Introduction
Cervical cancer is one of the most prevalent female
cancer types in developing countries, second only to breast cancer
(1). In China, it is estimated that
~61,691 women are newly diagnosed with invasive cervical cancer
(ICC) annually and 29,526 of these succumbed to this disease in
2012 (2). Shanxi province has been
identified as a high-incidence area, with an incidence rate of
23.04 per 100,000 (3,4). Cervical cancer develops from the
associated precursor lesion, cervical intraepithelial neoplasia
(CIN). Thus, it is necessary to develop effective strategies for
mass screening of CIN, in order to reduce the incidence of cervical
cancer.
The ThinPrep cytological test (TCT) and human
papillomavirus (HPV) detection are canonical screening methods for
cervical cancer currently (5–7). However, there are limitations to these
techniques. For example, TCT is only capable of identifying
patients with abnormal cell morphology, and cannot determine if
these patients are at high-risk of lesion progression.
Additionally, the results of TCT may be influenced by sampling and
slide quality, based on the skill and experience of the
practitioner (8). It has also been
reported that HPV infection is reversible in the reproductive
system and only 1–2% of persistent HPV infections progress to
neoplasia (9,10). Thus, it is beneficial to investigate
novel biomarkers for the diagnosis of CIN and evaluation of the
likelihood of lesion progression.
Recent studies have demonstrated that ICC invariably
possesses extra copies of the chromosome arm 3q, which contains the
human telomerase RNA component (h-TERC) gene in the 3q26 region
(11,12). h-TERC encodes the template for
telomerase RNA, corresponding to the repeat sequence which is added
in tandem to the ends of chromosomes, in order to maintain the
telomere length (13). Thus, abnormal
amplification of h-TERC may lead to the cells having increased
telomeres and enhanced proliferation ability, resulting in the
formation of cervical tumors (12,13).
Detection of h-TERC amplification may be beneficial for the
diagnosis of CIN and ICC (14).
c-MYC, located in chromosomal region 8q24, is reported to be the
most common integration site in the HPV genome (15). The c-MYC gene may be overexpressed
simultaneous to the amplification of HPV, which is associated with
the acquisition of a malignant phenotype in cervical cells
(16). Therefore, c-MYC may also be
an important oncogene involved in tumor progression, and hence may
be a potential biomarker for cervical cancer (17). Previously, there have been studies to
evaluate the diagnostic value of h-TERC (18) or c-MYC (19) alone in TCT and HPV, or a combination
of h-TERC and c-MYC (11) in CIN and
cancer. However, limited studies have been performed in order to
determine which is the most efficient and practical method based on
the four tests (TCT, HPV DNA, h-TERC and c-MYC) (8). Additionally, the majority of studies are
aimed at demonstrating the sensitivity and specificity of the
aforementioned biomarkers in distinguishing high-grade cervical
lesions (>CIN2+ or ≥CIN2) and invasive cancer types
from low-grade lesions (11,18,19);
consequently, studies rarely focus on the difference between normal
and all precursor lesions as well as ICC. Furthermore, the
conclusions remain inconsistent between different studies. For
example, Zheng et al (18)
observed that among three methods, h-TERC testing displayed the
highest specificity and positive predictive value, and HPV testing
displayed the highest sensitivity, while TCT was not optimal for
any of the diagnostic parameters. Jiang et al (20) demonstrated that for the detection of
advanced cervical lesions, cytological evaluations were optimal
(93.3%) in terms of specificity, followed by h-TERC amplification
(83.8%) and HPV DNA analyses (39.3%). Zhao et al (19) reported that the specificity of
fluorescence in situ hybridization (FISH) analysis with a
c-MYC-specific probe seemed to be higher compared with TCT and HPV
DNA testing; however, this was not consistent with the results from
Gao et al (8) or Li et
al (11). The diagnostic
performance of c-MYC [area under the curve (AUC)=0.865] was
slightly superior to that of h-TERC (AUC=0.843) in the study
performed by Gao et al (8),
although contrasting results were obtained from Li et al
(11) (c-MYC AUC=0.799 vs. h-TERC
AUC=0.838). Therefore, further studies are required in order to
confirm the diagnostic abilities of h-TERC and c-MYC compared with
TCT and HPV DNA testing.
The aim of the present study was to determine the
optimal programs for screening precancerous lesions and cervical
carcinoma among women in Shanghai, one of the largest cities in
China (21), by TCT, h-TERC- and
MYC-specific FISH and surface plasmon resonance (SPR)-HPV
genotyping.
Materials and methods
Patients and specimens
Residual liquid-based cytology PreservCyt specimens
were obtained from 1,000 women (aged 20–68 years) who were admitted
to the gynecological clinic of Ren Ji Hospital of Shanghai Jiao
Tong University (Shanghai, China) for routine screening between
August 2013 and December 2015. None of the patients had received
cytological tests within 6 months prior to cervical therapy,
including radical surgery (hysterectomy and bilateral pelvic
lymphadenectomy) with or without adjuvant radiotherapy or
chemotherapy. Furthermore, none of the patients had additional
neoplastic diseases. All specimens underwent HPV DNA, h-TERC, c-MYC
and TCT testing. Histopathological examination was performed for
patients who received positive results for any of the
aforementioned tests. The Ethics Committee of Ren Ji Hospital of
Shanghai Jiao Tong University approved this study. Written informed
consent was obtained from all patients when the biopsy was
performed.
Cytological examination
The liquid-based specimens were automatically
processed with the Cytyc T2000 ThinPrep® system (Cytic
Corp., Marlborough, MA, USA) and stained with the Papanicolaou
stain method [95% ethyl alcohol fixation for 10 min; Harris
hematoxylin (Papanicolaou solution A; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) for 3 min; running water, 2–3 times; 0.05%
hydrochloric acid, 30 sec; 95% ethyl alcohol rinsing 2–3 times;
orange G (Papanicolaou solution B, Sigma-Aldrich; Merck KGaA; 1
drop for 10 sec); 95% ethyl alcohol for 30 sec; 100% ethyl alcohol
for 30 sec; polychromatic solution EA50 for 3 min; 95% ethyl
alcohol for 2 min; 100% ethyl alcohol for 2 min; xylene for 2 min;
all at room temperature; and mounted in a mounting medium
(Permount; Thermo Fisher Scientific, Inc., Waltham, MA, USA)].
Cytological diagnoses were classified independently by two
cytopathology physicians, under double-blinded conditions according
to the 2001 Bethesda System (22).
Diagnoses were as follows: i) Negative for intraepithelial lesion
or malignancy; ii) epithelial cell abnormality e.g. atypical
squamous cells of undetermined significance (ASCUS); iii) atypical
squamous cells in which high-grade squamous intraepithelial lesion
cannot be excluded (ASCH); iv) low-grade squamous intraepithelial
lesion (LSIL); v) high-grade squamous intraepithelial lesion
(HSIL); vi) squamous cell carcinoma; and vii) atypical glandular
cells.
h-TERC and c-MYC gene detection
FISH was used in order to detect the amplification
of h-TERC and c-MYC genes as previously described (8), according to the following
procedures.
Preparation of slides
ThinPrep samples were enzymatically digested in 0.1%
collagenase (type B; 20–30 min), centrifuged (2,000 × g at 37°C for
10 min), re-suspended in deionized water (20 min) and fixed twice
with a methanol + acetic acid mixture (volume ratio, 3:1; 10 min)
to prepare one-layer slides.
Pretreatment
Following rinsing twice with 2X sodium chloride plus
sodium citrate (SSC; 5 min), the slides were placed in 0.1 M HCl
for 5 min and incubated with a pepsin solution (0.05% pepsin/0.01 M
HCl; (Sigma-Aldrich; Merck KGaA) for 10 min. Thereafter, the
samples were subjected to dehydration through a graded ethanol
series (70, 85 and 100%).
FISH
The prepared samples and probe mixture (consisting
of probe, 2 ml; hybridization buffer, 7 ml; and deionized water, 1
ml) were denatured in 70% formamide/2X SSC for 5 min and dehydrated
with the above graded ethanol series for 3 min each. Dual-color
fluorescence probes were used for the detection of h-TERC [red,
gene locus-specific probe (GLP) h-TERC probe; green, GLP chromosome
3 centromere-specific (CSP3) control probe], while the single probe
was performed for measurement of c-MYC (red) (GP Medical
Technologies, Beijing, China). The denatured probes were added onto
the denatured slides and hybridized overnight at 42°C. Following
removal of the cover slips, the slides were rinsed with 0.3%
NP-40/0.4X SSC for 2 min at 67°C, followed by washing with 0.1%
NP-40/2X SSC and 70% ethanol for 30 sec and 3 min, respectively.
The slides were mounted with DAPI at 37°C for 10–20 min to
counterstain.
FISH signal interpretation
Images of the slides were obtained on a fluorescence
microscope (DM 2500; Leica Microsystems GmbH, Wetzlar, Germany) and
analyzed using VideoTest-FISH software (version 2.0; VideoTest, St.
Petersburg, Russia). The cell areas were determined through a DAPI
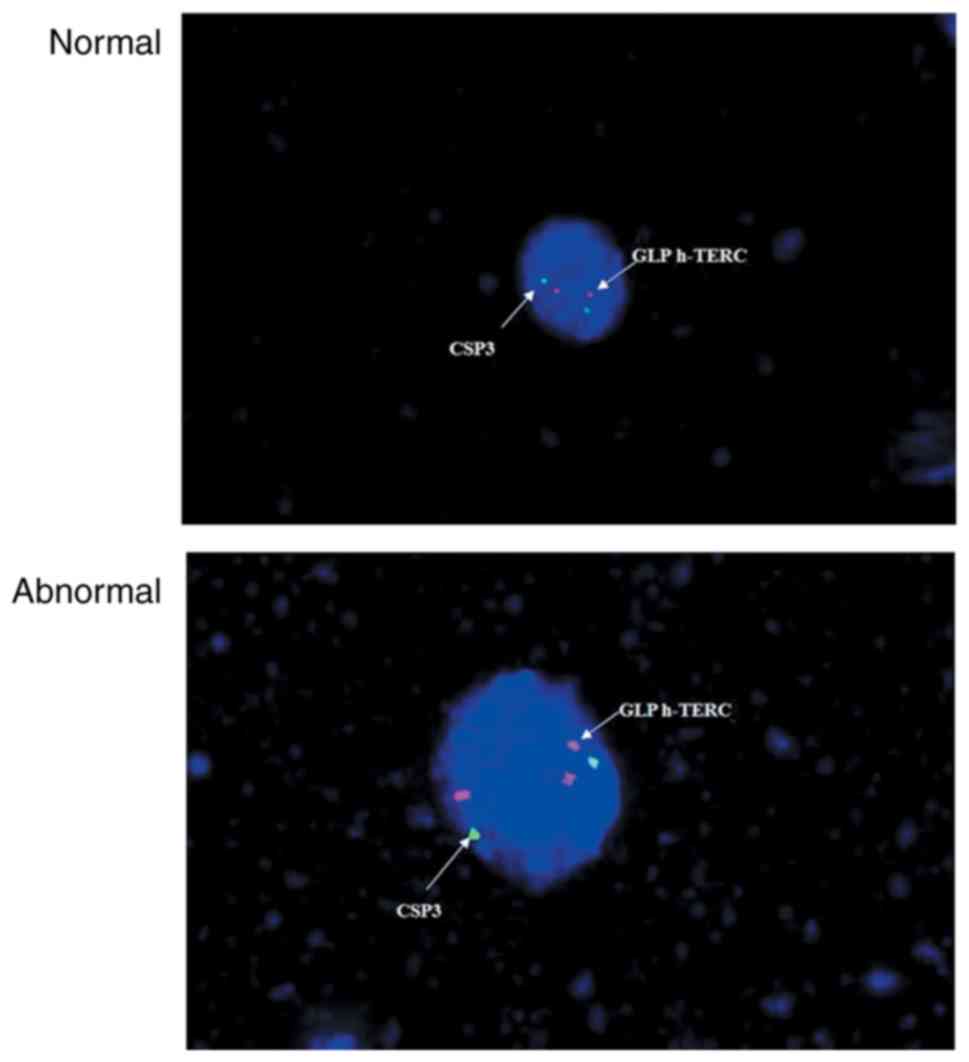
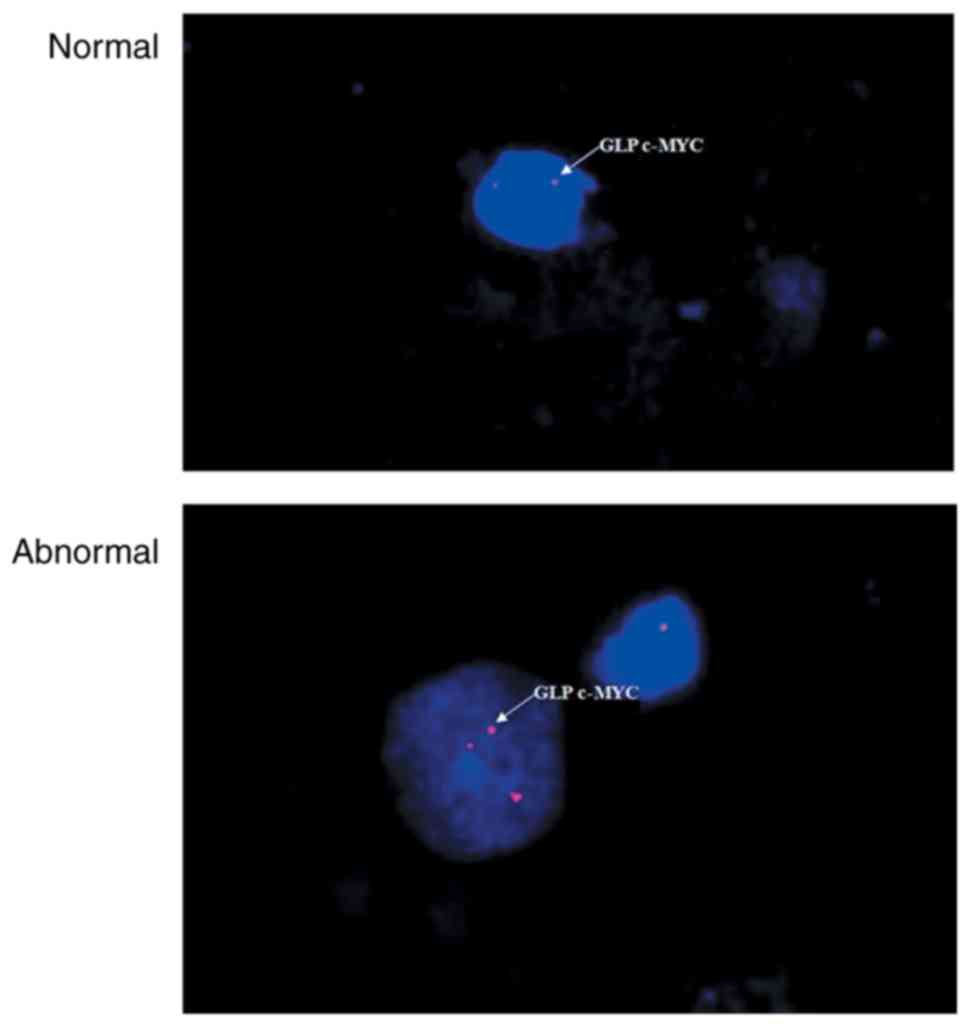
filter (×100 objective). A nucleus with >2h-TERC/CSP3 signals
(Fig. 1) or >2c-MYC signals
(Fig. 2) was scored positive, while
all remaining cells were considered normal.
HPV DNA detection and genotyping
SPR-based tests conducted at the Beijing Jinpujia
Medical Technology Co., Ltd. (Beijing, China) were used for HPV DNA
detection and genotyping (23,24).
Briefly, genomic DNA in ThinPrep cell suspension samples was
isolated using Fast Extract Solution and amplified on an Eppendorf
Mastercycler polymerase chain reaction (PCR) machine (model no.
5333; Eppendorf, Hamburg, Germany) with Taq polymerase (cat. no.
R001A; Takara, Inc., Otsu, Japan) using the following procedure: 2
min at 50°C, 4 min at 94°C; 28 cycles of 30 sec at 94°C, 45 sec at
48°C, and 20 sec at 72°C; followed by 25 cycles of 30 sec at 94°C,
45 sec at 65°C and 20 sec at 72°C. The HPV genotyping of the
amplified DNA product was performed using an SPR biosensor chip
reading instrument (W2600; Beijing Jinpujia Medical Technology Co.,
Ltd., Beijing, China) which is able to detect 16 high-risk (16, 18,
31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68 and 81) and
eight low-risk (6, 11, 40, 42, 43, 44, 54 and 70) HPV subtypes
simultaneously. The primers were provided by the Beijing Jinpujia
Medical Technology Co., Ltd. and not shown.
Histopathological evaluation
A cell block was prepared from the ThinPrep samples
by centrifugation at 1,500 × g for 10 min at 4°C and fixed in 10%
formaldehyde overnight at 4°C. Subsequently, the specimen were
transferred to graded ethanol for dehydration (70, 80, 90, 95 and
100%; each for 3 min), embedded in 5-µm-thick paraffin and stained
with hematoxylin (5 min) and eosin (5 min) at room temperature for
the pathological examination. The diagnosis was divided into four
categories: i) Normal/inflammation; ii) CIN I (mild atypical
hyperplasia and cellular atypism); iii) CIN II (moderate atypical
hyperplasia and distinct cellular atypism); and iv) CIN III (severe
atypical hyperplasia and significant cellular atypism) and ICC.
Statistical analysis
All the statistical analyses were performed with
SPSS software (version 18.0 for Windows; SPSS Inc., Chicago, IL,
USA). The enumerative results were expressed as n (%) and compared
using a χ2 test (or Fisher's exact test). Using the
histopathological diagnosis as the gold standard, the sensitivity
[a/(a+c) ×100%], specificity [d/(b+d) ×100%], positive predictive
value [a/(a+b) ×100%], negative predictive value [d/(c+d) ×100%],
misdiagnosis rate [b/(b+d) ×100%], omission diagnosis rate [c/(a+c)
×100%] and Youden's index (Y=sensitivity + specificity-1.0) of the
four screening programs for identification of CIN and ICC were
calculated, where ‘a’ referred to true positive, ‘b’ was the false
positive, ‘c’ was the false negative, and ‘d’ was true negative.
Combined diagnosis was evaluated in parallel (if any of the methods
were positive) or series (if all the methods were positive).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Correlation between histopathological
findings and TCT, h-TERC, c-MYC and HPV testing
Of the 1,000 cytological cases, 106 were reported to
be positive in the TCT test, including ASCUS in 57 patients, ASCH
in 13 patients, LSIL in 30 patients and HSIL in six patients. FISH
analysis revealed that 64 patients (6.40%) were h-TERC positive and
56 (5.60%) were c-MYC positive. SPR-HPV genotyping demonstrated
that 112 patients (11.20%) were HPV positive, of which 75 were
infected with high-risk types, 28 were infected with low-risk types
and nine were combined high- and low-risk types. In addition,
infection with a single HPV subtype was observed in 66 patients
(58.93%) and infection with multiple HPV subtypes was reported in
46 patients (41.07%).
Following screening, 213 patients were scheduled for
histopathological examination due to a positive result in any of
the aforementioned tests. The results reported inflammation in 159
patients, CIN I in 31 patients, CIN II in 14 patients, CIN III in 7
patients and ICC in two patients.
According to the histopathological findings, the
rate of h-TERC, c-MYC expression and high-risk HPV infection [with
the dominant genotypes of HPV16 (75.0%, 21/28), HPV18 (72.7%,
8/11), HPV58 (57.1%, 8/14), HPV52 (25.0%, 2/8) and HPV33 (20.0%,
1/5) (data not shown)], increased gradually with the increase in
the severity of lesions (P<0.05; Table
I). However, no significant differences in ASCH and HSIL of
cytological examination were observed among different
histopathological diagnoses. The LSIL ratio in the cytological
examination was significantly elevated, but ASCUS ratio was
significantly declined with the increase in the severity of lesions
(P<0.05; Table I).
 | Table I.Cytological, positive h-TERC, c-MYC
expression and HPV infection findings according to
histopathological diagnosis. |
Table I.
Cytological, positive h-TERC, c-MYC
expression and HPV infection findings according to
histopathological diagnosis.
|
|
| TCT, n (%) |
|
| HPV positive, n
(%) |
|---|
|
|
|
|
|
|
|
|---|
| Histopathological
classification | No. patients | ASCUS | ASCH | LSIL | HSIL | h-TERC positive, n
(%) | c-MYC positive, n
(%) | High risk | Low risk | Combined |
|---|
| Normal | 159 | 46 (28.93) | 1 (0.63) | 12 (7.55) | 0 (0.00) | 29 (18.24) | 32 (20.13) | 41 (25.79) | 25 (15.72) | 6 (3.77) |
| CIN I | 31 | 10 (32.26) | 7 (22.58) | 8 (25.81) | 0 (0.00) | 18 (58.06) | 11 (35.48) | 16 (51.61) | 2 (6.45) | 3 (9.68) |
| CIN II | 14 | 1 (7.14) | 5 (35.71) | 6 (42.86) | 1 (7.14) | 9 (64.29) | 7 (50.00) | 10 (71.43) | 1 (7.14) | 0 (0.00) |
| CIN III | 7 | 0 (0.00) | 0 (0.00) | 3 (42.86) | 4 (57.14) | 6 (85.71) | 4 (57.14) | 6 (85.71) | 0 (0.0) | 0 (0.00) |
| ICC | 2 | 0 (0.00) | 0 (0.00) | 1 (50.00) | 1 (50.00) | 2 (100.00) | 2 (100.00) | 2 (100.00) | 0 (0.00) | 0 (0.00) |
| P-value | − | <0.001 | 0.076 | 0.003 | 0.342 | 0.01 | <0.001 | 0.02 | 0.52 | 0.552 |
Diagnostic value of each method for
cervical lesions
Using the pathological result of ‘normal’ as the
criterion, the sensitivity, specificity, positive predictive value,
negative predictive value, misdiagnosis rate, omission diagnosis
rate and Youden's index of TCT, HPV, h-TERC and c-MYC for the
detection of cervical lesions, including CIN and ICC, were
determined. The results demonstrated that TCT exhibited the highest
sensitivity (87.04%) and lowest omission diagnosis rate (12.96%),
while h-TERC gene analysis had the highest specificity (81.76%) and
the lowest misdiagnosis rate (18.24%), suggesting these two methods
may be important when screening cervical lesions (Table II). These results were confirmed with
the Youden's index, with a score of 0.5 and 0.47 for TCT and
h-TERC, respectively. Although no significant difference in the
sensitivity was observed between TCT and HPV, as well as between
h-TERC and c-MYC (P>0.05), Youden's indices of HPV and c-MYC
were approximately 0.2, suggesting their poor utility for case
detection (Table II).
 | Table II.Diagnostic value of TCT, h-TERC,
c-MYC and HPV for cervical lesions. |
Table II.
Diagnostic value of TCT, h-TERC,
c-MYC and HPV for cervical lesions.
| Test | Results | CIN and ICC | Normal | Sensitivity
(%) | Specificity
(%) | Positive predictive
value (%) | Negative predictive
value (%) | Misdiagnosis rate
(%) | Omission diagnosis
rate (%) | Youden's index |
|---|
| TCT | + | 47 | 59 | 87.04 | 62.89 | 44.34 | 93.46 | 37.11 | 12.96 | 0.50 |
|
| − | 7 | 100 | − | − | − | − | − | − | − |
| h-TERC | + | 35 | 29 | 64.81a | 81.76a | 54.69 | 87.25 | 18.24a | 35.19a | 0.47 |
|
| − | 19 | 130 | − | − | − | − | − | − | − |
| c-MYC | + | 24 | 32 | 44.44a | 79.87a | 42.86 | 80.89a | 20.13a | 55.56a | 0.24 |
|
| − | 30 | 127 | − | − | − | − | − | − | − |
| HPV | + | 40 | 72 | 74.07c | 54.72b,c | 35.71b | 86.14 | 45.28b,c | 25.93a,c | 0.29 |
|
| − | 14 | 87 | − | − | − | − | − | − | − |
Furthermore, the combined diagnostic values of the
above tests were analyzed. The results indicated that the
sensitivity increased from 87.04 to >90% when the TCT was
combined with the h-TERC, c-MYC and/or HPV test (any positive;
Table III), however the specificity
decreased from 62.89 to <50%, with the highest specificity
observed in combined h-TERC and c-MYC testing (62.89%).
Nevertheless, the Youden's index of TCT and h-TERC remained
relatively high (0.49), further implying their importance.
 | Table III.Parallel and serial tests of combined
TCT, h-TERC, c-MYC and HPV testing for diagnosis of cervical
lesions. |
Table III.
Parallel and serial tests of combined
TCT, h-TERC, c-MYC and HPV testing for diagnosis of cervical
lesions.
| A, Parallel |
|---|
|
|---|
| Method | Sensitivity
(%) | Specificity
(%) | Positive predictive
value (%) | Negative predictive
value (%) | Misdiagnosis rate
(%) | Omission diagnosis
rate (%) | Youden's index |
|---|
| TCT + h-TERC | 100.00 | 49.06 | 40.00 | 100.00 | 50.94 | 0.00 | 0.49 |
| TCT + c-MYC | 94.44 | 46.54 | 37.50 | 96.10 | 53.46 | 5.56 | 0.41 |
| TCT + HPV | 98.15 | 27.67 | 31.55 | 97.78 | 72.33 | 1.85 | 0.26 |
| h-TERC + c-MYC | 75.93 | 62.89 | 41.00 | 88.50 | 37.11 | 24.07 | 0.39 |
| h-TERC + HPV | 88.89 | 38.99 | 33.10 | 91.18 | 61.01 | 11.11 | 0.28 |
| c-MYC + HPV | 85.19 | 37.11 | 31.51 | 88.06 | 62.89 | 14.81 | 0.22 |
| TCT + h-TERC +
c-MYC | 100.00 | 33.33 | 33.75 | 100.00 | 66.67 | 0.00 | 0.33 |
| TCT + h-TERC +
HPV | 100.00 | 14.47 | 28.42 | 100.00 | 85.53 | 0.00 | 0.14 |
| TCT + c-MYC +
HPV | 98.15 | 14.47 | 28.04 | 95.83 | 85.53 | 1.85 | 0.13 |
| h-TERC + c-MYC +
HPV | 90.74 | 21.38 | 28.16 | 87.18 | 78.62 | 9.26 | 0.12 |
|
| B,
Serial |
|
| Method | Sensitivity
(%) | Specificity
(%) | Positive
predictive value (%) | Negative
predictive value (%) | Misdiagnosis
rate (%) | Omission
diagnosis rate (%) | Youden's
index |
|
| TCT + h-TERC | 51.85 | 95.60 | 80.00 | 85.39 | 4.40 | 48.15 | 0.47 |
| TCT + c-MYC | 37.04 | 96.23 | 76.92 | 81.82 | 3.77 | 62.96 | 0.33 |
| TCT + HPV | 62.96 | 89.94 | 68.00 | 87.73 | 10.06 | 37.04 | 0.53 |
| h-TERC + c-MYC | 33.33 | 98.74 | 90.00 | 81.35 | 1.26 | 66.67 | 0.32 |
| h-TERC + HPV | 50.00 | 97.48 | 87.10 | 85.16 | 2.52 | 50.00 | 0.47 |
| c-MYC + HPV | 33.33 | 97.48 | 81.82 | 81.15 | 2.52 | 66.67 | 0.31 |
| TCT + h-TERC +
c-MYC | 25.93 | 99.37 | 93.33 | 79.80 | 0.63 | 74.07 | 0.25 |
| TCT + h-TERC +
HPV | 38.89 | 98.11 | 87.50 | 82.54 | 1.89 | 61.11 | 0.37 |
| TCT + c-MYC +
HPV | 27.78 | 99.37 | 93.75 | 80.20 | 0.63 | 72.22 | 0.27 |
| h-TERC + c-MYC +
HPV | 24.07 | 98.74 | 86.67 | 79.29 | 1.26 | 75.93 | 0.23 |
In addition, serial tests (both or all positive)
were also performed. As a result, the specificity of any
combination was notably higher compared with the parallel test, yet
the sensitivity was decreased, with the highest sensitivity being
62.96%. According to the Youden's index, TCT + HPV (0.53) may be
the most effective for diagnosis of cervical lesions, followed by
TCT + h-TERC (0.47) and h-TERC + HPV (0.47) (Table III). Comprehensively, dual positive
TCT and HPV were suggested to be an efficient approach for the
basic screening of cervical lesions. h-TERC amplification may serve
as an auxiliary test for TCT and HPV testing, to improve the
specificity. This conclusion may be credible due to the highest
Youden's index in TCT + HPV + h-TERC (0.37) among the three
combination groups (TCT + h-TERC + c-MYC, 0.25; TCT + c-MYC + HPV,
0.27; h-TERC + c-MYC + HPV, 0.23).
Discussion
In the present study, for the first time to the best
of our knowledge, the optimal programs for screening CIN and
cervical carcinoma among women in Shanghai were demonstrated
(21), through simultaneous detection
of TCT, h-TERC, MYC and HPV. The results demonstrated that although
TCT (87.04%) and HPV testing (74.07%) alone had higher sensitivity
for screening CIN and cervical carcinoma, their specificities
(62.89 and 54.72%, respectively) were significantly lower compared
with the alternative tests analyzed, suggesting that there may be
limitations when using these tests alone to screen precancerous
cervical lesions. This trend seemed to be in accordance with
previous studies (19,25); however, the quantitative values were
slightly different, which may be attributed to the differences in
the sample size (25), regional
location of the population (21) and
detection methods (18,19). In particular, it has been reported
that there are discordant results between SPR and hybrid capture II
(HC2) tests or direct DNA sequencing assays (23,24).
Nevertheless, the SPR test has the incomparable advantages of a
low-cost, rapid detection and an easy-to-use method with the
potential for automation (23,24). In
addition, the SPR method is able to test 24 HPV subtypes, which is
higher compared with the 13 subtypes which are currently able to be
tested using the HC2 test (21), or
the 21 subtypes which are able to be detected via the CAPE HPV
genotyping assay kit (19). The
results from the present study suggested that the SPR test may be a
superior approach for HPV genotyping in a clinical setting. When
the TCT and HPV tests were combined and one or both of the markers
amplified to be positive, the sensitivity and the specificity were
increased to 98.15 and 89.94%, respectively, displaying the highest
Youden's index values (0.53) compared with all alternative
combinations. These results implied that the combination of the TCT
and HPV tests may improve the sensitivity of cervical disease
screening and reduce the rate of misdiagnosis, leading to the
timely detection of this disease in high-risk populations. These
results further demonstrated the necessity of detection of TCT and
HPV in the clinic as previously reported (25,26).
Although combined TCT and HPV tests may improve the
sensitivity and the specificity to 98.15 and 89.94% respectively, a
number of patients may still be misdiagnosed and missed. Therefore,
alternative biomarkers are required to serve as supplementary
detection methods. The present study reported that h-TERC gene
analysis had the highest specificity (81.76%) and the lowest
misdiagnosis rate (18.24%). Additionally, the combination of TCT,
HPV and h-TERC displayed 100% sensitivity and 98.11% specificity,
suggesting h-TERC may be the principal gene in cervical
carcinogenesis, which is in line with results from previous studies
(19,27). It has been reported that the h-TERC
gene is significantly upregulated in numerous cancer types
(28,29). Inhibition of h-TERC blocks tumor
growth and promotes cell apoptosis, including cervical cancer
(30). This phenomenon was also
observed in the present study, with the h-TERC-positive rate
significantly increased from the histological diagnoses of normal
(18.24%), CIN I (58.06%), CIN II (64.29%), CIN III (85.71%) to ICC
(100%). These results indicate that h-TERC amplification may be a
useful genetic testing marker that might assist in the clinical
histopathological analysis for the differential diagnosis of normal
or CIN cases, and may even distinguish between low-grade (<CIN
I) and high-grade (>CIN II) cervical lesions.
Furthermore, extensive studies have demonstrated
that the transcription factor c-MYC may serve an important role in
carcinogenesis, due to its effect on basic cell processes including
cell proliferation, the cell cycle and apoptosis (31–33). Thus,
c-MYC amplification may also be a potential diagnostic indicator
for cervical cancer (8,11,19). In
the present study, it was observed that the c-MYC-positive rates
increased with increasing severity of histological diagnosis.
However, the fact that Youden's index of c-MYC itself was lower and
the addition of c-MYC did not increase the Youden's index for the
alternative methods, suggests that c-MYC may be not the most ideal
biomarker in the screening of cervical lesions.
There are a number of limitations to the present
study. First, the study was a single center study and the sample
size was relatively small, which may have led to the
underestimation or overestimation of the diagnostic values of the
biomarkers. Second, the aim of the study was to demonstrate the
diagnostic performance of the biomarkers for differentiating CIN
and ICC from healthy controls; therefore, further investigations
are required in order to validate the utility of the biomarkers for
screening different cervical lesion severities (11). Third, beyond h-TERC and c-MYC genes,
numerous studies have reported that the epigenetically-regulated
genes, including paired box 1 (34)
and zinc finger protein 582 (35),
may serve as genetic biomarkers for detecting high-grade CIN
lesions and cervical cancer. Therefore, it may be beneficial for
further studies to also consider these epigenetic factors.
In conclusion, the results of this preliminary study
revealed that a combination of TCT and HPV testing may be the most
efficient approach for the basic screening of cervical lesions in
Shanghai. h-TERC amplification may serve as an auxiliary test to
improve the specificity. However, further investigation with larger
samples collected from a multicenter study is required in order to
confirm these results.
Acknowledgements
Not applicable.
Funding
This study was funded by The Shanghai Municipal
Health and Family Planning Commission Fund (grant no.
15GWZK0701).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Author's contributions
WJ and WD participated in the design of this study.
WL and ZH were involved in the sample collection. WJ and LQ served
important roles in statistical analyses. WJ, WL and WD contributed
to the acquisition and interpretation of data. WJ and WD drafted
and revised the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The Ethics Committee of Ren Ji Hospital of Shanghai
Jiao Tong University approved this study. Written informed consent
was obtained from all patients when the biopsy was performed.
Patient consent for publication
All patients agreed with their data used for
publication purposes.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Di J, Rutherford S and Chu C: Review of
the cervical cancer burden and population-based cervical cancer
screening in China. Asian Pac J Cancer Prev. 16:7401–7407. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang XZ, Liu KR, Yuan FM, Hou JZ, Zhang JJ
and Zhang YZ: An analysis of both high incidence of esophageal and
cervical cancer in Yangcheng County, Shanxi Province. China Cancer.
4:259–261. 2011.
|
|
4
|
Wang Z, Wang J, Fan J, Zhao W, Yang X, Wu
L, Li D, Ding L, Wang W, Xu J, et al: Risk factors for cervical
intraepithelial neoplasia and cervical cancer in Chinese women:
Large study in Jiexiu, Shanxi Province, China. J Cancer. 8:924–932.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu Y, Zhang L, Zhao G, Che L, Zhang H and
Fang J: The clinical research of Thinprep Cytology Test (TCT)
combined with HPV-DNA detection in screening cervical cancer. Cell
Mol Biol (Noisy-le-grand). 63:92–95. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Wang Y, Liu L, Guo C, Liu Z and
Nie S: Prevalence of human papillomavirus infection and genotyping
for population-based cervical screening in developed regions in
China. Oncotarget. 7:62411–62424. 2016.PubMed/NCBI
|
|
7
|
Wang JL, Yang YZ, Dong WW, Sun J, Tao HT,
Li RX and Hu Y: Application of human papillomavirus in screening
for cervical cancer and precancerous lesions. Asian Pac J Cancer
Prev. 14:2979–2982. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gao K, Eurasian M, Zhang J, Wei Y, Zheng
Q, Ye H and Li L: Can genomic amplification of human telomerase
gene and C-MYC in liquid-based cytological specimens be used as a
method for opportunistic cervical cancer screening? Gynecol Obstet
Invest. 80:153–163. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Szarewski A: HPV vaccination and cervical
cancer. Curr Oncol Rep. 14:559–567. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chandra R: Relevance of persistent
infection with high-risk HPV genotypes in cervical cancer
progression. MLO Med Lab Obs. 45:40, 42, 44. 2013.PubMed/NCBI
|
|
11
|
Li T, Tang L, Bian D, Jia Y, Huang X and
Zhang X: Detection of hTERC and c-MYC genes in cervical epithelial
exfoliated cells for cervical cancer screening. Int J Mol Med.
33:1289–1297. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kuglik P, Kasikova K, Smetana J, Vallova
V, Lastuvkova A, Moukova L, Cvanova M and Brozova L: Molecular
cytogenetic analyses of hTERC (3q26) and MYC (8q24) genes
amplifications in correlation with oncogenic human papillomavirus
infection in Czech patients with cervical intraepithelial neoplasia
and cervical carcinomas. Neoplasma. 62:130–139. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cao Y, Bryan TM and Reddel RR: Increased
copy number of the TERT and TERC telomerase subunit genes in cancer
cells. Cancer Sci. 99:1092–1099. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bin H, Ruifang W, Ruizhen L, Yiheng L,
Zhihong L, Juan L, Chun W, Yanqiu Z and Leiming W: Detention of HPV
L1 capsid protein and hTERC gene in screening of cervical cancer.
Iran J Basic Med Sci. 16:797–802. 2013.PubMed/NCBI
|
|
15
|
Ferber MJ, Thorland EC, Brink AA, Rapp AK,
Phillips LA, Mcgovern R, Gostout BS, Cheung TH, Chung TK, Fu WY and
Smith DI: Preferential integration of human papillomavirus type 18
near the c-myc locus in cervical carcinoma. Oncogene. 22:7233–7242.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Abba MC, Laguens RM, Dulout FN and Golijow
CD: The c-myc activation in cervical carcinomas and HPV 16
infections. Mutat Res. 557:151–158. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kübler K, Heinenberg S, Rudlowski C,
Keyver-Paik MD, Abramian A, Merkelbach-Bruse S, Büttner R, Kuhn W
and Schildhaus HU: c-myc copy number gain is a powerful
prognosticator of disease outcome in cervical dysplasia.
Oncotarget. 6:825–835. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zheng X, Liang P, Zheng Y, Yi P, Liu Q,
Han J, Huang Y, Zhou Y, Guo J and Li L: Clinical significance of
hTERC gene detection in exfoliated cervical epithelial cells for
cervical lesions. Int J Gynecol Cancer. 23:785–790. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao WH, Hao M, Cheng XT, Yang X, Wang ZL,
Cheng KY, Liu FL and Bai YX: c-myc gene copy number variation in
cervical exfoliated cells detected on fluorescence in situ
hybridization for cervical cancer screening. Gynecol Obstet Invest.
81:416–423. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang J, Wei LH, Li YL, Wu RF, Xie X, Feng
YJ, Zhang G, Zhao C, Zhao Y and Chen Z: Detection of TERC
amplification in cervical epithelial cells for the diagnosis of
high-grade cervical lesions and invasive cancer: A multicenter
study in China. J Mol Diagn. 12:808–817. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gu Y, Ma C, Zou J, Zhu Y, Yang R, Xu Y and
Zhang Y: Prevalence characteristics of high-risk human
papillomaviruses in women living in Shanghai with cervical
precancerous lesions and cancer. Oncotarget. 7:24656–24663. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Solomon D, Davey D, Kurman R, Moriarty A,
O'Connor D, Prey M, Raab S, Sherman M, Wilbur D, Wright T Jr, et
al: The 2001 Bethesda System: Terminology for reporting results of
cervical cytology. JAMA. 287:2114–2119. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He X, Yang P, Wang H, Wang Y and Liu S:
Human papillomavirus genotyping by surface plasmon resonance-based
test. Clin Lab. 62:2079–2084. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang S, Yang H, Zhang H, Yang F, Zhou M,
Jia C, Lan Y, Ma Y, Zhou L, Tian S, et al: A surface plasmon
resonance--based system to genotype human papillomavirus. Cancer
Genet Cytogenet. 200:100–105. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pan QJ, Hu SY, Guo HQ, Zhang WH, Zhang X,
Chen W, Cao J, Jiang Y, Zhao FH and Qiao YL: Liquid-based cytology
and human papillomavirus testing: A pooled analysis using the data
from 13 population-based cervical cancer screening studies from
China. Gynecol Oncol. 133:172–179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Castle PE, Aslam S and Behrens C: Cervical
precancer and cancer risk by human papillomavirus status and
cytologic interpretation: Implications for risk-based management.
Cancer Epidemiol Biomarkers Prev. 25:1595–1599. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zappacosta R, Ianieri MM, Buca D, Repetti
E, Ricciardulli A and Liberati M: Clinical role of the detection of
human telomerase RNA component gene amplification by fluorescence
in situ hybridization on liquid-based cervical samples: Comparison
with human papillomavirus-DNA testing and histopathology. Acta
Cytol. 59:345–354. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Baena-Del Valle JA, Zheng Q, Esopi DM,
Rubenstein M, Hubbard GK, Moncaliano MC, Hruszkewycz A, Vaghasia A,
Yegnasubramanian S, Wheelan SJ, et al: MYC drives overexpression of
telomerase RNA (hTR/TERC) in prostate cancer. J Pathol. 244:11–24.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Penzo M, Ludovini V, Treré D, Siggillino
A, Vannucci J, Bellezza G, Crinò L and Montanaro L: Dyskerin and
TERC expression may condition survival in lung cancer patients.
Oncotarget. 6:21755–21760. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Y, Li H, Yao G, Li W, Wang F, Jiang Z
and Li M: Inhibition of telomerase RNA (hTR) in cervical cancer by
adenovirus-delivered siRNA. Cancer Gene Ther. 14:748–755. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yuan Y, Zhang J, Cai L, Ding C, Wang X,
Chen H, Wang X, Yan J and Lu J: Leptin induces cell proliferation
and reduces cell apoptosis by activating c-myc in cervical cancer.
Oncol Rep. 29:2291–2296. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liao LM, Sun XY, Liu AW, Wu JB, Cheng XL,
Lin JX, Zheng M and Huang L: Low expression of long noncoding
XLOC_010588 indicates a poor prognosis and promotes proliferation
through upregulation of c-Myc in cervical cancer. Gynecol Oncol.
133:616–623. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cui F, Hou J, Huang C, Sun X, Zeng Y,
Cheng H, Wang H and Li C: C-Myc regulates radiation-induced G2/M
cell cycle arrest and cell death in human cervical cancer cells. J
Obstet Gynaecol Res. 43:729–735. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Luan T, Hua Q, Liu X, Xu P, Gu Y, Qian H,
Yan L, Xu X, Geng R and Zeng X and Zeng X: PAX1 methylation as a
potential biomarker to predict the progression of cervical
intraepithelial neoplasia: A meta-analysis of related studies. Int
J Gynecol Cancer. 27:1480–1488. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liou YL, Zhang Y, Liu Y, Cao L, Qin CZ,
Zhang TL, Chang CF, Wang HJ, Lin SY, Chu TY, et al: Comparison of
HPV genotyping and methylated ZNF582 as triage for women with
equivocal liquid-based cytology results. Clin Epigenetics.
7:502015. View Article : Google Scholar : PubMed/NCBI
|