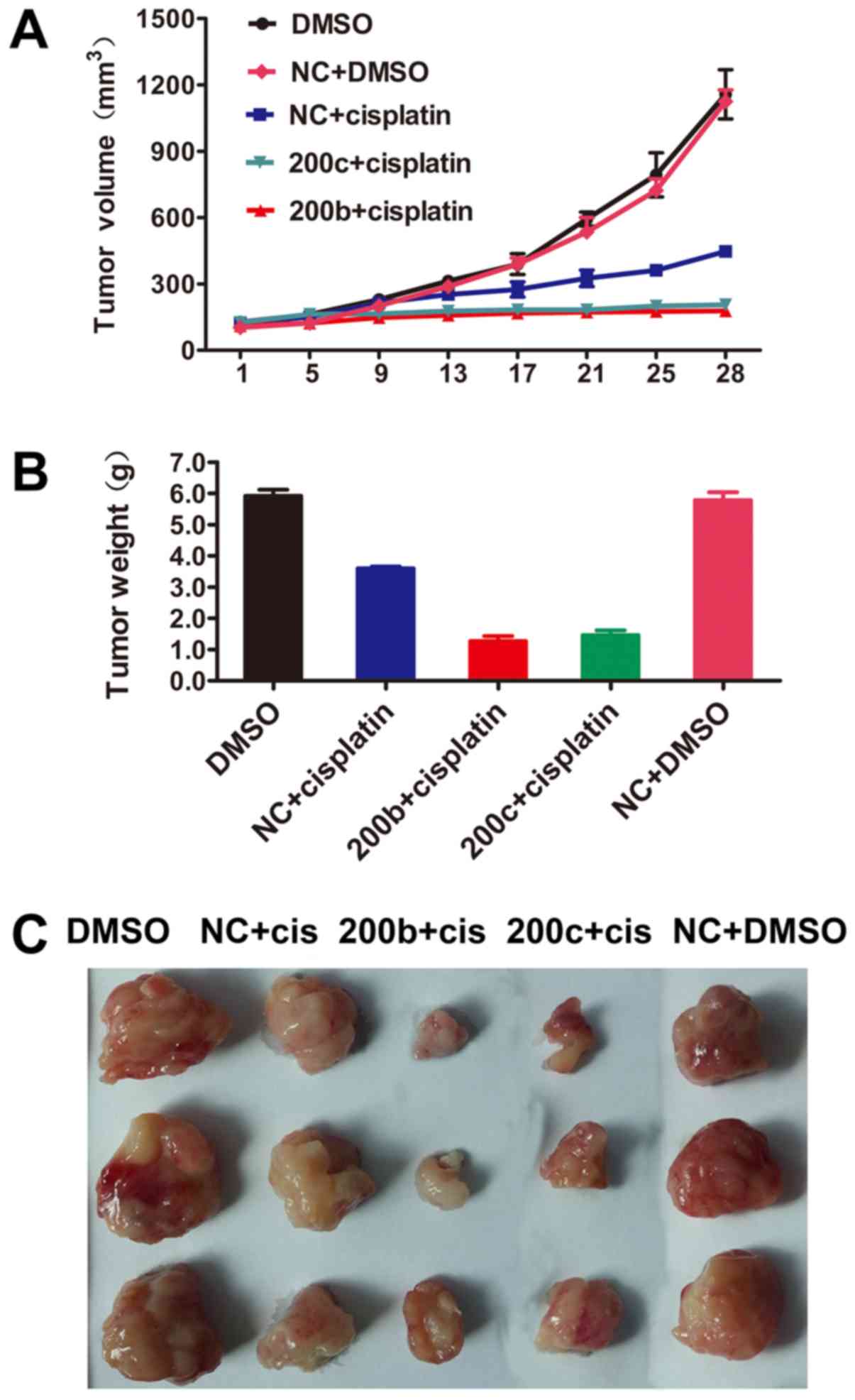
As the overexpression of miR-200b and miR-200c
significantly decreased the IC50 of cisplatin, the
therapeutic potential of miR-200b and miR-200c was investigated
in vivo. Nude mice were subcutaneously injected with A2780CP
cells transfected with miR-200b, miR-200c mimic or NC. Tumor
volumes were determined. Transduction of miR-200b and miR-200c did
not affect tumor growth in vivo. Cisplatin therapy was
administered at a dose of 5 mg/kg twice weekly. After 4 consecutive
weeks of treatment, the mice were sacrificed. The tumors were
excised, and the wet weights of the tumors were determined
(Fig. 3A-C). These results suggested
that miR-200b and miR-200c markedly sensitized tumor cells to
cisplatin treatment.
miR-200b and miR-200c directly target
DNMT3A and DNMT3B, and indirectly target DNMT1 in ovarian cancer
cells
Bioinformatics analyses predicted that DNMT3A and
DNMT3B are the potential targets of miR-200b and miR-200c, and it
has been demonstrated that DNMT3A and DNMT3B are true targets of in
human gastric cancer (24). A
dual-luciferase reporter assay was performed in A2780CP cells.
Co-transfection of miR-200b or miR-200c and the reporter plasmids
revealed that the introduction of miR-200b/miR-200c significantly
suppressed the luciferase activity of the vectors containing the
3′-untranslated region (UTR) of DNMT3A and DNMT3B, but not of those
containing mutations in the miRNA-binding site of DNMT3A/DNMTB. It
was identified that miR-200b and miR-200c directly targeted the
3′-UTR of DNMT3A and DNMT3B, respectively (Fig. 4A). Previous studies have revealed that
Sp1 positively regulates DNMT1 by increasing the activity of the
DNMT1 promoter (24). In the present
study, it was confirmed whether miR-200b and miR-200c downregulated
DNMT1 indirectly via Sp1 in ovarian cancer cells. To this end, the
3′-UTR of Sp1 was cloned into a luciferase reporter vector, and the
results revealed that miR-200b and miR-200c bound directly to Sp1
and markedly decreased luciferase activity (Fig. 4A). Therefore, the transfection of
miR-200b mimic and miR-200c mimic into A2780CP cells resulted in a
marked decrease in DNMT1, DNMT3A and DNMT3B protein levels compared
with the transfection of NC (Fig.
4B).
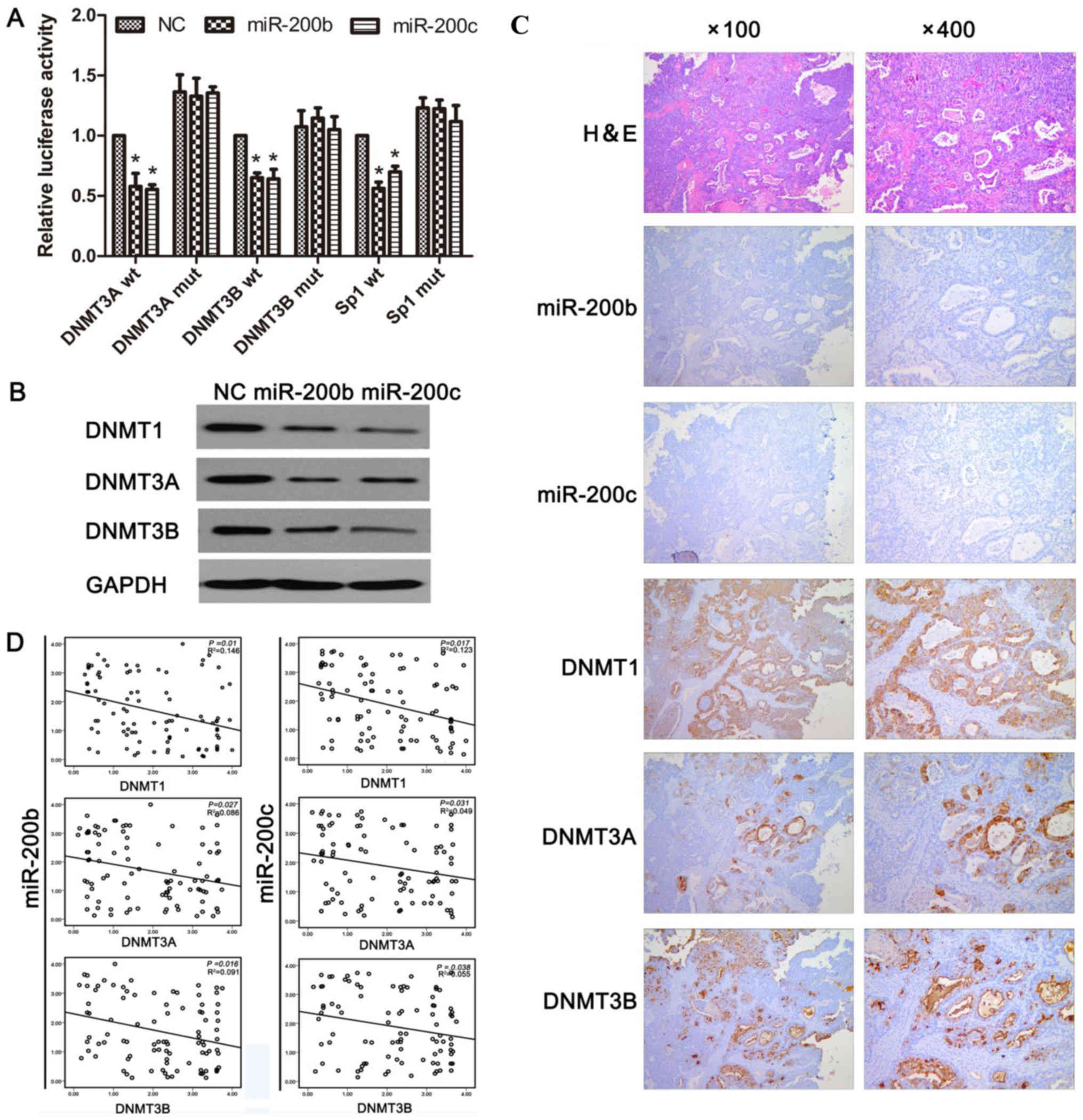 | Figure 4.miR-200b and miR-200c target DNMT1
and DNMT3A/3B in human ovarian cancer. (A) miR-200b and miR-200c
inhibit the reporter activity of wt, but not mut
DNMT1/3A/3B-3′-UTR-untranslated region. An empty luciferase
reporter construct was used as the negative control. *P<0.05 vs.
NC. (B) A2780CP cells was transfected with NC, miR-200b and
miR-200c mimics for 24 h. Expression of DNMT1, DNMT3A and DNMT3B in
the cells was determined by western blotting (normalized to GAPDH).
(C) Ovarian cancer specimens were analyzed by ISH and
immunohistochemical staining, and the representative miR-200b,
miR-200c, DNMT1, DNMT3A, and DNMT3B expression are presented. (D)
Analysis of immunohistochemical data by linear regressions and
inverse correlations of miR-200b/c with DNMT1, DNMT3A, and DNMT3B
in human ovarian cancer. miR, microRNA; DNMT, DNA
methyltransferase; wt, wild-type; mut, mutated; NC, negative
control mimics. |
To investigate whether miR-200b and miR-200c
negatively regulate their target genes in clinical samples,
endogenous DNMT1, DNMT3A and DNMT3B expression was determined in
human ovarian cancer tissues. Immunohistochemical staining of
DNMT1, DNMT3A and DNMT3B revealed an increased expression of these
DNMTs in EOC compared with in the neighboring normal tissues
(Fig. 4C). An inverse correlation
between miR-200b/miR-200c and DNMT1/DNMT3A/DNMT3B was identified in
randomly selected human ovarian cancer sections (Fig. 4D). These results further supported the
regulation of DNMT1, DNMT3A and DNMT3B by miR-200b and miR-200c,
and indicated the significance of miR-200b and miR-200c as
biomarkers in the progression of EOC.
Discussion
Cisplatin is one of the most widely used first-line
chemotherapy drugs for treating advanced-stage malignancies, such
as testicular, cervical and non-small cell lung cancer. It has been
called a ‘platinum chemotherapeutic agent’ (25). Numerous studies with cisplatin-based
combination therapies have been performed over the last 30 years.
Despite a marked initial response, the efficacy of cisplatin is
significantly hindered by the development of resistance during
treatment. Therefore, patients with advanced carcinoma are not
eligible for standard treatment with cisplatin-based chemotherapy
(26). Furthermore, combination
therapies of cisplatin with other drugs or mechanisms of resistance
have been considered to overcome drug resistance and to decrease
toxicity.
Although multiple mechanisms that mediate intrinsic
or acquired resistance to cisplatin have been recognized,
alterations in the DNA repair capacity of damaged cells are now
being recognized as being important in regulating the resistance to
cisplatin (27–29). Consequently, alterations in DNA repair
pathways have been implicated in cisplatin resistance. Strategies
for overcoming cisplatin resistance are urgently required in cancer
therapy. Substantial changes in DNA methylation have previously
been reported to occur during the acquisition of cisplatin
resistance. DNA methylation is an epigenetic modification that is
mediated by DNMTs. DNMT1 is the most abundant DNMT in mammalian
cells and the key enzyme for the maintenance of hemimethylated DNA
during DNA replication and tumorigenesis. DNMTs have also been
identified to be overexpressed in a number of malignancies
(30–32). In previous studies, DNMT1 and
DNMT3A/DNMT3B were identified to be upregulated in a
cisplatin-resistant ovarian cancer cell line, suggesting a common
regulatory pathway for the expression of DNMT genes (25–26).
Several human miRNAs, including those of the miR-29 family, miR-148
and miR-143, have been identified to be frequently downregulated in
human cancers, and to lead to the increased expression of DNMT1 and
DNMT3A or DNMT3B because they directly target the 3′-UTR of DNMTs
(33). miR-200a, miR-200b, miR-200c,
miR-141 and miR-429 belong to a cluster of miRNAs that are markedly
associated with epithelial-mesenchymal transition (EMT), where
miR-200b and miR-200c are identified as critical regulators of
tumor invasion, metastasis and chemosensitivity (34). miR-200b and miR-200c are known to
inhibit the translation of EMT activators, particularly zinc finger
E-box-binding homeobox factors, and EMT activators, thereby
inducing mesenchymal-epithelial transition (MET) (35–38). Thus,
miR-200 family members, in particular miR-200b and miR-200c,
control crucial cellular processes, such as motility and stemness,
and their own regulators also serve an important function in these
processes. The results of the present study suggested that the
upregulation of miR-200b and miR-200c may lead to the repression of
DNMT1 and DNMT3A/DNMT3B and in turn contribute to the sensitivity
of EOC cells to cisplatin.
In the present study, the function of miR-200b and
miR-200c in cisplatin resistance was characterized, and it was
identified that miR-200b and miR-200c increased cisplatin
sensitivity principally through the direct downregulation of
DNMT3A/DNMT3B and the indirect downregulation of DNMT1 by targeting
Sp1. Sp1 and Sp3 have been reported to increase the activity of the
DNMT1 promoter by physically binding to it in mouse NIH3T3 cells
(39). In addition, previous studies
have identified that wild-type p53 was able to negatively regulate
the DNMT1 gene by binding to the Sp1 protein at their binding sites
on the DNMT1 promoter; furthermore, wild-type p53 was identified to
modulate Sp1 to act as a co-repressor with histone deacetylase
(HDAC)1, HDAC6 and retinol-binding protein 2 lysine demethylase to
suppress the expression of the DNMT1 gene when the level of Sp1
protein was low (12). On the other
hand, p53 has been confirmed to be a transcriptional activator of
the gene encoding miR-200c, and its clinical relevance, as
validated in human breast cancer, revealed an association of mutant
p53 expression with decreased levels of miR-200c (40). Results from a previous study (40) also support the hypothesis that
alterations in p53 may influence the sensitivity to cisplatin-based
chemotherapy because p53 interacts with cisplatin-damaged DNA
molecules.
In conclusion, the results of the present study
suggested a possible mechanism by which miR-200b and miR-200c
enhance cisplatin sensitivity by promoting apoptosis, and suggested
their potential use as therapeutic targets for overcoming cisplatin
resistance in ovarian cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed in the present study
are included in this article.
Authors' contributions
JL and XXW conceived the study and wrote the
manuscript. JL, XBZ, YLH, QFZ, JBZ and XDZ performed experiments,
collected data, and analyzed data. XXW, YLH, QFZ and JBZ reviewed,
and revised manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of The Second Affiliated Hospital of South China
University (Hengyang, China). All tissues were obtained following
written informed consent from the patients.
Patient consent for publication
Written informed consent was obtained by the
patients and/or guardians.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ahmad S, Qureshi AN, Kazmi A, Rasool A,
Gul M, Ashfaq M, Batool L, Rehman RA, Ahmad J and Muniba: First
cancer statistics report from Hazara division. J Ayub Med Coll
Abbottabad. 25:71–73. 2013.
|
|
3
|
Bradley A, Zheng H, Ziebarth A, Sakati W,
Branham-O'Connor M, Blumer JB, Liu Y, Kistner-Griffin E,
Rodriguez-Aguayo C, Lopez-Berestein G, et al: EDD enhances cell
survival and cisplatin resistance and is a therapeutic target for
epithelial ovarian cancer. Carcinogenesis. 35:1100–1109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cortés-Sempere M, de Miguel MP, Pernia O,
Rodriguez C, de Castro Carpeño J, Nistal M, Conde E, López-Ríos F,
Belda-Iniesta C, Perona R and Ibanez de Caceres I: IGFBP-3
methylation-derived deficiency mediates the resistance to cisplatin
through the activation of the IGFIR/Akt pathway in non-small cell
lung cancer. Oncogene. 32:1274–1283. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zeller C, Dai W, Steele NL, Siddiq A,
Walley AJ, Wilhelm-Benartzi CS, Rizzo S, van der Zee A, Plumb JA
and Brown R: Candidate DNA methylation drivers of acquired
cisplatin resistance in ovarian cancer identified by methylome and
expression profiling. Oncogene. 31:4567–4576. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gebhard C, Benner C, Ehrich M,
Schwarzfischer L, Schilling E, Klug M, Dietmaier W, Thiede C,
Holler E, Andreesen R and Rehli M: General transcription factor
binding at CpG islands in normal cells correlates with resistance
to de novo DNA methylation in cancer cells. Cancer Res.
70:1398–1407. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nojima M, Maruyama R, Yasui H, Suzuki H,
Maruyama Y, Tarasawa I, Sasaki Y, Asaoku H, Sakai H, Hayashi T, et
al: Genomic screening for genes silenced by DNA methylation
revealed an association between RASD1 inactivation and
dexamethasone resistance in multiple myeloma. Clin Cancer Res.
15:4356–4364. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Du J and Zhang L: Integrated analysis of
DNA methylation and microRNA regulation of the lung adenocarcinoma
transcriptome. Oncol Rep. 34:585–594. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kwon MJ and Shin YK: Epigenetic regulation
of cancer-associated genes in ovarian cancer. Int J Mol Sci.
12:983–1008. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sui C, Meng F, Li Y and Jiang Y: miR-148b
reverses cisplatin-resistance in non-small cell cancer cells via
negatively regulating DNA (cytosine-5)-methyltransferase 1(DNMT1)
expression. J Transl Med. 13:1322015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fumagalli C, Della Pasqua S, Bagnardi V,
Cardillo A, Sporchia A, Colleoni M, Viale G, Barberis M and Pruneri
G: Prevalence and clinicopathologic correlates of
O′-methylguanine-DNA methyltransferase methylation status in
patients with triple-negative breast cancer treated preoperatively
by alkylating drugs. Clin Breast Cancer. 14:285–290. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lin RK, Wu CY, Chang JW, Juan LJ, Hsu HS,
Chen CY, Lu YY, Tang YA, Yang YC, Yang PC and Wang YC:
Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1
overexpression in lung cancer. Cancer Res. 70:5807–5817. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qin T, Jelinek J, Si J, Shu J and Issa JP:
Mechanisms of resistance to 5-aza-2′-deoxycytidine in human cancer
cell lines. Blood. 113:659–667. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lynch SM, O'Neill KM, McKenna MM, Walsh CP
and McKenna DJ: Regulation of miR-200c and miR-141 by methylation
in prostate cancer. Prostate. 76:1146–1159. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu S, Liu Z, Xie Z, Pang J, Yu J, Lehmann
E, Huynh L, Vukosavljevic T, Takeki M, Klisovic RB, et al:
Bortezomib induces DNA hypomethylation and silenced gene
transcription by interfering with Sp1/NF-kappaB-dependent DNA
methyltransferase activity in acute myeloid leukemia. Blood.
111:2364–2373. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang H, Ou CC, Feldman RI, Nicosia SV,
Kruk PA and Cheng JQ: Aurora-A kinase regulates telomerase activity
through c-Myc in human ovarian and breast epithelial cells. Cancer
Res. 64:463–467. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tang H, Wang Z, Liu X, Liu Q, Xu G, Li G
and Wu M: LRRC4 inhibits glioma cell growth and invasion through a
miR-185-dependent pathway. Curr Cancer Drug Targets. 12:1032–1042.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang L, Tang H, Kong Y and Xie X, Chen J,
Song C, Liu X, Ye F, Li N, Wang N and Xie X: LGR5 promotes breast
cancer progression and maintains stem-like cells through activation
of Wnt/β-catenin signaling. Stem Cells. 33:2913–2924. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ding W, Dang H, You H, Steinway S,
Takahashi Y, Wang HG, Liao J, Stiles B, Albert R and Rountree CB:
miR-200b restoration and DNA methyltransferase inhibitor block lung
metastasis of mesenchymal-phenotype hepatocellular carcinoma.
Oncogenesis. 1:e152012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tang H, Deng M, Tang Y and Xie X, Guo J,
Kong Y, Ye F, Su Q and Xie X: miR-200b and miR-200c as prognostic
factors and mediators of gastric cancer cell progression. Clin
Cancer Res. 19:5602–5612. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stathopoulos GP: Cisplatin: Process and
future. J BUON. 18:564–569. 2013.PubMed/NCBI
|
|
26
|
Roossink F, de Jong S, Wisman GB, van der
Zee AG and Schuuring E: DNA hypermethylation biomarkers to predict
response to cisplatin treatment, radiotherapy or chemoradiation:
The present state of art. Cell Oncol (Dordr). 35:231–241. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kelland L: The resurgence of
platinum-based cancer chemotherapy. Nat Rev Cancer. 7:573–584.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Helleday T, Petermann E, Lundin C, Hodgson
B and Sharma RA: DNA repair pathways as targets for cancer therapy.
Nat Rev Cancer. 8:193–204. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
O'Grady S, Finn SP, Cuffe S, Richard DJ,
O'Byrne KJ and Barr MP: The role of DNA repair pathways in
cisplatin resistant lung cancer. Cancer Treat Rev. 40:1161–1170.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Weis B, Schmidt J, Maamar H, Raj A, Lin H,
Tóth C, Riedmann K, Raddatz G, Seitz HK, Ho AD, et al: Inhibition
of intestinal tumor formation by deletion of the DNA
methyltransferase 3a. Oncogene. 34:1822–1830. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu B, Wang Z, Zhang L, Ghosh S, Zheng H
and Zhou Z: Depleting the methyltransferase Suv39h1 improves DNA
repair and extends lifespan in a progeria mouse model. Nat Commun.
4:18682013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hayette S, Thomas X, Jallades L, Chabane
K, Charlot C, Tigaud I, Gazzo S, Morisset S, Cornillet-Lefebvre P,
Plesa A, et al: High DNA methyltransferase DNMT3B levels: A poor
prognostic marker in acute myeloid leukemia. PLoS One.
7:e515272012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fabbri M, Garzon R, Cimmino A, Liu Z,
Zanesi N, Callegari E, Liu S, Alder H, Costinean S,
Fernandez-Cymering C, et al: MicroRNA-29 family reverts aberrant
methylation in lung cancer by targeting DNA methyltransferases 3A
and 3B. Proc Natl Acad Sci USA. 104:15805–15810. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hur K, Toiyama Y, Takahashi M, Balaguer F,
Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR and Goel A:
MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT)
in human colorectal cancer metastasis. Gut. 62:1315–1326. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Burk U, Schubert J, Wellner U, Schmalhofer
O, Vincan E, Spaderna S and Brabletz T: A reciprocal repression
between ZEB1 and members of the miR-200 family promotes EMT and
invasion in cancer cells. EMBO Rep. 9:582–589. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Park SM, Gaur AB, Lengyel E and Peter ME:
The miR-200 family determines the epithelial phenotype of cancer
cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes
Dev. 22:894–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gregory PA, Bert AG, Paterson EL, Barry
SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y and Goodall GJ:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bracken CP, Gregory PA, Kolesnikoff N,
Bert AG, Wang J, Shannon MF and Goodall GJ: A double-negative
feedback loop between ZEB1-SIP1 and the microRNA-200 family
regulates epithelial-mesenchymal transition. Cancer Res.
68:7846–7854. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kishikawa S, Murata T, Kimura H, Shiota K
and Yokoyama KK: Regulation of transcription of the Dnmt1 gene by
Sp1 and Sp3 zinc finger proteins. Eur J Biochem. 269:2961–2970.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cicalese A, Bonizzi G, Pasi CE, Faretta M,
Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP and Pelicci
PG: The tumor suppressor p53 regulates polarity of self-renewing
divisions in mammary stem cells. Cell. 138:1083–1095. 2009.
View Article : Google Scholar : PubMed/NCBI
|