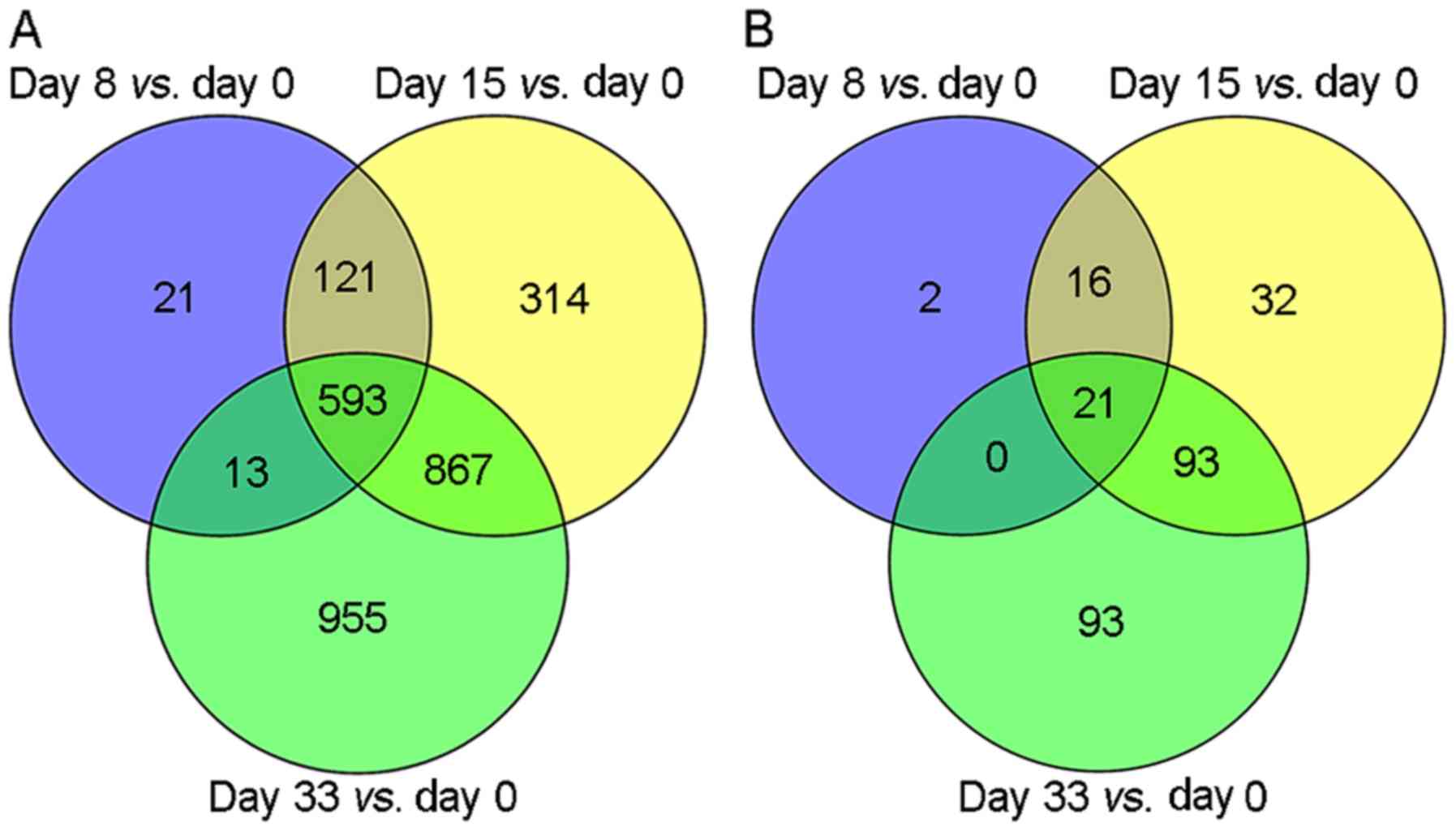
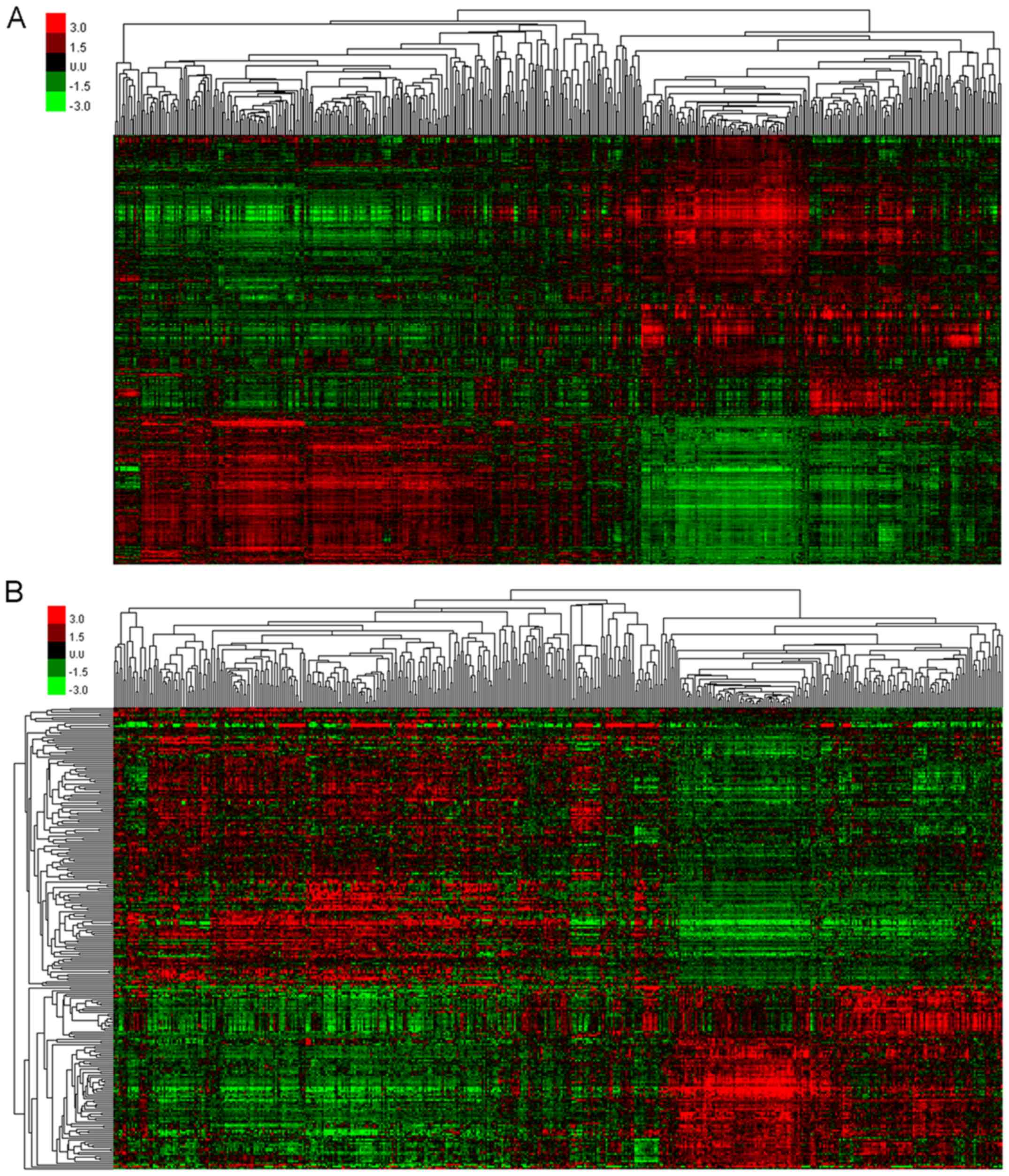
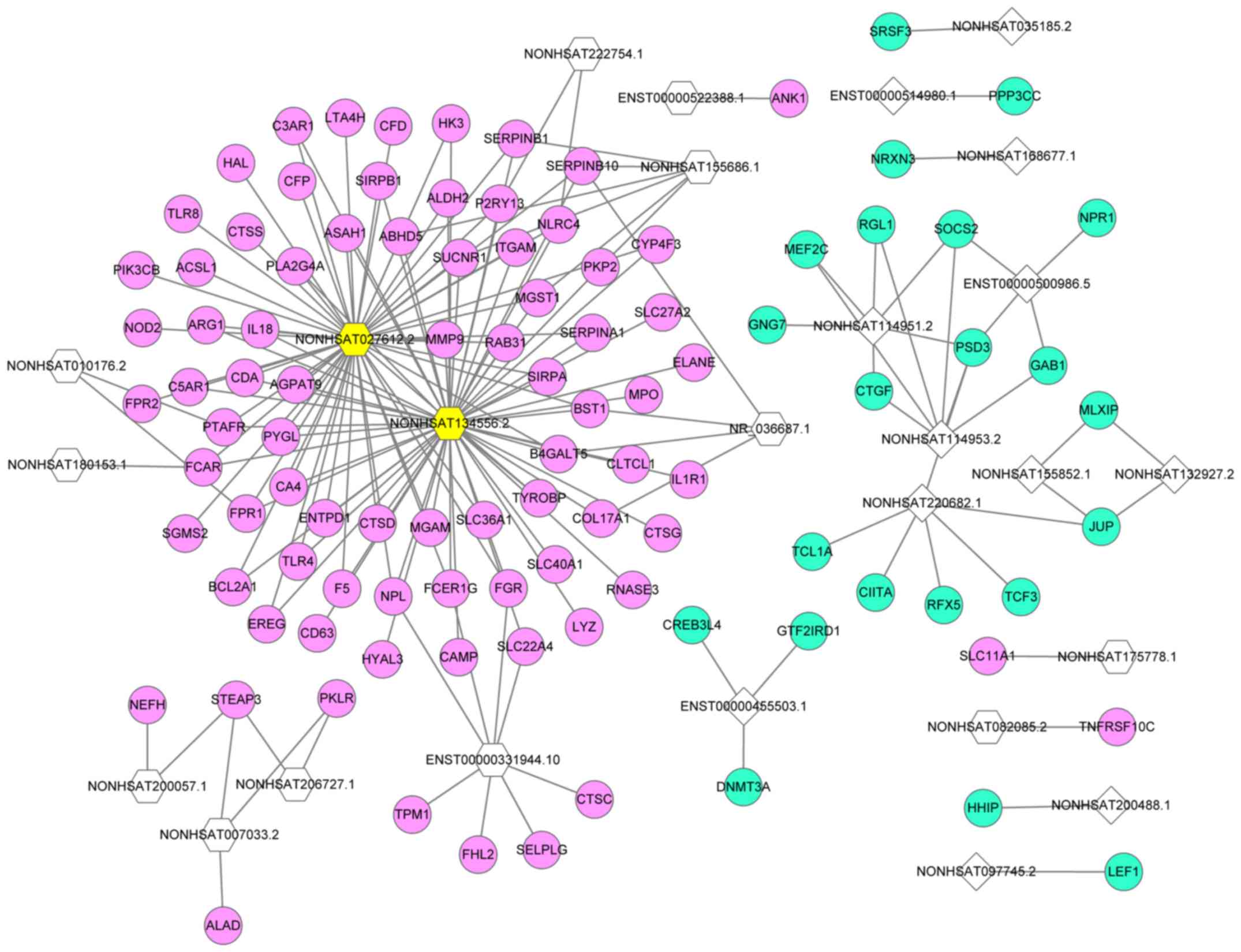
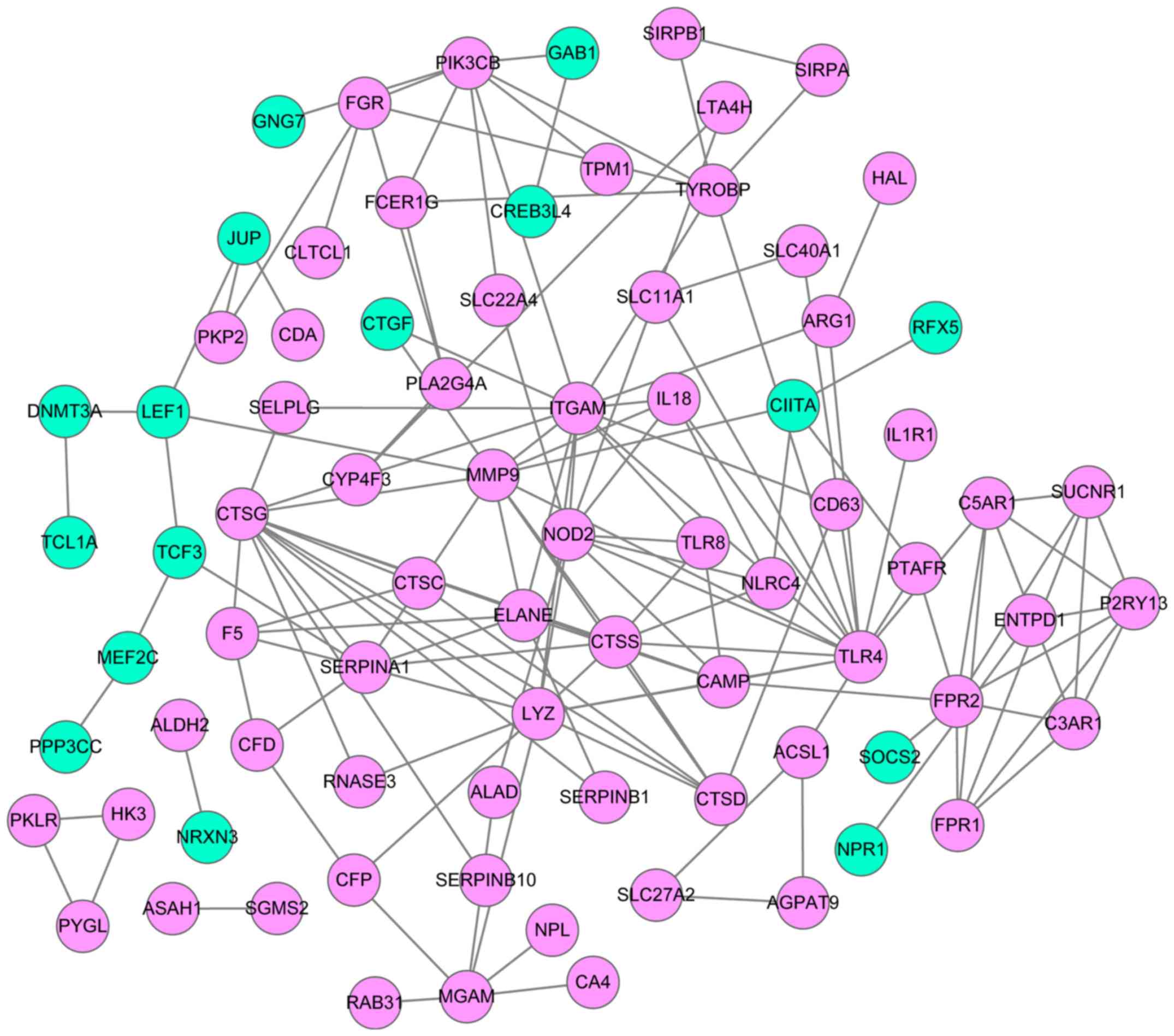
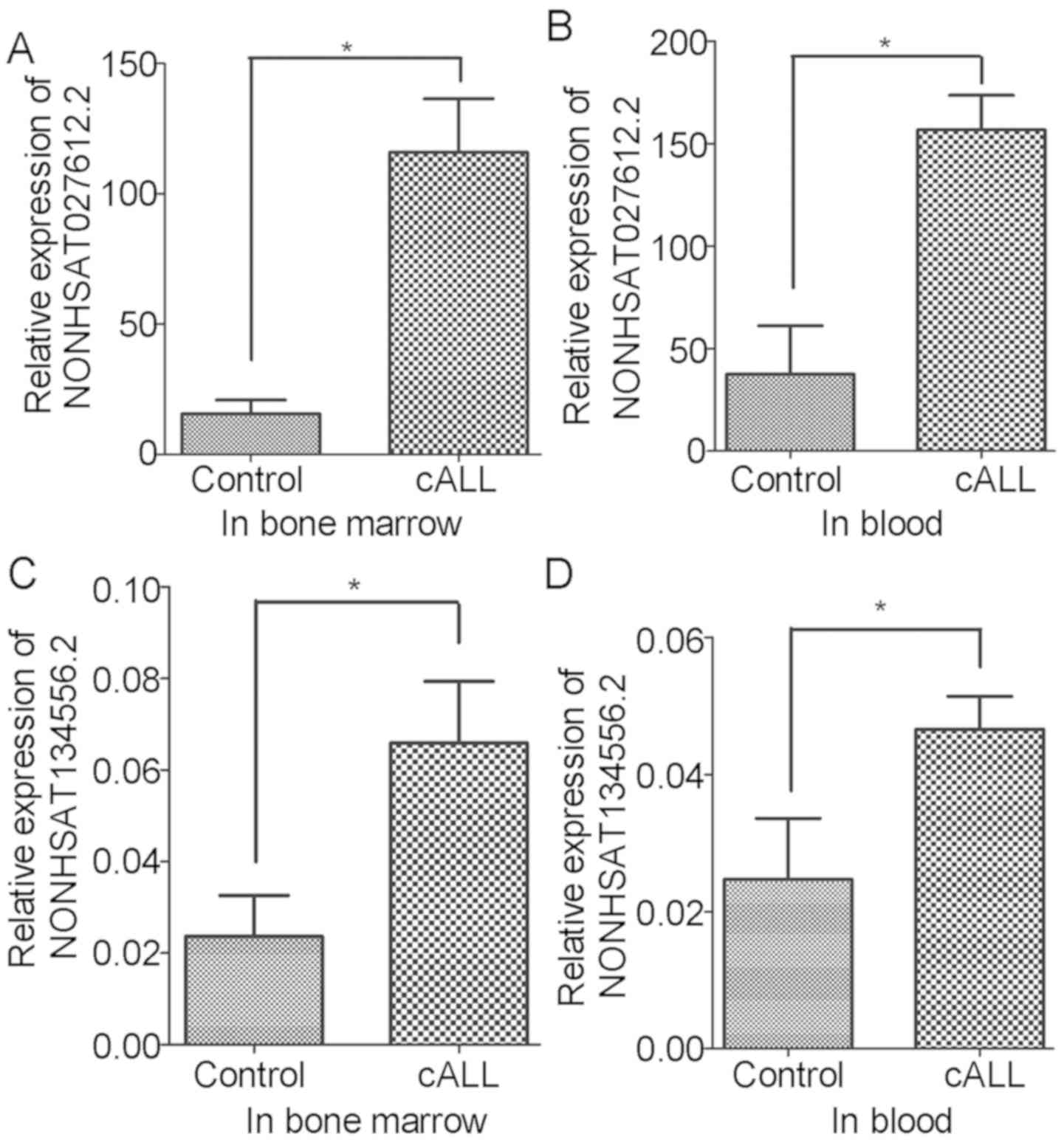
|
1
|
Roberts KG, Li Y, Payne-Turner D, Harvey
RC, Yang YL, Pei D, McCastlain K, Ding L, Lu C, Song G, et al:
Targetable kinase-activating lesions in Ph-like acute lymphoblastic
leukemia. N Engl J Med. 371:1005–1015. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stock W: Adolescents and young adults with
acute lymphoblastic leukemia. Hematology Am Soc Hematol Educ
Program. 2010:21–29. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hunger SP, Lu X, Devidas M, Camitta BM,
Gaynon PS, Winick NJ, Reaman GH and Carroll WL: Improved survival
for children and adolescents with acute lymphoblastic leukemia
between 1990 and 2005: A report from the children's oncology group.
J Clin Oncol. 30:1663–1669. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hoelzer D: Treatment of acute
lymphoblastic leukemia. Semin Hematol. 31:1–15. 1994.PubMed/NCBI
|
|
5
|
Eapen M, Zhang MJ, Devidas M, Raetz E,
Barredo JC, Ritchey AK, Godder K, Grupp S, Lewis VA, Malloy K, et
al: Outcomes after HLA-matched sibling transplantation or
chemotherapy in children with acute lymphoblastic leukemia in a
second remission after an isolated central nervous system relapse:
A collaborative study of the Children's Oncology Group and the
center for international blood and marrow transplant research.
Leukemia. 22:281–286. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liz J and Esteller M: lncRNAs and
microRNAs with a role in cancer development. Biochim Biophys Acta.
1859:169–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou X, Ye F, Yin C, Zhuang Y, Yue G and
Zhang G: The Interaction between MiR-141 and lncRNA-H19 in
regulating cell proliferation and migration in gastric cancer. Cell
Physiol Biochem. 36:1440–1452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fang Z, Wu L, Wang L, Yang Y, Meng Y and
Yang H: Increased expression of the long non-coding RNA UCA1 in
tongue squamous cell carcinomas: A possible correlation with cancer
metastasis. Oral Surg Oral Med Oral Pathol Oral Radiol. 117:89–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shi SJ, Wang LJ, Yu B, Li YH, Jin Y and
Bai XZ: LncRNA-ATB promotes trastuzumab resistance and
invasion-metastasis cascade in breast cancer. Oncotarget.
6:11652–11663. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang K, Long B, Zhou LY, Liu F, Zhou QY,
Liu CY, Fan YY and Li PF: CARL lncRNA inhibits anoxia-induced
mitochondrial fission and apoptosis in cardiomyocytes by impairing
miR-539-dependent PHB2 downregulation. Nat Commun. 5:35962014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ding C, Yang Z, Lv Z, DU C, Xiao H, Peng
C, Cheng S, Xie H, Zhou L, Wu J and Zheng S: Long non-coding RNA
PVT1 Is associated with tumor progression and predicts recurrence
in hepatocellular carcinoma patients. Oncol Lett. 9:955–963. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fang K, Han BW, Chen ZH, Lin KY, Zeng CW,
Li XJ, Li JH, Luo XQ and Chen YQ: A distinct set of long non-coding
RNAs in childhood MLL-rearranged acute lymphoblastic leukemia:
Biology and epigenetic target. Hum Mol Genet. 23:3278–3288. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Trimarchi T, Bilal E, Ntziachristos P,
Fabbri G, Dalla-Favera R, Tsirigos A and Aifantis I: Genome-wide
mapping and characterization of Notch-regulated long noncoding RNAs
in acute leukemia. Cell. 158:593–606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Y, Wu P, Lin R, Rong L, Xue Y and
Fang Y: LncRNA NALT interaction with NOTCH1 promoted cell
proliferation in pediatric T cell acute lymphoblastic leukemia. Sci
Rep. 5:137492015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yeoh A, Li ZH, Dong DF, Lu Y, Jiang N,
Trka J, Tan AM, Lin HP, Quah TC, Ariffin H and Wong L: Effective
response metric: A novel tool to predict relapse in childhood acute
lymphoblastic leukaemia using time-series gene expression
profiling. Br J Haematol. 181:653–663. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Miao MH, Ji XQ, Zhang H, Xu J, Zhu H and
Shao XJ: miR-590 promotes cell proliferation and invasion in T-cell
acute lymphoblastic leukaemia by inhibiting RB1. Oncotarget.
7:39527–39534. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Miller LD, Long PM, Wong L, Mukherjee S,
Mcshane LM and Liu ET: Optimal gene expression analysis by
microarrays. Cancer Cell. 2:353–361. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ramoni MF, Sebastiani P and Kohane IS:
Cluster analysis of gene expression dynamics. Proc Natl Acad Sci
USA. 99:9121–9126. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang CF, Zhao CC, Weng WJ, Lei J, Lin Y,
Mao Q, Gao GY, Feng JF and Jiang JY: Alteration in long non-coding
RNA expression after traumatic brain injury in rats. J Neurotrauma.
34:2100–2108. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Monnier P, Martinet C, Pontis J, Stancheva
I, Ait-Si-Ali S and Dandolo L: H19 lncRNA controls gene expression
of the Imprinted Gene Network by recruiting MBD1. Proc Natl Acad
Sci USA. 110:20693–20698. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nie L, Wu HJ, Hsu JM, Chang SS, Labaff AM,
Li CW, Wang Y, Hsu JL and Hung MC: Long non-coding RNAs: Versatile
master regulators of gene expression and crucial players in cancer.
Am J Transl Res. 4:127–150. 2012.PubMed/NCBI
|
|
23
|
Mrózek K, Radmacher MD, Bloomfield CD and
Marcucci G: Molecular signatures in acute myeloid leukemia. Curr
Opin Hematol. 16:64–69. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hayashi EA, Akira S and Nobrega A: Role of
TLR in B cell development: Signaling through TLR4 promotes B cell
maturation and is inhibited by TLR2. J Immunol. 174:6639–6647.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Miedema KG, te Poele EM, Tissing WJ,
Postma DS, Koppelman GH, de Pagter AP, Kamps WA, Alizadeh BZ,
Boezen HM and de Bont ES: Association of polymorphisms in the TLR4
gene with the risk of developing neutropenia in children with
leukemia. Leukemia. 25:995–1000. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He W, Liu Q, Wang L, Chen W, Li N and Cao
X: TLR4 signaling promotes immune escape of human lung cancer cells
by inducing immunosuppressive cytokines and apoptosis resistance.
Mol Immunol. 44:2850–2859. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
McGettrick AF and O'Neill LA: Toll-like
receptors: Key activators of leucocytes and regulator of
haematopoiesis. Br J Haematol. 139:185–193. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hewamana S, Alghazal S, Lin TT, Clement M,
Jenkins C, Guzman ML, Jordan CT, Neelakantan S, Crooks PA, Burnett
AK, et al: The NF-kappaB subunit Rel A is associated with in vitro
survival and clinical disease progression in chronic lymphocytic
leukemia and represents a promising therapeutic target. Blood.
111:4681–4689. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ntoufa S, Vardi A, Papakonstantinou N,
Anagnostopoulos A, Aleporou-Marinou V, Belessi C, Ghia P,
Caligaris-Cappio F, Muzio M and Stamatopoulos K: Distinct innate
immunity pathways to activation and tolerance in subgroups of
chronic lymphocytic leukemia with distinct immunoglobulin
receptors. Mol Med. 18:1281–1291. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mayor NP, Shaw BE, Hughes DA,
Maldonado-Torres H, Madrigal JA, Keshav S and Marsh SG: Single
nucleotide polymorphisms in the NOD2/CARD15 gene are associated
with an increased risk of relapse and death for patients with acute
leukemia after hematopoietic stem-cell transplantation with
unrelated donors. J Clin Oncol. 25:4262–4269. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Muzio M, Fonte E and Caligaris-Cappio F:
Toll-like receptors in chronic lymphocytic leukemia. Mediterr J
Hematol Infect Dis. 4:e20120552012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Muzio M, Scielzo C, Bertilaccio MT,
Frenquelli M, Ghia P and Caligaris-Cappio F: Expression and
function of toll like receptors in chronic lymphocytic leukaemia
cells. Br J Haematol. 144:507–516. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rahman MK, Midtling EH, Svingen PA, Xiong
Y, Bell MP, Tung J, Smyrk T, Egan LJ and Faubion WA Jr: The
pathogen recognition receptor NOD2 regulates human
FOXP3+ T cell survival. J Immunol. 184:7247–7256. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gomes S, Marques PI, Matthiesen R and
Seixas S: Adaptive evolution and divergence of SERPINB3: A young
duplicate in great Apes. PLoS One. 9:e1049352014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chang WT, Pan CY, Rajanbabu V, Cheng CW
and Chen JY: Tilapia (Oreochromis mossambicus) antimicrobial
peptide, hepcidin 1–5, shows antitumor activity in cancer cells.
Peptides. 32:342–352. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zöller M: CD44: Can a cancer-initiating
cell profit from an abundantly expressed molecule? Nat Rev Cancer.
11:254–267. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Korkmaz B, Horwitz MS, Jenne DE and
Gauthier F: Neutrophil elastase, proteinase 3 and cathepsin G as
therapeutic targets in human diseases. Pharmacol Rev. 62:726–759.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gocheva V, Zeng W, Ke D, Klimstra D,
Reinheckel T, Peters C, Hanahan D and Joyce JA: Distinct roles for
cysteine cathepsin genes in multistage tumorigenesis. Genes Dev.
20:543–556. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang B, Sun J, Kitamoto S, Yang M, Grubb
A, Chapman HA, Kalluri R and Shi GP: Cathepsin S controls
angiogenesis and tumor growth via matrix-derived angiogenic
factors. J Biol Chem. 281:6020–6029. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gounaris E, Tung CH, Restaino C, Maehr R,
Kohler R, Joyce JA, Ploegh HL, Barrett TA, Weissleder R and Khazaie
K: Live imaging of cysteine-cathepsin activity reveals dynamics of
focal inflammation, angiogenesis and polyp growth. PLoS One.
3:e29162008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang Y, Lim SK, Choong LY, Lee H, Chen Y,
Chong PK, Ashktorab H, Wang TT, Salto-Tellez M, Yeoh KG and Lim YP:
Cathepsin S mediates gastric cancer cell migration and invasion via
a putative network of metastasis-associated proteins. J Proteome
Res. 9:4767–4778. 2010. View Article : Google Scholar : PubMed/NCBI
|