Introduction
Hepatocellular carcinoma (HCC), which represents an
overwhelming majority of liver cancer, is the sixth most widespread
cancer all over the world and the third most common cause of
cancer-related deaths (1). As a
health threat to people worldwide and, in particular, to developing
countries, its overall 5-year survival rate is 5–9% (2). However, with early diagnosis and
curative resection, the 5-year survival rate can be increased to
69% (3). Hence, a diagnostic
biomarker with high efficacy is urgently needed. In current
clinical work, HCC detection often hinges on a-fetoprotein (AFP),
which is the most general biomarker for HCC detection. An aberrant
high AFP expression level is frequently observed in HCC patients,
with a sensitivity of 39–65% and a specificity of 76–94% (4). In addition, some researchers have
reported that the ability of AFP to identify and differentiate HCC
from non-cancerous hepatopathy is unsatisfying (5–7).
MicroRNAs (miRNAs) are a sequence of short noncoding
RNAs that are ~20–22 nucleotides in length. They
post-transcriptionally regulate the gene level by combining with
their target mRNAs and play important administrative roles in a
variety of biological processes (8–12). During
the past few years, aberrant expression of miRNAs for the early
discovery of HCC has been widely verified with inconsistent
results. Since then, the significant high expression of miR-15b-5p
in HCC patients has been demonstrated by multiple studies (13,14). Liu
et al (15) detected
miR-15b-5p in HCC with an AUC of 0.485 (98.25% sensitivity, 15.25%
specificity). In another study, Chen et al (16) revealed that the AUC value of
miR-15b-5p for HCC detection was 0.654 (68.1% sensitivity, 79.0%
specificity), 0.871 (87.2% sensitivity, 74.2% specificity), and
0.765 (68.1% sensitivity, 80.0% specificity), respectively, in
subgroups of HCC vs. liver cirrhosis patients, HCC vs. healthy
controls, as well as HCC vs. liver cirrhosis and healthy controls.
In fact, the clinical effects of miR-15b-5p on HCC have been
reported by only three research groups: i) Hung et al
(14) first analyzed the role of
miR-15b-5p in the early diagnosis of HCC. They found that when
dysplastic nodules (DN) progressed to HCC (n=10), miR-15b-5p levels
significantly increased and the serum level of miR-15b-5p in 120
patients with early HCC was also upregulated as compared to that of
30 patients with chronic hepatitis B using reverse
transcription-quantitative polymerase chain reaction (RT-qPCR). ii)
Liu et al (15) detected the
serum levels of 29 hepatitis B carriers, 57 patients with HCC and
30 healthy controls also using RT-qPCR. They revealed that the
expression of miR-15b-5p was significantly higher in all HCC
samples. iii) Chen et al (16)
detected the expression level of miR-15b-5p in 37 patients with
HCC, 29 patients with cirrhosis, and 31 healthy controls by
RT-qPCR, and the results revealed that the plasma levels of
miR-15b-5p in HCC patients were higher than in the other 2 groups
(P<0.05). However, these studies were inconsistent their
miR-15b-5p multiple ability indexes. To improve this issue, a
meta-analysis of miR-15b-5p in patients with HCC grounded in data
gathered from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and The Cancer
Genome Atlas (TCGA; http://cancergenome.nih.gov/) were utilized to
evaluate the clinical effectiveness of miR-15b-5p. Bioinformatics
analyses were also utilized to investigate the mechanism of
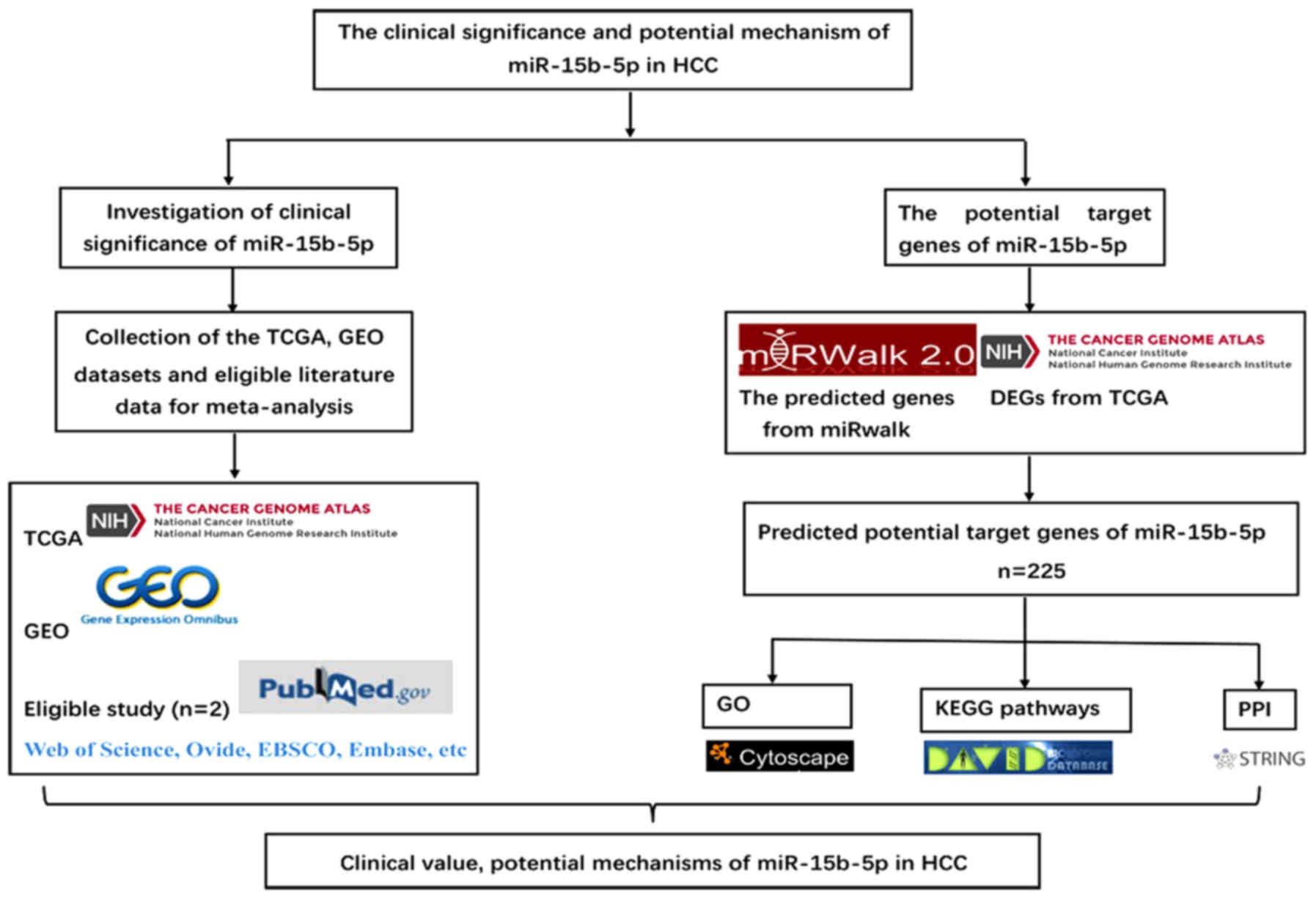
miR-15b-5p in HCC. The framework of this study is displayed in
Fig. 1.
Materials and methods
Excavation of TCGA and GEO
In TCGA, HCC-related resources with entries named
liver hepatocellular carcinoma (LIHC) were downloaded, correlated
miRNA-Seq profiles were provided and miR-15b-5p levels were
extracted. To normalize the expression level of miR-15b-5p in
different trials, data were log2-scaled afterwards.
Expression data sets of miR-15b-5p were attained from the GEO
database, and the expression levels of miR-15b-5p in other types of
hepatic tissues were also gathered as the control group as well as
miR-15b-5p in HCC. Aimed at more precisely assessing the potential
value of miR-15b-5p, the inclusion criteria extended to various
non-cancerous samples. Microarrays concerning cell lines and other
species were excluded because they did not conform to our study.
Based on the expression levels of miR-15b-5p in GEO and TCGA,
GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA) was
used to generate the scatter plots and ROC. Additionally, HCC
patients obtained from TCGA were utilized to analyze the
corresponding clinical information.
Literature search
Relevant literature was read and gathered from
PubMed (https://www.ncbi.nlm.nih.gov/pubmed/), Web of Science
(https://clarivate.com/products/web-of-science/), Ovid
(http://www.ovid.com/site/index.jsp),
EBSCO (https://www.ebsco.com/products/research-databases),
Embase (https://www.elsevier.com/solutions/embase-biomedical-research),
Cochrane Library (https://www.cochranelibrary.com/), Chinese CNKI
(http://www.cnki.net/), China Biology Medicine
disc, the Chinese Chong Qing VIP (http://en.cqvip.com/), and the Chinese Wan Fang
(http://eng.med.wanfangdata.com.cn/).
No language limitations were imposed. The following combination of
terms with two sets of keywords were screened by combining the
underlying searching strategies: (miR-15b OR miRNA-15b OR
micrORNA-15b OR miR15b OR miRNA15b OR microRNA15b OR ‘miR 15b’ OR
‘miRNA 15b’ OR ‘microRNA 15b’ OR miR-15b-5p OR miRNA-15b-5p OR
micrORNA-15b-5p OR micrORNA-15b OR miR-15b-5p) AND (malignan* OR
cancer OR tumor OR tumour OR neoplas* OR carcinoma) AND
(hepatocellular cancer OR hepatocellular tumor OR hepatocellular
carcinoma OR hepatocellular neoplasm OR liver cancer OR liver tumor
OR liver carcinoma OR liver neoplasm OR HCC). Human studies were
limited to our literature searches. To confirm the qualified
studies, the relevant studies and other references of the review
papers were included to avoid missing related studies. Sufficient
HCC data and control groups were required to obtain true positives
(TPs), false positives (FPs), false negatives (FNs), and true
negatives (TNs).
Potential target gene collection and
bioinformatics analyses
To further investigate the regulatory mechanism of
miR-15b-5p in HCC, MiRWalk2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/),
which combines 12 online prediction programs, was used to provide
comprehensive potential targets for miR-15b-5p. Genes identified by
>4 prediction software programs for miR-15b-5p were selected to
obtain more reliable targets. The selected predicted target genes
were further intersected with TCGA differentially expressed genes.
The overlapping genes were considered to be potential target genes
of miR-15b-5p. We combined the two parts of the target genes of
miR-15b-5p for further gene functional enrichment analyses. In
addition, the genes were input to the STRING version 10.0 online
tool (http://string-db.org/) to construct the
protein-protein interaction (PPI) network.
DAVID 6.8 (https://david.ncifcrf.gov/) was applied for gene
ontology (GO; http://geneontology.org/) and Kyoto Encyclopedia of
Genes and Genomes (KEGG; http://www.genome.jp/kegg/) pathway analysis. GO was
composed of three sections: Molecular function (MF), cellular
component (CC) and biological process (BP). The top ten terms of
each GO category and marked KEGG pathways were visualized as GO
maps and KEGG maps (17,18). Protein expression of hub genes was
validated by The Human Protein Atlas (HPA; www.proteinatlas.org), an immunohistochemistry (IHC)
database. The IHC images are publicly available.
Diagnostic test and statistical
analysis
Diagnostic tests were conducted to assess the
efficacy of miR-15b-5p in HCC. To thoroughly determine its clinical
potential, analyses were carried out between HCC patients and
controls, including healthy controls, non-cancerous controls,
adjacent non-neoplastic hepatic tissues,
HBV+/HCV+ controls and liver cirrhosis
controls. The efficiency of miR-15b-5p in the serum/plasma and
tissues was also examined. Stata 12.0 (https://www.stata.com/stata12/) was used to detect
publication bias, and the standard mean difference (SMD) was used
to calculate the outcome from GEO and TCGA. The remaining analyses
were accomplished by Meta-DiSc 1.4. P<0.05 was recognized as
statistically significant. The summary receiver operator
characteristic (SROC) curve was plotted according to the included
studies. A meta-analysis was carried out using a random effects
model, including SEN, SEP, diagnostic odds ratio (DOR), positive
likelihood ratio (PLR), and negative likelihood ratio (NLR). In
addition, the I2 index and χ2 test were used to evaluate
the heterogeneity in the study. If the I2 value was over
50% or P-values of the χ2 test were <0.05,
heterogeneity would be shown. Finally, Deek's funnel plot was
displayed to assess the publication bias.
Results
Qualified studies and dataset
A total of 3,310 relevant articles were acquired
from the aforementioned online databases by a primary search. After
removing the duplicated articles as well as screening titles,
abstracts and full texts, 2 studies were eventually included, and
both were published in English (15,19). The
chosen studies provided data from 114 HCC patients and 119 people
as controls.
According to our criteria, 11 microarray datasets
from GEO were evaluated as eligible, consisting of 512 HCC tissue
samples and 287 control samples. Sequencing data in TCGA were based
upon 425 samples, with 375 diagnosed HCC samples and 50 control
samples, as shown in Table I
(20–27).
 | Table I.Basic information and clinical data
of the included studies. |
Table I.
Basic information and clinical data
of the included studies.
| Accession | Author | Year | Country | Experiment
type | Platform | HCC numbers | Control
numbers | Sample type | TP | FP | FN | TN | (Refs.) |
|---|
| GSE57555 | Murakami et
al | 2015 | Japan | Non-coding RNA
profiling by array | GPL18044 | 5 | 16 | Tissue | 2 | 1 | 3 | 15 | (20) |
| GSE69580 | Hung et
al | 2015 | Taiwan | Non-coding RNA
profiling by array | GPL10850 | 5 | 5 | Tissue | 2 | 0 | 3 | 5 | Citation
missing |
| GSE67882 | Ghosh et
al | 2015 | India | Non-coding RNA
profiling by array | GPL10850 | 4 | 8 | Tissue | 5 | 3 | 0 | 4 | Citation
missing |
| GSE54751 | Shen et
al | 2015 | USA | Expression
profiling by RT-PCR | GPL18262 | 10 | 10 | Tissue | 4 | 1 | 6 | 9 | (21) |
| GSE21362 | Sato et
al | 2011 | Japan | Non-coding RNA
profiling by array | GPL10312 | 73 | 73 | Tissue | 49 | 24 | 26 | 47 | (22) |
| GSE22058 | Burchard et
al | 2010 | USA | Expression
profiling by | GPL10457 | 96 | 96 | Tissue | 72 | 9 | 24 | 87 | (23) |
| GSE10694 | Li et
al | 2008 | China | Non-coding RNA
profiling by array | GPL6542 | 78 | 10 | Tissue | 44 | 5 | 34 | 83 | (24) |
| GSE12717 | Su et
al | 2008 | China | Non-coding RNA
profiling by array | GPL7274 | 5 | 3 | Tissue | 7 | 1 | 2 | 5 | (25) |
| GSE40744 | Diaz G et
al | 2013 | USA | Non-coding RNA
profiling by array | GPL14613 | 10 | 33 | Tissue | 7 | 30 | 2 | 37 | (26) |
| GSE74618 | Martinez-Quetglas I
et al | 2016 | Spain | Non-coding RNA
profiling by array | GPL14613 | 218 | 20 | Tissue | 139 | 0 | 79 | 20 | (27) |
| GSE41874 | Morita et
al | 2013 | Japan | Non-coding RNA
profiling by array | GPL7722 | 6 | 4 | Tissue | 5 | 0 | 1 | 4 | Citation
missing |
| TCGA |
| 2018 | USA | miRNA-Seq | Illumina | 375 | 50 | Tissue | 285 | 46 | 90 | 4 |
|
| − | Liu et
al | 2012 | China | RT-qPCR | / | 57 | 59 | Serum | 27 | 0 | 10 | 31 | (15) |
| − | Chen et
al | 2015 | China | RT-qPCR | / | 47 | 60 | Plasma | 35 | 4 | 4 | 21 | (16) |
Overall assessment of the diagnostic
value and diagnostic meta-analysis
For a more comprehensive understanding of the
efficiency of miR-15b-5p in HCC, the eligible 11 GSE chips that we
searched were included in our meta-analysis. The expression levels
that correlated with miR-15b-5p in GEO and TCGA were also displayed
in Fig. 2. TCGA and 11 GEO profiles
with an AUC were presented in Fig. 3.
To further explore the clinicopathological features of miR-15b-5p
in TCGA, all of the clinicopathological features mentioned in the
chips were collected to investigate their correlation with the
miR-15b-5p expression level, the results of which are provided in
Table II. No noteworthy
relationships were observed between miR-15b-5p expression and the
clinicopathological characteristics. Compared with the
non-neoplastic controls, the miR-15b-5p levels in HCC revealed that
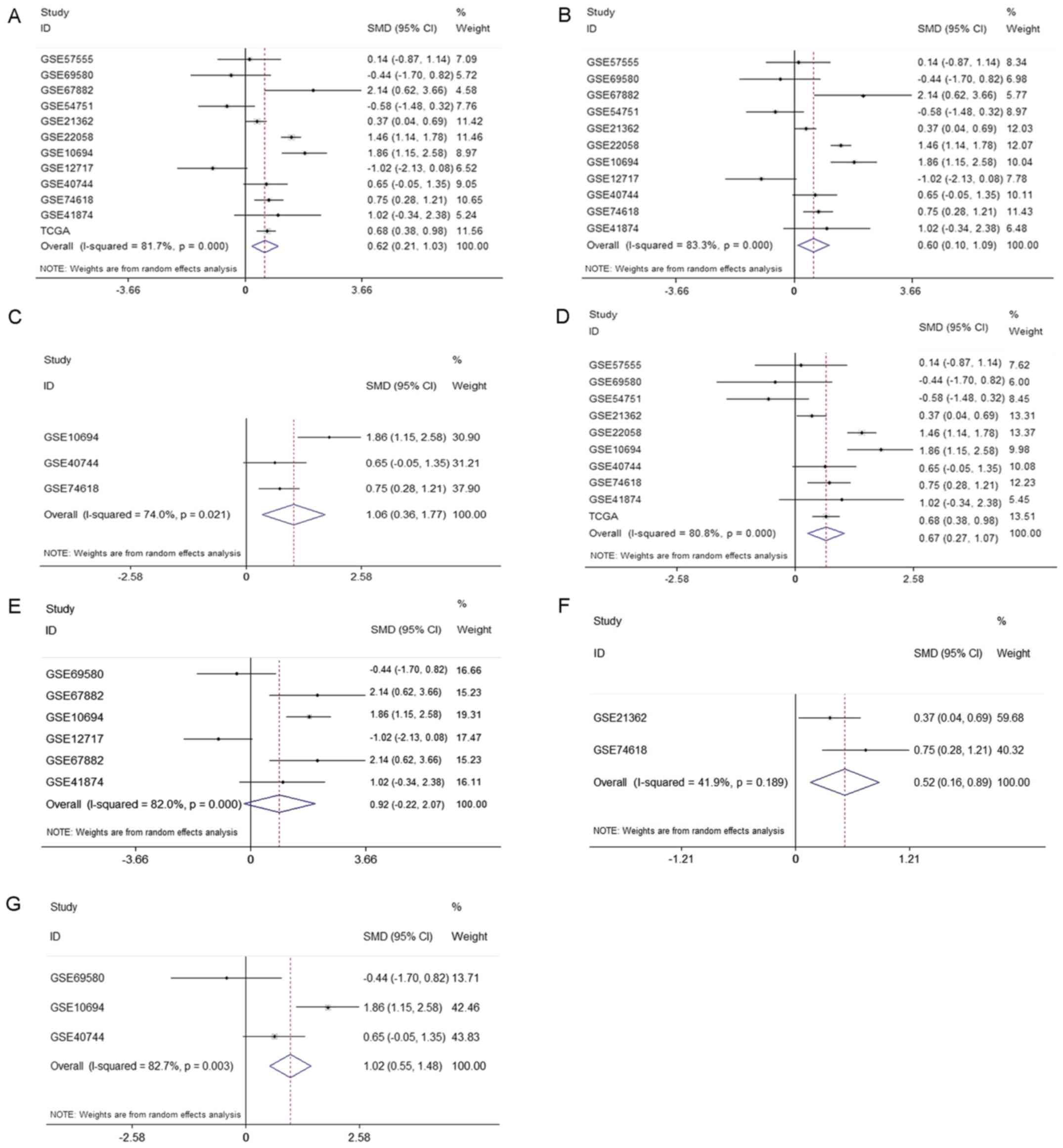
the pooled AUC, SEN, SPE, PLR, NLR and DOR were 0.81 (Q*=0.74),
0.72 (95% CI: 0.69–0.75), 0.68 (95% CI: 0.65–0.72), 3.18 (95% CI:
1.83–5.51), 0.43 (95% CI: 0.35–0.54), and 8.98 (95% CI: 4.30–18.76)
(Fig. 4), Furthermore, subgroup
analyses with both SMD and sROC methods were performed. The
subgroups included sample sources (tissues and serum/plasma) and
control types (healthy controls, adjacent non-cancerous hepatic
tissues, HBV+ or HCV+ tissues, liver
cirrhosis tissues, and those combining
HBV+/HCV+ and cirrhosis). The results were
presented from Fig. 5 to Fig. 12. The control types in healthy people
and HBV+ or HCV+ patients had a favorable
diagnostic accuracy with AUC-SROC over 0.9, respectively, when in
tissues, serum/plasma, adjacent non-cancerous hepatic tissues,
liver cirrhosis and combined HBV+/HCV+ and
cirrhosis were used as the control. The AUC in the ROC curve was
over 0.7.
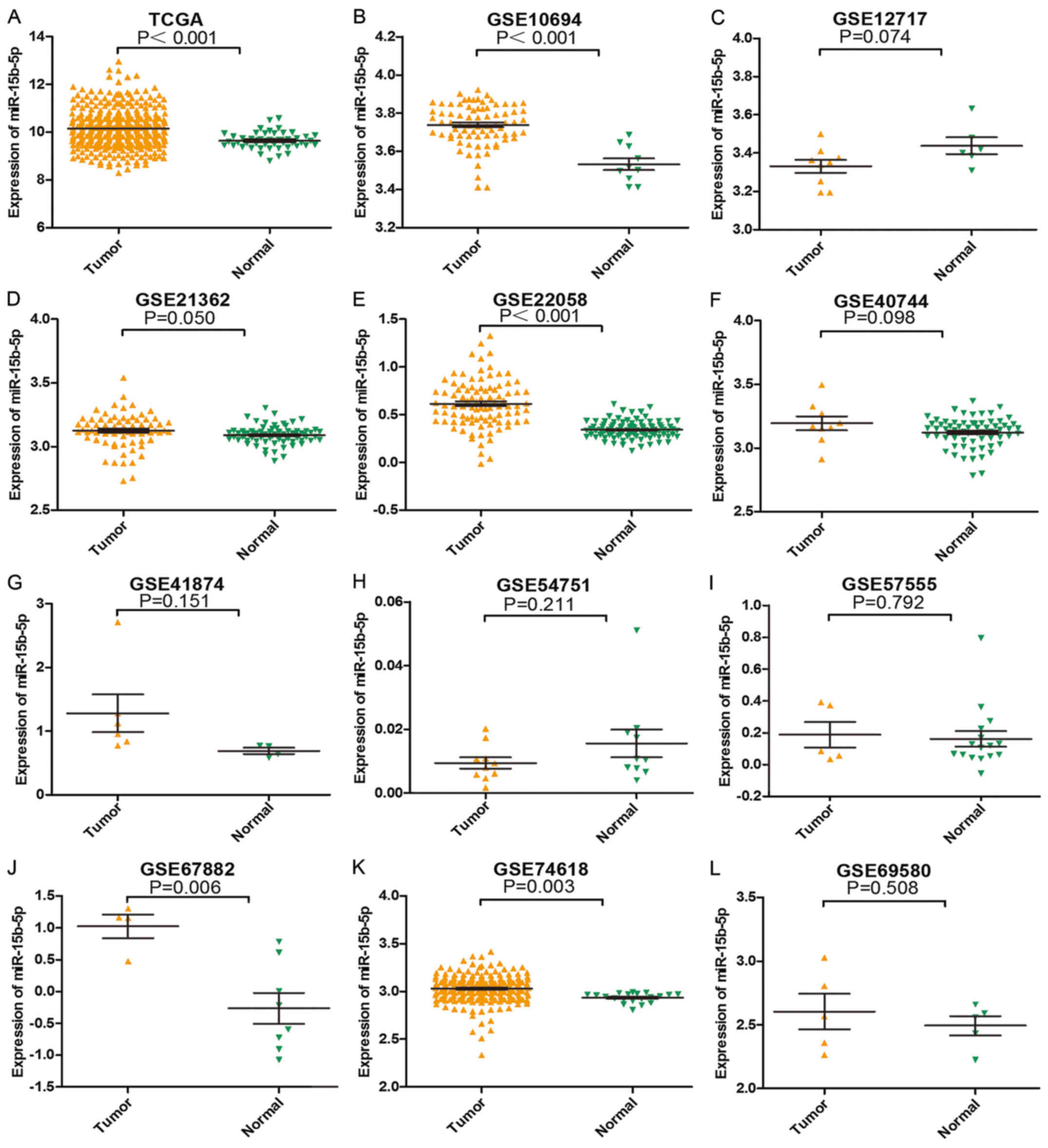 | Figure 2.Expression level of each study from
TCGA and GEO. (A) TCGA, (B) GSE10694, (C) GSE12717, (D) GSE21362,
(E) GSE22058, (F) GSE40744, (G) GSE41874, (H) GSE54751, (I)
GSE57555, (J) GSE67882, (K) GSE74618 and (L) GSE69580. |
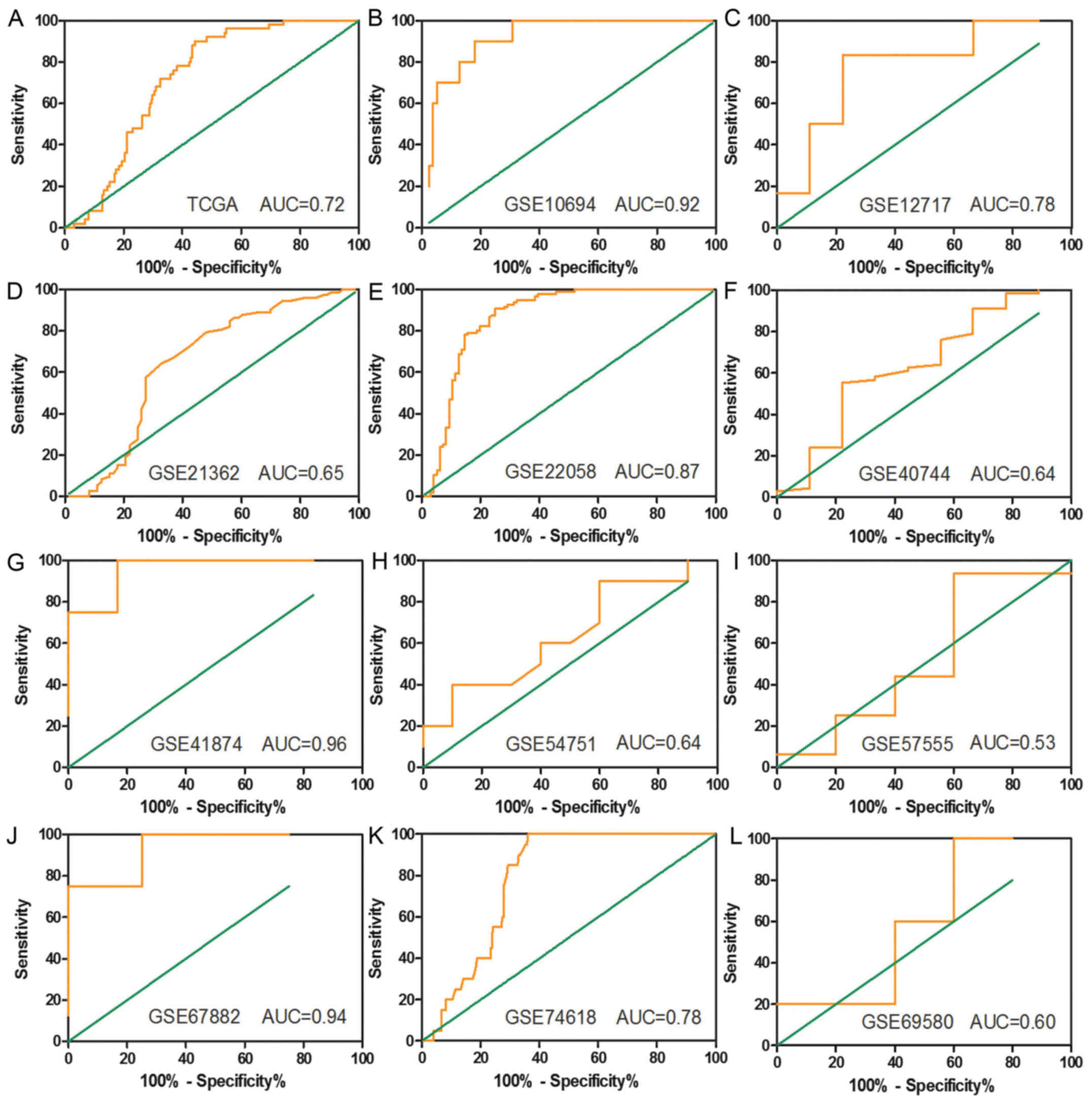 | Figure 3.Receiver operating characteristic
(ROC) curves based on the TCGA and GEO datasets. (A) TCGA, (B)
GSE10694, (C) GSE12717, (D) GSE21362, (E) GSE22058, (F) GSE40744,
(G) GSE41874, (H) GSE54751, (I) GSE57555, (J) GSE67882, (K)
GSE74618 and (L) GSE69580. |
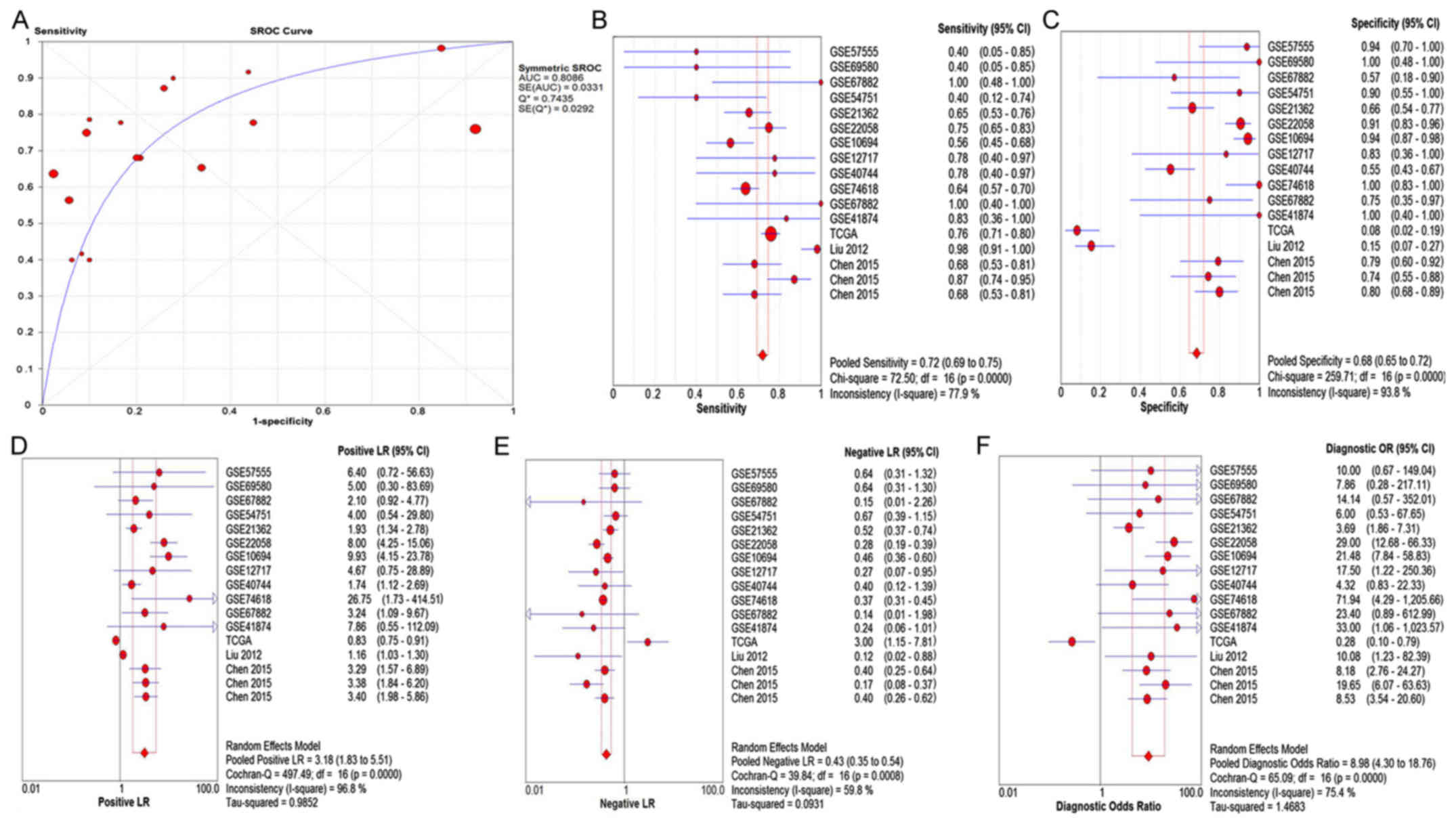 | Figure 4.The pooled (A) SROC curve, (B) SEN,
(C) SPE, (D) PLR, (E) NLR and (F) DOR analysis of the qualified
studies of miR-15b-5p in the HCC group compared with the control
group. SROC, summarized receiver operating characteristic; SEN,
sensitivity; SPE, specificity; PLR, positive likelihood ratio; NLR;
negative likelihood ratio; DOR, diagnostic odds ratio; HCC,
hepatocellular carcinoma. |
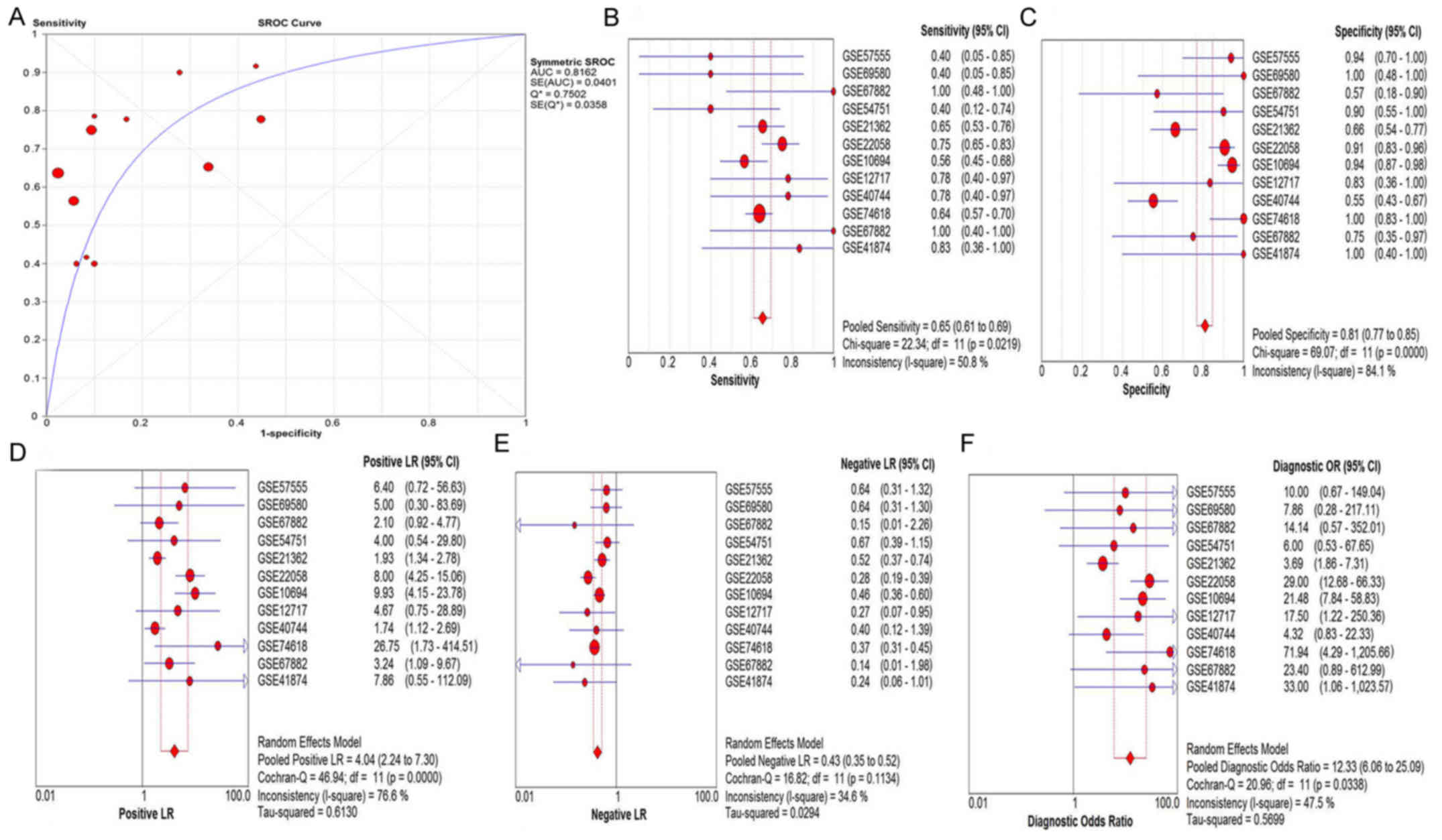 | Figure 5.The pooled (A) SROC curve, (B) SEN,
(C) SPE, (D) PLR, (E) NLR and (F) DOR of miR-15b-5p in tissues.
SROC, summarized receiver operating characteristic; SEN,
sensitivity; SPE, specificity; PLR, positive likelihood ratio; NLR,
negative likelihood ratio; DOR, diagnostic odds ratio. |
 | Table II.Relationship between the levels of
miR-15b-5p and clinicopathological variables in HCC from the TCGA
database. |
Table II.
Relationship between the levels of
miR-15b-5p and clinicopathological variables in HCC from the TCGA
database.
| Parameters | N | Mean value | T-value | P-value |
|---|
| Group |
|
HCC | 375 | 10.128±0.796 | −4.331 | <0.001 |
|
Normal | 50 | 9.823±0.402 |
|
|
| Sex |
|
Male | 254 | 10.072±0.973 | 1.069 | 0.009 |
|
Female | 122 | 9.886±1.802 |
|
|
| Tumor status |
| With
tumor | 152 | 10.207±0.818 | −1.673 | 0.527 |
|
Tumor-free | 201 | 10.066±0.764 |
|
|
| Age (years) |
|
<60 | 170 | 10.100±0.778 | −0.424 | 0.711 |
|
≥60 | 201 | 10.134±0.799 |
|
|
| Race |
|
Caucasian | 182 | 10.074±0.819 | −1.199 | 0.562 |
|
Asian | 161 | 10.176±0.742 |
|
|
| TNM Stage |
|
I–II | 258 | 10.119±0.767 | 0.346 | 0.203 |
|
III–IV | 90 | 10.085±0.859 |
|
|
| Pathological
Stage |
|
G1-2 | 231 | 10.054±0.754 | −2.037 | 0.922 |
|
G3-4 | 137 | 10.226±0.832 |
|
|
| T stage |
|
T1-2 | 276 | 10.111±0.775 | −0.471 | 0.477 |
|
T3-4 | 93 | 10.156±0.833 |
|
|
With the random-effects model, forest-plots were
generated to represent significant differences in expression
between HCC and non-neoplastic control tissues. The pooled SMD
(0.62, 95% CI: 0.21, 1.03) is presented in Fig. 12A. The results of pooled SMD between
HCC and tissues, healthy subjects, adjacent non-cancerous hepatic
tissues, HBV+ or HCV+ patients, liver
cirrhosis, and HBV+/HCV+ combined with
cirrhosis were displayed in Fig. 12.
The expression level of miR-15b-5p in HCC samples was markedly
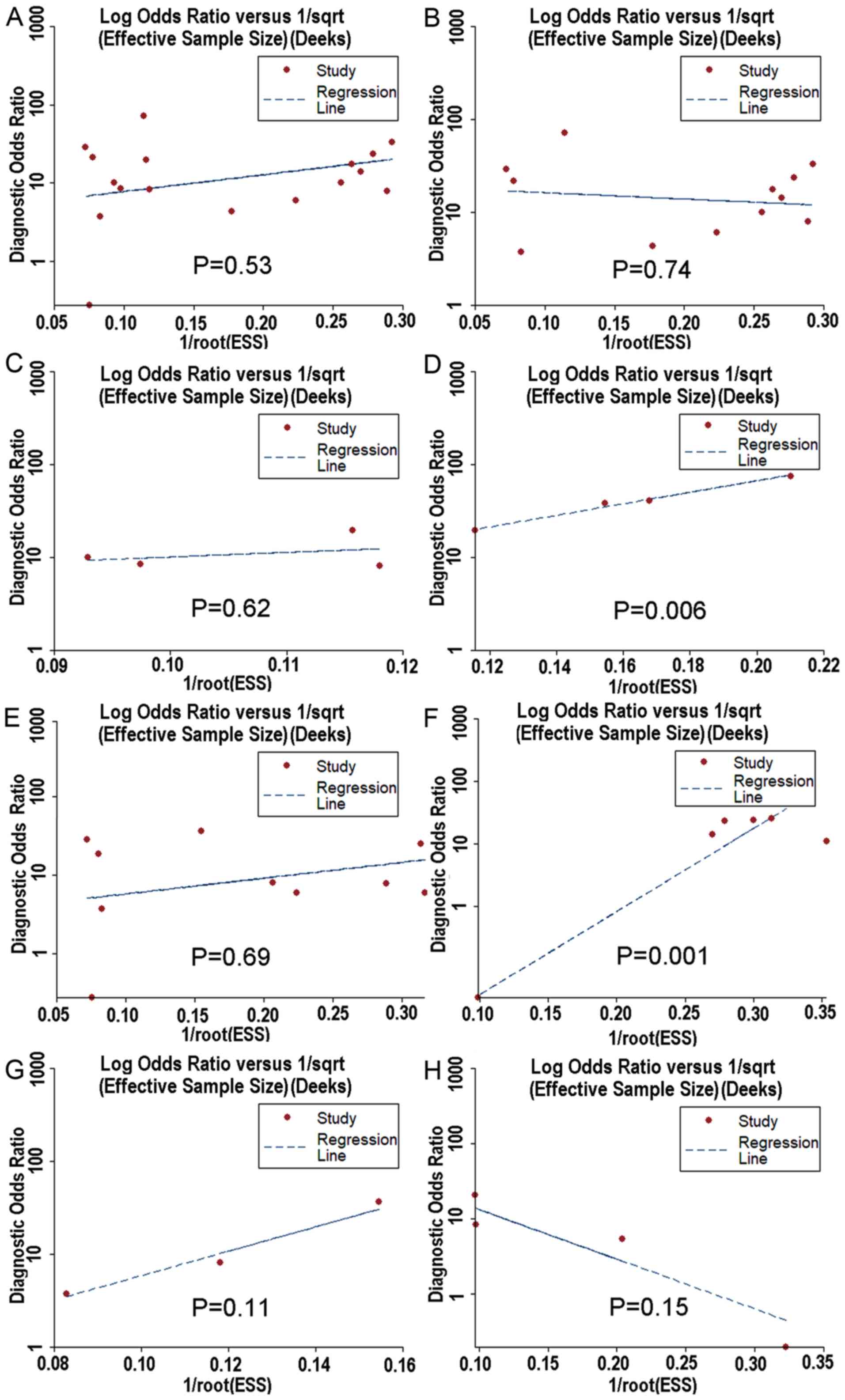
overexpressed than in non-HCC tissues samples. Moreover, the Deek's
funnel plot asymmetry test was carried out with STATA 12.0, and no
publication bias was detected apart from healthy people and
HBV+ or HCV+ patients. (P<0.05) (Fig. 13).
Potential target genes and
bioinformatics annotation
Nine thousand seven-hundred-eighty target genes
appearing ≥4 times in 12 prediction methods were regarded as
probable target genes of miR-15b-5p from miRWalk. Furthermore,
downregulated expressed genes assembled from TCGA were integrated
to generate the intersection of target genes, which had more
potential to be the real targets of miR-15b-5p in HCC. After 9,780
potential target genes and 1,123 TCGA differentially expressed
genes were analyzed for intersection, 225 co-predicted genes from
the following bioinformatics analyses were established. In respect
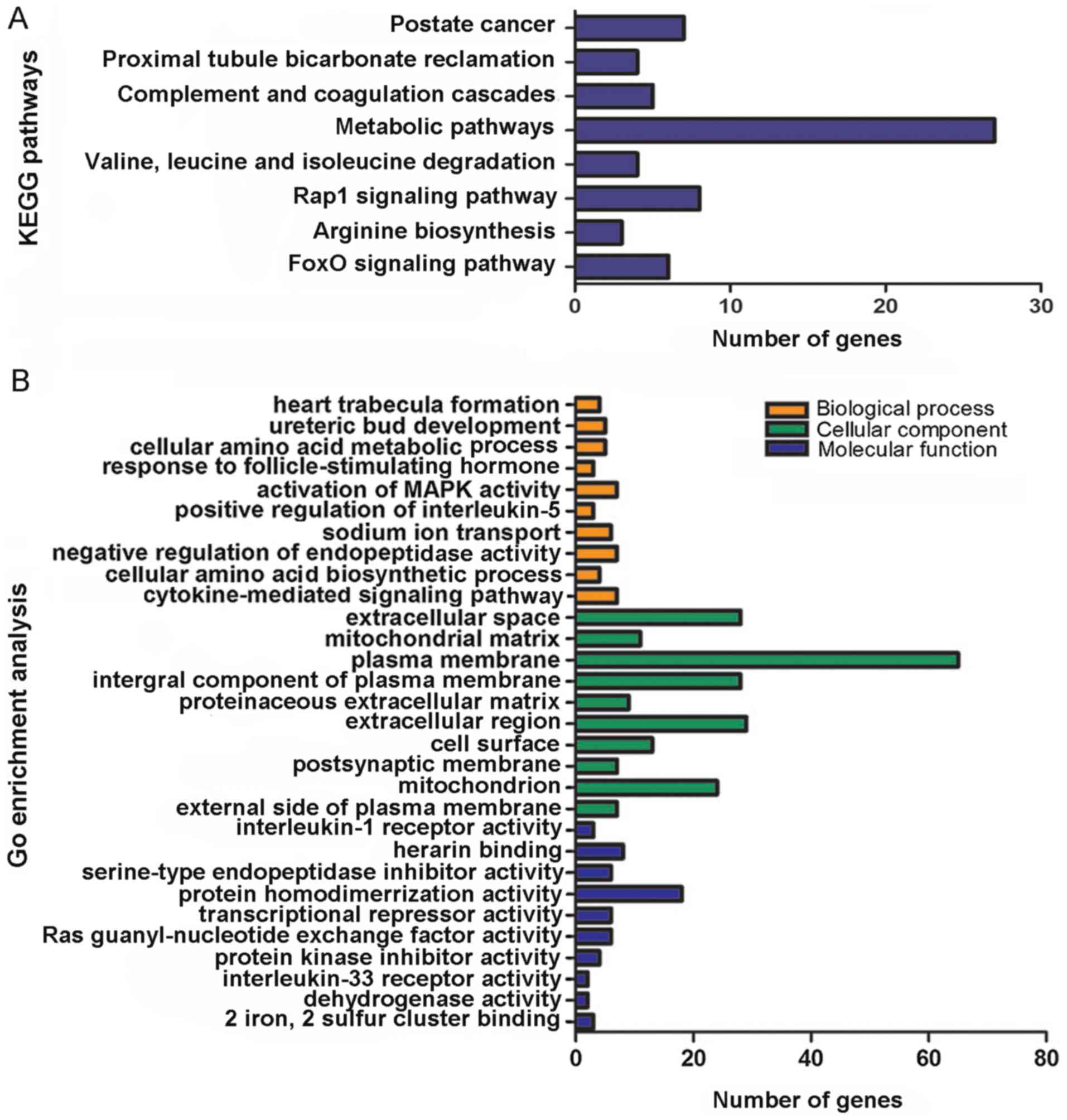
to the bioinformatics analyses, ‘Prostate cancer’
(P=1.56×10−3), ‘Proximal tubule bicarbonate reclamation’
(P=4.10×10−3), ‘Complement and coagulation cascades’
(P=1.68×10−2), ‘Metabolic pathways’
(P=2.18×10−2), ‘Valine, leucine and isoleucine
degradation’ (P=2.94×10−2), ‘Rap1 signaling pathway’
(P=3.06×10−2), ‘Arginine biosynthesis’
(P=3.26×10−2) and ‘FoxO signaling pathway’
(P=4.28×10−2) (Table
III, Fig. 14A) were recognized
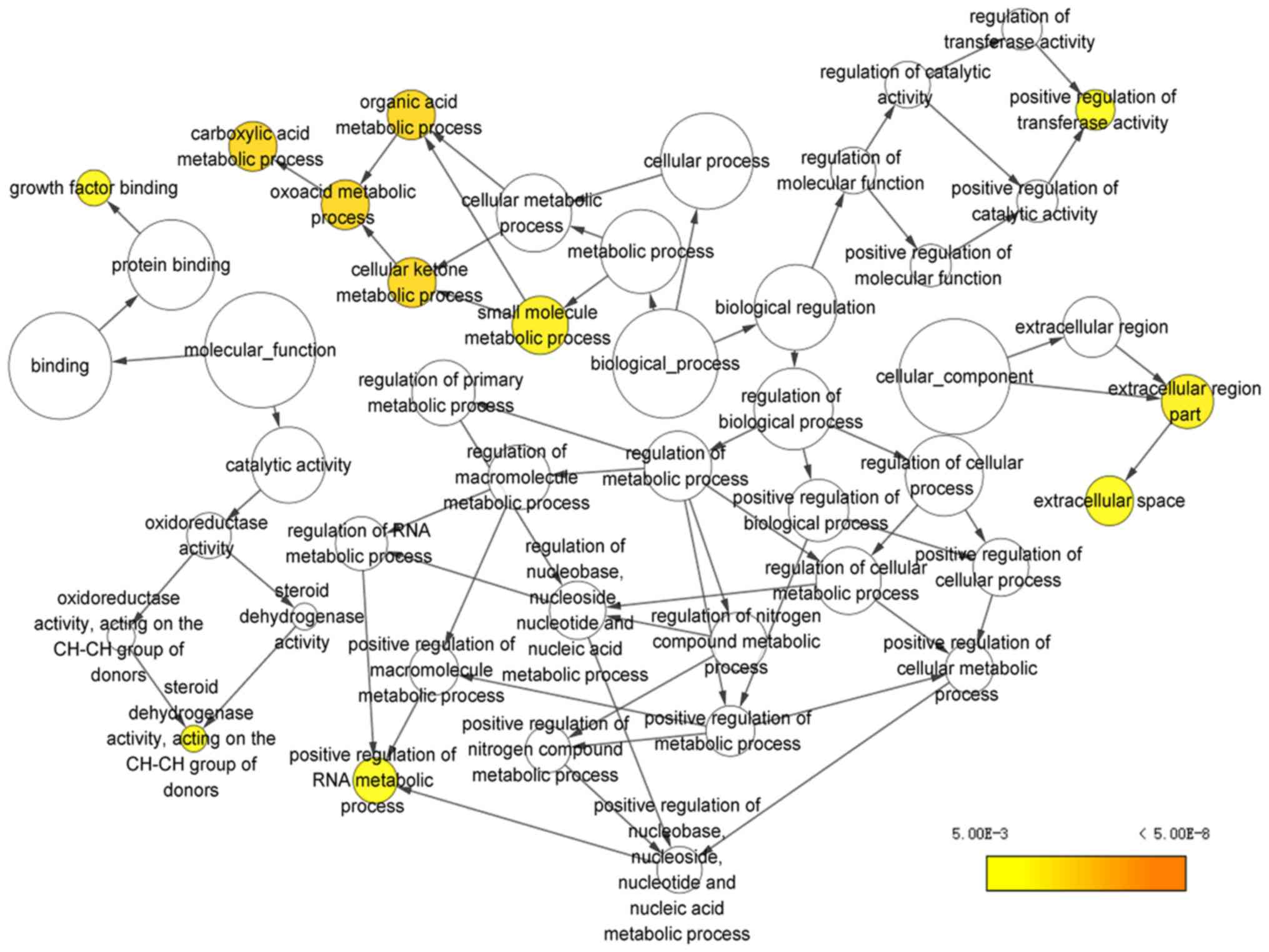
as the most enriched KEGG pathways. For the results of GO pathway
analysis in DAVID, the potential targets of miR-15b-5p were notably
associated with ‘heart trabecula formation’
(P=5.26×10−4), ‘extracellular space’
(P=4.20×10−3) and ‘interleukin-1 receptor activity’
(P=2.79×10−3) (Table IV;
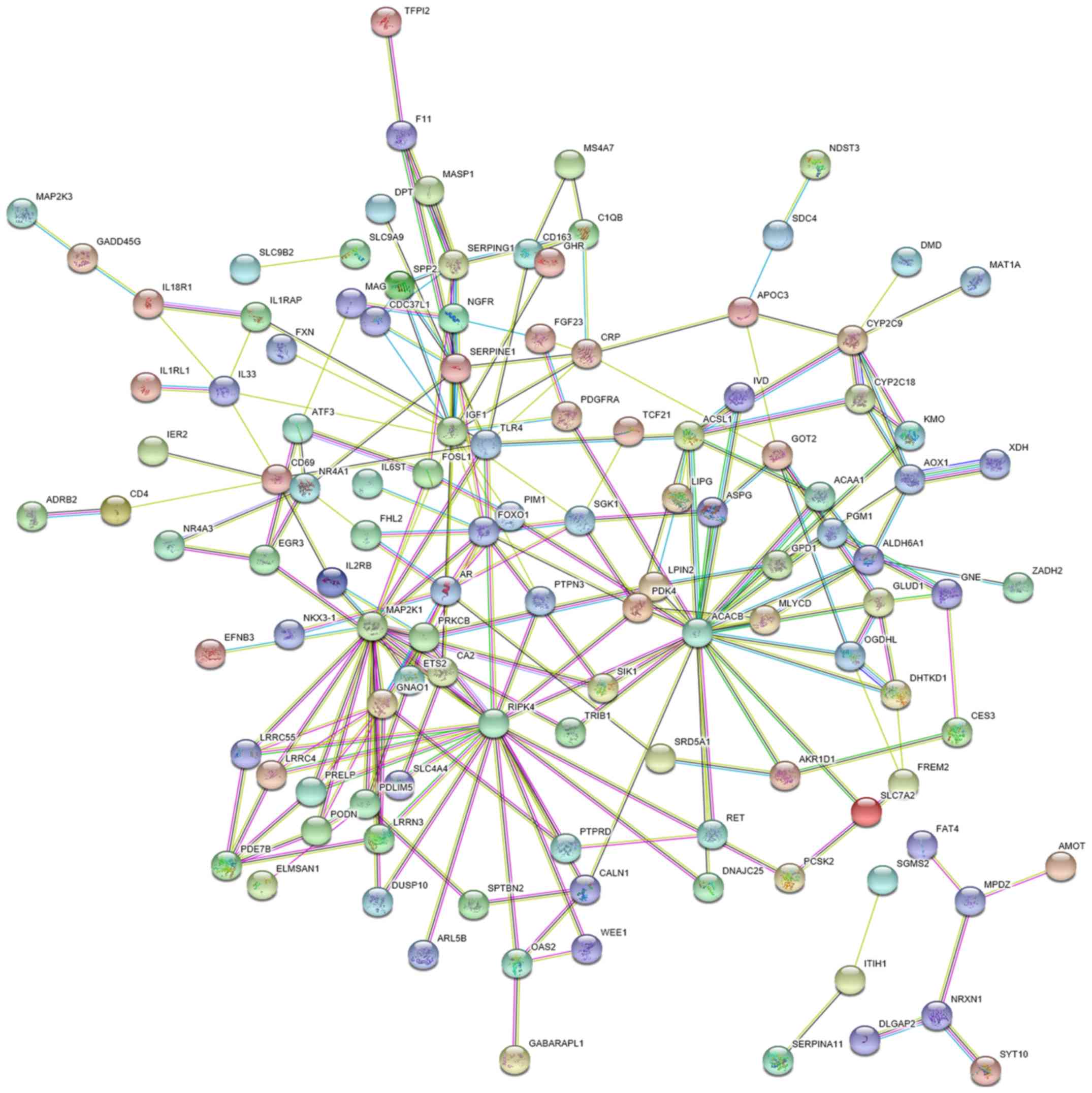
Figs. 14B and 15). A PPI network of the 225 genes was
constructed in the present study with 224 nodes and 221 edges. In
the network, ACACB, RIPK4, MAP2K1, TLR4, and IGF1 were identified
as the hub target genes of miR-15b-5p due to the highest
significance (Figs. 16–18). In the respect to the bioinformatics
analyses with these hub genes, the ‘activation of MAPK activity’
(P=2.39×10−4), ‘ATP binding’ (P=4.17×10−2)
were regarded as the most significant GO categories. The pathways
of ‘HIF-1 signaling pathway’ (P=5.92×10−4),
‘Proteoglycans in cancer’ (P=2.45×10−3) and ‘PI3K-Akt
signaling pathway’ (P=7.21×10−3) were considered to be
the most significant pathways as assessed by KEGG (Tables V and VI). We also ascertained the downregulation
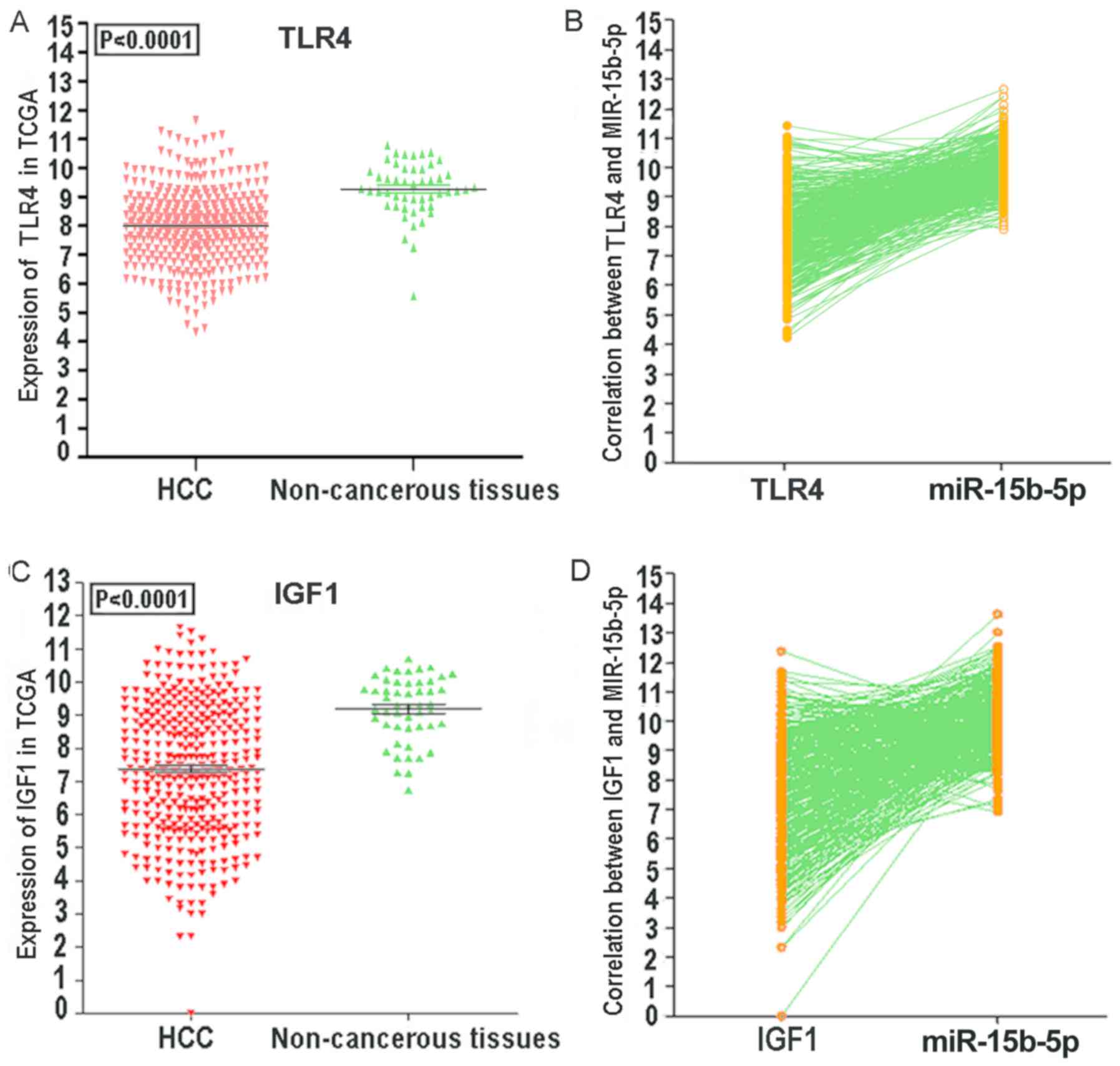
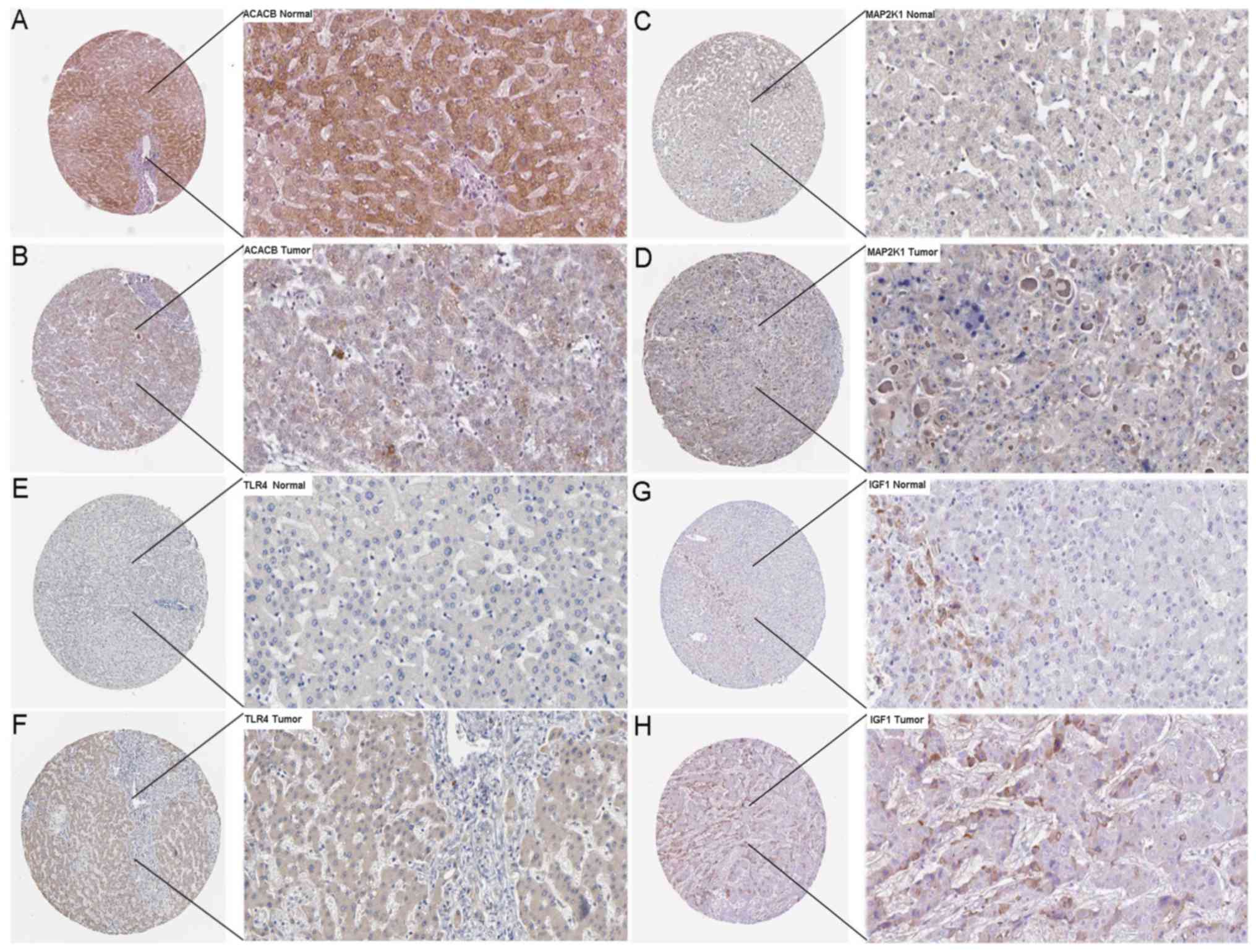
of ACACB, RIPK4, MAP2K1, TLR4 and IGF1 in HCC tissues via TCGA
data. To verify the possibility that these hub genes were targeted
by miR-15b-5p, we further revealed the protein levels of ACACB,
RIPK4, MAP2K1, TLR4 and IGF1 in HCC tissues and normal tissues. As
revealed in Fig. 19, ACACB had
medium staining and moderate intensity in cytoplasmic/membranous
normal liver tissues. MAP2K1, TLR4 and IGF1 exhibited low staining
and weaker intensity in cytoplasmic/membranous normal liver
tissues. In addition, all these hub genes had a lower staining and
weaker intensity in HCC tissues. These findings warrant further
validation, as limited sample size is provided by the HPA.
 | Table III.KEGG functional annotation for most
significantly related targets of miR-15b-5p. |
Table III.
KEGG functional annotation for most
significantly related targets of miR-15b-5p.
| Category | Term | Count | P-value |
|---|
| KEGG_PATHWAY | hsa05215: Prostate
cancer | 7 |
1.56×10−3 |
|
| hsa04964: Proximal
tubule bicarbonate reclamation | 4 |
4.10×10−3 |
|
| hsa04610:
Complement and coagulation cascades | 5 |
1.68×10−2 |
|
| hsa01100: Metabolic
pathways | 27 |
2.18×10−2 |
|
| hsa00280: Valine,
leucine and isoleucine degradation | 4 |
2.94×10−2 |
|
| hsa04015: Rap1
signaling pathway | 8 |
3.06×10−2 |
|
| hsa00220: Arginine
biosynthesis | 3 |
3.26×10−2 |
|
| hsa04068: FoxO
signaling pathway | 6 |
4.28×10−2 |
 | Table IV.GO functional annotation of the
target genes of miR-15b-5p. |
Table IV.
GO functional annotation of the
target genes of miR-15b-5p.
| GO ID | Category | GO term | P-value | Count |
|---|
| GO:0060347 | GO_Biological
process | Heart trabecula
formation |
5.26×10−4 | 4 |
| GO:0001657 | GO_Biological
process | Ureteric bud
development |
9.93×10−4 | 5 |
| GO:0006520 | GO_Biological
process | Cellular amino acid
metabolic process |
1.21×10−3 | 5 |
| GO:0032354 | GO_Biological
process | Response to
follicle-stimulating hormone |
1.34×10−3 | 3 |
| GO:0000187 | GO_Biological
process | Activation of MAPK
activity |
1.64×10−3 | 7 |
| GO:0032754 | GO_Biological
process | Positive regulation
of interleukin-5 production |
1.99×10−3 | 3 |
| GO:0006814 | GO_Biological
process | Sodium ion
transport |
2.63×10−3 | 6 |
| GO:0010951 | GO_Biological
process | Negative regulation
of endopeptidase activity |
3.04×10−3 | 7 |
| GO:0008652 | GO_Biological
process | Cellular amino acid
biosynthetic process |
3.39×10−3 | 4 |
| GO:0019221 | GO_Biological
process | Cytokine-mediated
signaling pathway |
4.49×10−3 | 7 |
| GO:0005615 | GO_Cellular
component | Extracellular
space |
4.20×10−3 | 28 |
| GO:0005759 | GO_Cellular
component | Mitochondrial
matrix |
5.27×10−3 | 11 |
| GO:0005886 | GO_Cellular
component | Plasma
membrane |
6.97×10−3 | 65 |
| GO:0005887 | GO_Cellular
component | Integral component
of plasma membrane |
7.97×10−3 | 28 |
| GO:0005578 | GO_Cellular
component | Proteinaceous
extracellular matrix |
1.37×10−2 | 9 |
| GO:0005576 | GO_Cellular
component | Extracellular
region |
2.18×10−2 | 29 |
| GO:0009986 | GO_Cellular
component | Cell surface |
2.59×10−2 | 13 |
| GO:0045211 | GO_Cellular
component | Postsynaptic
membrane |
3.79×10−2 | 7 |
| GO:0005739 | GO_Cellular
component | Mitochondrion |
3.85×10−2 | 24 |
| GO:0009897 | GO_Cellular
component | External side of
plasma membrane |
3.93×10−2 | 7 |
| GO:0004908 | GO_Molecular
function | Interleukin-1
receptor activity |
2.79×10−3 | 3 |
| GO:0008201 | GO_Molecular
function | Heparin
binding |
2.91×10−3 | 8 |
| GO:0004867 | GO_Molecular
function | Serine-type
endopeptidase inhibitor activity |
5.81×10−3 | 6 |
| GO:0042803 | GO_Molecular
function | Protein
homodimerization activity |
5.89×10−3 | 18 |
| GO:0001078 | GO_Molecular
function | Transcriptional
repressor activity, RNA polymerase II core promoter proximal region
sequence-specific binding |
1.01×10−2 | 6 |
| GO:0005088 | GO_Molecular
function | Ras
guanyl-nucleotide exchange factor activity |
1.17×10−2 | 6 |
| GO:0004860 | GO_Molecular
function | Protein kinase
inhibitor activity |
2.34×10−2 | 4 |
| GO:0002114 | GO_Molecular
function | Interleukin-33
receptor activity |
2.34×10−2 | 2 |
| GO:0004854 | GO_Molecular
function | Xanthine
dehydrogenase activity |
2.34×10−2 | 2 |
| GO:0051537 | GO_Molecular
function | 2 iron, 2 sulfur
cluster binding |
3.47×10−2 | 3 |
 | Table V.GO functional annotation of the hub
genes of miR-15b-5p in HCC. |
Table V.
GO functional annotation of the hub
genes of miR-15b-5p in HCC.
| GO ID | Category | Term | P-value | Count |
|---|
| GO:0000187 | GOTERM_BP | Activation of MAPK
activity |
2.39×10−4 | 3 |
| GO:0070371 | GOTERM_BP | ERK1 and ERK2
cascade |
5.71×10−3 | 2 |
| GO:0006928 | GOTERM_BP | Movement of cell or
subcellular component |
2.03×10−2 | 2 |
| GO:0051092 | GOTERM_BP | Positive regulation
of NF-κB transcription factor activity |
3.13×10−2 | 2 |
| GO:0010629 | GOTERM_BP | Negative regulation
of gene expression |
3.22×10−2 | 2 |
| GO:0070374 | GOTERM_BP | Positive regulation
of ERK1 and ERK2 cascade |
4.10×10−2 | 2 |
| GO:0010628 | GOTERM_BP | Positive regulation
of gene expression |
6.10×10−2 | 2 |
| GO:0005524 | GOTERM_MF | ATP binding |
4.17×10−2 | 3 |
| GO:0005515 | GOTERM_MF | Protein
binding |
7.33×10−2 | 5 |
| GO:0004672 | GOTERM_MF | Protein kinase
activity |
8.24×10−2 | 2 |
| GO:0004674 | GOTERM_MF | Protein
serine/threonine kinase activity |
8.62×10−2 | 2 |
 | Table VI.KEGG pathway analysis of the hub
genes of miR-15b-5p in HCC. |
Table VI.
KEGG pathway analysis of the hub
genes of miR-15b-5p in HCC.
| Category | Term | Count | P-value | Genes |
|---|
| KEGG_PATHWAY | hsa04066: HIF-1
signaling pathway | 3 |
5.92×10−4 | MAP2K1, IGF1,
TLR4 |
| KEGG_PATHWAY | hsa05205:
Proteoglycans in cancer | 3 |
2.45×10−3 | MAP2K1, IGF1,
TLR4 |
| KEGG_PATHWAY | hsa04151: PI3K-Akt
signaling pathway | 3 |
7.21×10−3 | MAP2K1, IGF1,
TLR4 |
| KEGG_PATHWAY | hsa04730: Long-term
depression | 2 |
2.58×10−2 | MAP2K1, IGF1 |
| KEGG_PATHWAY | hsa05214:
Glioma | 2 |
2.80×10−2 | MAP2K1, IGF1 |
| KEGG_PATHWAY | hsa05218:
Melanoma | 2 |
3.05×10−2 | MAP2K1, IGF1 |
| KEGG_PATHWAY | hsa04914:
Progesterone-mediated oocyte maturation | 2 |
3.73×10−2 | MAP2K1, IGF1 |
| KEGG_PATHWAY | hsa05215: Prostate
cancer | 2 |
3.77×10−2 | MAP2K1, IGF1 |
| KEGG_PATHWAY | hsa04620: Toll-like
receptor signaling pathway | 2 |
4.53×10−2 | MAP2K1, TLR4 |
| KEGG_PATHWAY | hsa04114: Oocyte
meiosis | 2 |
4.66×10−2 | MAP2K1, IGF1 |
Discussion
In the present study, the results of the
investigation demonstrated an overall moderate test performance of
miR-15b-5p with respect to its clinical value. The summary
sensitivity of the plasma/serum miR-15b-5p (81%) revealed
superiority compared to AFP despite an overall sensitivity of less
than 60%. The AUC value was 0.83 in serum and 0.82 in tissue. Data
comparing HCC with non-HCC tissue in the same liver or others
controls indicated different factors. In our study, detection of
miR-15b-5p in serum/plasma sample had a more favorable diagnostic
accuracy than that in tissues. The results also revealed via SMD
analysis, that the miR-15b-5p expression level in HCC was markedly
overexpressed when compared to non-HCC tissues samples. In detail,
the random-effects model was used for the pooled SMD of miR-15b-5p
to resolve the problem of heterogeneity. Furthermore, to decrease
the heterogeneity, we also performed subgroup analyses with both
SMD and sROC methods. The subgroups included sample sources
(tissues and serum/plasma) and control types (healthy controls,
adjacent non-cancerous hepatic tissues, HBV+ or
HCV+ tissues, liver cirrhosis tissues, and those
combining HBV+/HCV+ and cirrhosis), which
revealed that miR-15b-5p may be a prospective biomarker to
distinguish HCC patients from healthy people. Numerous HCC patients
have a background of liver cirrhosis and chronic HBV and/or HCV.
Whether circulating miR-15b-5p can be used to differentiate HCC
from benign hepatic lesions has been studied (16). Furthermore, our analysis of the
studies indicated that the summary sensitivity and specificity of
miR-15b-5p for distinguishing HCC from chronic HBV and/or HCV as
well as liver cirrhosis were 60 and 80%, respectively, indicating a
potential value that was worth exploring. It is regrettable that
there is no evidence that miR-15b-5p is related to the progression
of HCC, which is worthy of further study.
Some studies have revealed that circulating miRNAs
can be potential markers for HCC determination. These miRNAs have
been reported to be deregulated in cirrhosis and during development
of hepatic malignancy. Abdalla et al summarized the tested
diagnostic accuracy of miR-618 and miR-650 in HCC which were
revealed to be 0.71 and 0.70, and may be of great value for the
early diagnosis of HCC (28). Tan
et al reported that serum miR-206, miR-141-3p, miR-433-3p,
miR-1228-5p, miR-199a-5p, miR-122-5p, miR-192-5p and miR-26a-5p
were potential circulating markers for the diagnosis of HCC with
AUC from 0.53–0.73 (29). In the
current study, compared with the non-neoplastic controls, the
miR-15b-5p levels in HCC revealed a favorable diagnostic value with
a pooled AUC of 0.81 which indicated that miR-15b-5p may be a
prospective biomarker to distinguish HCC patients from non-HCC
controls. Our studies on miR-15b-5p in HCC are limited and the role
of miR-15b-5p remains largely unknown. The main limitation of this
study was that the sample size was small and present findings
should be validated in trials with more cases.
With regards to the existing related literature,
there were 3 studies that addressed the clinical potential of
miR-15b-5p. Hung et al (14)
first indicated that miR-15b-5p could be utilized for the early
detection of HCC. However, no explicit diagnostic test was
conducted in this study. The study of Liu et al (15) noted that binding of miR-130b and
miR-15b-5p could improve the accuracy of HCC diagnosis based on
expression data collected from HCC patients, HBV carriers and
healthy controls. Chen et al (16) designed three subgroups including HCC
patients, cirrhosis patients and healthy individuals to evaluate
the clinical potential of miR-15b-5p. Only two genes: OIP5 and
Rab1A have been revealed to be the direct targets of miR-15b-5p in
HCC, however, the existing studies solely concentrated on the
performance of circulating miR-15b-5p. Therefore, we collected
expression profiles in hepatic tissues to broaden the spectrum of
research and carried out a comprehensive analysis.
In bioinformatics analyses, KEGG analysis
highlighted the insulin signaling pathway, which was connected with
other enriched pathways in our analysis. Insulin can induce
phosphorylation of the insulin receptor substrate (IRS), thus
allowing IRS to interact with the PI3K/Akt signaling pathway and
MAPK signaling pathway, which are involved in biological
mechanisms, such as glycogen synthesis, cell glucose intake,
protein synthesis and gene transcription (30). In HCC, aberrantly elevated insulin
receptor and IRS-1 were demonstrated to have a correlation with
tumorigenesis contributing to precancerous liver glycogenosis and
hepatocellular proliferation (31–33).
Tanaka et al also reported that IRS-1 could impede
transforming growth factor β1-induced cell apoptosis, which may
increase the risk of HCC (34). In
association with IRS-1, the PI3K/Akt signaling pathway and MAPK
signaling pathway were also considered to play roles in the
molecular mechanism of HCC, and the joint effect of these pathways
was also investigated by researchers. Liang et al (35) reported that aconitum coreanum
polysaccharide inhibited the expression of pituitary tumor
transforming gene 1, an oncogene, by suppressing the PI3K/Akt
signaling pathway and upregulating the MAPK signaling pathway. A
study from Gedaly et al (36)
also discovered that targeting the PI3K/Akt signaling pathway and
MAPK signaling pathway could attenuate the proliferation of HCC
cells. Moreover, these two signaling pathways were also revealed to
have independent correlations with HCC cell growth (37,38)
apoptosis (39,40) migration, and invasion, as well as
adhesion (41–43), which affects the tumorigenesis and
progression of HCC (44,45) via various genes. Therefore, we
concluded that miR-15b-5p may participate in hepatocellular
carcinogenesis via diverse pathways.
However, certain limitations still exist in our
study. Significant heterogeneity was considered in our
meta-analysis. The sample types, study design, population, sample
size and some of the clinical characteristics may contribute to the
increased heterogeneity. Furthermore, overexpression of miR-15b-5p
has been previously reported in other malignancies, such as
colorectal cancer (46), endometrial
endometrioid adenocarcinoma (47) and
non-small cell lung cancer (48). In
summary, we confirmed that increased miR-15b-5p may not be
particularly connected with HCC itself, however it affects the
progression of different cancers. For this reason, in daily
clinical practice, miR-15b-5p alone may not play an essential role
for HCC, however if it is used in combination with other markers,
this biomarker may improve test performance.
To conclude, a high miR-15b-5p level may be one the
probable causes of HCC tumorigenesis, however, it could also be the
consequence after HCC has already occurred, which warrants further
investigation. Based on the marked upregulation of the level of
miR-15b-5p in HCC, its potential clinical value is anticipated,
which also requires practical validation. Notably, the prospective
role and signaling pathways of miR-15b-5p have been revealed by
in silico methods only, and thus, the specific mechanism of
miR-15b-5p in HCC requires further study.
Acknowledgements
Not applicable.
Funding
The present study was supported by a fund from the
National Natural Science Foundation of China (NSFC81560386).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
WYP collected the public data and drafted the
present study. JHZ and PPW performed the statistical analysis and
constructed enrichment pathways. DYW and JYW re-analyzed the data
of the results and drafted the section of the results, including
writing and modifying tables and figures. GC and ZBF participated
in the design and revision of the study. All authors read and
approved the final manuscript
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
miR-15b-5p
|
microRNA-15b-5p
|
|
HCC
|
hepatocellular carcinoma
|
|
GEO
|
gene expression omnibus
|
|
TCGA
|
The cancer genome atlas
|
|
ROC
|
receiver operating characteristic
|
|
AUC
|
area under curves
|
|
GO
|
gene ontology
|
|
KEGG
|
Kyoto encyclopedia of genes and
genomes
|
|
PPI
|
protein-protein interactions
|
References
|
1
|
Bosetti C, Turati F and La Vecchia C:
Hepatocellular carcinoma epidemiology. Best Pract Res Clin
Gastroenterol. 28:753–770. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schwartz M, Roayaie S and Konstadoulakis
M: Strategies for the management of hepatocellular carcinoma. Nat
Clin Pract Oncol. 4:424–432. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Trevisani F, Cantarini MC, Wands JR and
Bernardi M: Recent advances in the natural history of
hepatocellular carcinoma. Carcinogenesis. 29:1299–1305. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Daniele B, Bencivenga A, Megna AS and
Tinessa V: Alpha-fetoprotein and ultrasonography screening for
hepatocellular carcinoma. Gastroenterology. 127 (5 Suppl
1):S108–S112. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li JJ, Luo J, Lu JN, Liang XN, Luo YH, Liu
YR, Yang J, Ding H, Qin GH, Yang LH, et al: Relationship between
TRAF6 and deterioration of HCC: An immunohistochemical and in vitro
study. Cancer Cell Int. 16:762016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Luo Y, Ren F, Liu Y, Shi Z, Tan Z, Xiong
H, Dang Y and Chen G: Clinicopathological and prognostic
significance of high Ki-67 labeling index in hepatocellular
carcinoma patients: A meta-analysis. Int J Clin Exp Med.
8:10235–10247. 2015.PubMed/NCBI
|
|
7
|
Huang Z, Huang L, Shen S, Li J, Lu H, Mo
W, Dang Y, Luo D, Chen G and Feng Z: Sp1 cooperates with Sp3 to
upregulate MALAT1 expression in human hepatocellular carcinoma.
Oncol Rep. 34:2403–2412. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang X, Tang W, Li R, He R, Gan T, Luo Y,
Chen G and Rong M: Downregulation of microRNA-132 indicates
progression in hepatocellular carcinoma. Exp Ther Med.
12:2095–2101. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang WT, Wang HL, Yang H, Ren FH, Luo YH,
Huang CQ, Liang YY, Liang HW, Chen G and Dang YW: Lower expressed
miR-198 and its potential targets in hepatocellular carcinoma: A
clinicopathological and in silico study. Onco Targets Ther.
9:5163–5180. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang X, Tang W, Chen G, Ren F, Liang H,
Dang Y and Rong M: An encapsulation of gene signatures for
hepatocellular carcinoma, MicroRNA-132 predicted target genes and
the corresponding overlaps. PLoS One. 11:e01594982016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He R, Yang L, Lin X, Chen X, Lin X, Wei F,
Liang X, Luo Y, Wu Y, Gan T, et al: MiR-30a-5p suppresses cell
growth and enhances apoptosis of hepatocellular carcinoma cells via
targeting AEG-1. Int J Clin Exp Pathol. 8:15632–15641.
2015.PubMed/NCBI
|
|
12
|
Liu Y, Ren F, Luo Y, Rong M, Chen G and
Dang Y: Down-regulation of MiR-193a-3p dictates deterioration of
HCC: A clinical real-time qRT-PCR study. Med Sci Monit.
21:2352–2360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang YH, Lin KH, Chen HC, Chang ML, Hsu
CW, Lai MW, Chen TC, Lee WC, Tseng YH and Yeh CT: Identification of
postoperative prognostic microRNA predictors in hepatocellular
carcinoma. PLoS One. 7:e371882012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hung CH, Hu TH, Lu SN, Kuo FY, Chen CH,
Wang JH, Huang CM, Lee CM, Lin CY, Yen YH and Chiu YC: Circulating
microRNAs as biomarkers for diagnosis of early hepatocellular
carcinoma associated with hepatitis B virus. Int J Cancer.
138:714–720. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu AM, Yao TJ, Wang W, Wong KF, Lee NP,
Fan ST, Poon RT, Gao C and Luk JM: Circulating miR-15b and miR-130b
in serum as potential markers for detecting hepatocellular
carcinoma: A retrospective cohort study. BMJ Open. 2:e0008252012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen Y, Chen J, Liu Y, Li S and Huang P:
Plasma miR-15b-5p, miR-338-5p, and miR-764 as biomarkers for
hepatocellular carcinoma. Med Sci Monit. 21:1864–1871. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang Y, Dang YW, Wang X, Yang X, Zhang R,
Lv ZL and Chen G: Comprehensive analysis of long non-coding RNA
PVT1 gene interaction regulatory network in hepatocellular
carcinoma using gene microarray and bioinformatics. Am J Transl
Res. 9:3904–3917. 2017.PubMed/NCBI
|
|
18
|
Zhang Y, He RQ, Dang YW, Zhang XL, Wang X,
Huang SN, Huang WT, Jiang MT, Gan XN, Xie Y, et al: Comprehensive
analysis of the long noncoding RNA HOXA11-AS gene interaction
regulatory network in NSCLC cells. Cancer Cell Int. 16:892016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kew MC: Epidemiology of chronic hepatitis
B virus infection, hepatocellular carcinoma, and hepatitis B
virus-induced hepatocellular carcinoma. Pathol Biol (Paris).
58:273–277. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Murakami Y, Kubo S, Tamori A, Itami S,
Kawamura E, Iwaisako K, Ikeda K, Kawada N, Ochiya T and Taguchi YH:
Comprehensive analysis of transcriptome and metabolome analysis in
Intrahepatic Cholangiocarcinoma and Hepatocellular Carcinoma. Sci
Rep. 5:162942015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shen J, LeFave C, Sirosh I, Siegel AB,
Tycko B and Santella RM: Integrative epigenomic and genomic
filtering for methylation markers in hepatocellular carcinomas. BMC
Med Genomics. 8:282015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sato F, Hatano E, Kitamura K, Myomoto A,
Fujiwara T, Takizawa S, Tsuchiya S, Tsujimoto G, Uemoto S and
Shimizu K: MicroRNA profile predicts recurrence after resection in
patients with hepatocellular carcinoma within the Milan Criteria.
PLoS One. 6:e164352011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Burchard J, Zhang C, Liu AM, Poon RT, Lee
NP, Wong KF, Sham PC, Lam BY, Ferguson MD, Tokiwa G, et al:
microRNA-122 as a regulator of mitochondrial metabolic gene network
in hepatocellular carcinoma. Mol Syst Biol. 6:4022010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li W, Xie L, He X, Li J, Tu K, Wei L, Wu
J, Guo Y, Ma X, Zhang P, et al: Diagnostic and prognostic
implications of microRNAs in human hepatocellular carcinoma. Int J
Cancer. 123:1616–1622. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y
and Zhuang SM: MicroRNA-101, down-regulated in hepatocellular
carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer
Res. 69:1135–1142. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Diaz G, Melis M, Tice A, Kleiner DE,
Mishra L, Zamboni F and Farci P: Identification of microRNAs
specifically expressed in hepatitis C virus-associated
hepatocellular carcinoma. Int J Cancer. 133:816–824. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Martinez-Quetglas I, Pinyol R, Dauch D,
Torrecilla S, Tovar V, Moeini A, Alsinet C, Portela A,
Rodriguez-Carunchio L, Solé M, et al: IGF2 Is Up-regulated by
epigenetic mechanisms in hepatocellular carcinomas and is an
actionable oncogene product in experimental models.
Gastroenterology. 151:1192–1205. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Abdalla MA and Haj-Ahmad Y: Promising
candidate urinary MicroRNA biomarkers for the early detection of
hepatocellular carcinoma among high-risk Hepatitis C virus Egyptian
patients. J Cancer. 3:19–31. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tan Y, Ge G, Pan T, Wen D, Chen L, Yu X,
Zhou X and Gan J: A serum microRNA panel as potential biomarkers
for hepatocellular carcinoma related with hepatitis B virus. PLoS
One. 9:e1079862014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bevan P: Insulin signalling. J Cell Sci.
114:1429–1430. 2001.PubMed/NCBI
|
|
31
|
Aleem E, Nehrbass D, Klimek F, Mayer D and
Bannasch P: Upregulation of the insulin receptor and type I
insulin-like growth factor receptor are early events in
hepatocarcinogenesis. Toxicol Pathol. 39:524–543. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nehrbass D, Klimek F and Bannasch P:
Overexpression of insulin receptor substrate-1 emerges early in
hepatocarcinogenesis and elicits preneoplastic hepatic
glycogenosis. Am J Pathol. 152:341–345. 1998.PubMed/NCBI
|
|
33
|
Mohr L, Banerjee K, Kleinschmidt M,
Bartolomé Rodriguez MM and Wands JR: Transgenic overexpression of
insulin receptor substrate 1 in hepatocytes enhances hepatocellular
proliferation in young mice only. Hepatol Res. 38:1233–1240.
2008.PubMed/NCBI
|
|
34
|
Tanaka S and Wands JR: Insulin receptor
substrate 1 overexpression in human hepatocellular carcinoma cells
prevents transforming growth factor beta1-induced apoptosis. Cancer
Res. 56:3391–3394. 1996.PubMed/NCBI
|
|
35
|
Liang M, Liu J, Ji H, Chen M, Zhao Y, Li
S, Zhang X and Li J: A Aconitum coreanum polysaccharide fraction
induces apoptosis of hepatocellular carcinoma (HCC) cells via
pituitary tumor transforming gene 1 (PTTG1)-mediated suppression of
the P13K/Akt and activation of p38 MAPK signaling pathway and
displays antitumor activity in vivo. Tumour Biol. 36:7085–7091.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gedaly R, Angulo P, Hundley J, Daily MF,
Chen C and Evers BM: PKI-587 and sorafenib targeting PI3K/AKT/mTOR
and Ras/Raf/MAPK pathways synergistically inhibit HCC cell
proliferation. J Surg Res. 176:542–548. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu R, Duan L, Cui F, Cao J, Xiang Y, Tang
Y and Zhou L: S100A9 promotes human hepatocellular carcinoma cell
growth and invasion through RAGE-mediated ERK1/2 and p38 MAPK
pathways. Exp Cell Res. 334:228–238. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fang X, Yang D, Luo H, Wu S, Dong W, Xiao
J, Yuan S, Ni A, Zhang KJ, Liu XY and Chu L: SNORD126 promotes HCC
and CRC cell growth by activating the PI3K-AKT pathway through
FGFR2. J Mol Cell Biol. 9:243–255. 2017.PubMed/NCBI
|
|
39
|
Bao H, Liu P, Jiang K, Zhang X, Xie L,
Wang Z and Gong P: Huaier polysaccharide induces apoptosis in
hepatocellular carcinoma cells through p38 MAPK. Oncol Lett.
12:1058–1066. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jiang S, Wang Q, Feng M, Li J, Guan Z, An
D, Dong M, Peng Y, Kuerban K and Ye L: C2-ceramide enhances
sorafenib-induced caspase-dependent apoptosis via PI3K/AKT/mTOR and
Erk signaling pathways in HCC cells. Appl Microbiol Biotechnol.
101:1535–1546. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhu B, Shi S, Ma YG, Fan F and Yao ZZ:
Lysophosphatidic acid enhances human hepatocellular carcinoma cell
migration, invasion and adhesion through P38 MAPK pathway.
Hepatogastroenterology. 59:785–789. 2012.PubMed/NCBI
|
|
42
|
Hsieh YH, Hsieh SC, Lee CH, Yang SF, Cheng
CW, Tang MJ, Lin CL, Lin CL and Chou RH: Targeting EMP3 suppresses
proliferation and invasion of hepatocellular carcinoma cells
through inactivation of PI3K/Akt pathway. Oncotarget.
6:34859–34874. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zheng Y, Wang X, Wang H, Yan W, Zhang Q
and Chang X: Bone morphogenetic protein 2 inhibits hepatocellular
carcinoma growth and migration through downregulation of the
PI3K/AKT pathway. Tumour Biol. 35:5189–5198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Qiu FN, Huang Y, Chen DY, Li F, Wu YA, Wu
WB and Huang XL: Eukaryotic elongation factor-1α 2 knockdown
inhibits hepatocarcinogenesis by suppressing PI3K/Akt/NF-κB
signaling. World J Gastroenterol. 22:4226–4237. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhao J, Dong QZ, Zhong F, Cai LL, Qin ZY,
Liu Y, Lin CZ, Qin LX and He FC: NMI promotes hepatocellular
carcinoma progression via BDKRB2 and MAPK/ERK pathway. Oncotarget.
8:12174–12185. 2017.PubMed/NCBI
|
|
46
|
Li J, Chen Y, Guo X, Zhou L, Jia Z, Tang
Y, Lin L, Liu W and Ren C: Inhibition of miR-15b decreases cell
migration and metastasis in colorectal cancer. Tumour Biol.
37:8765–8773. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang L, Chen YJ, Xu K, Xu H, Shen XZ and
Tu RQ: Circulating microRNAs as a fingerprint for endometrial
endometrioid adenocarcinoma. PLoS One. 9:e1107672014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fan L, Qi H, Teng J, Su B, Chen H, Wang C
and Xia Q: Identification of serum miRNAs by nano-quantum dots
microarray as diagnostic biomarkers for early detection of
non-small cell lung cancer. Tumour Biol. 37:7777–7784. 2016.
View Article : Google Scholar : PubMed/NCBI
|