Introduction
Glioma is the most common malignant brain tumor in
the human central nervous system (1,2). The World
Health Organization (3) has defined 4
grades of glioma based on histological features: Grade I, pilocytic
astrocytoma; grade II, diffuse astrocytoma; grade III, anaplastic
astrocytoma; and grade IV, glioblastoma multiforme (GBM). Despite
attempts to combine surgery, radiotherapy and chemotherapy, grade
III and IV gliomas recur in >90% of cases, typically within 2 cm
of the original location, and 10–20% may develop novel distant
lesions (4). Therefore,
neuro-oncological research has become focused on overcoming glioma
cell resilience against the majority of aggressive treatments, in
addition to providing an explanation for the high recurrence rate,
particularly in GBM.
Recently, a number of studies have described the
presence of a tumor cell subpopulation with stem cell-like
properties, known as cancer stem cells (CSCs) (5–9). This
population is characterized by self-renewal, a high migration rate,
unlimited growth, chemotherapy and radiotherapy resistance, and
tumor formation (8). It has been
suggested that the clinical behavior of a tumor is largely
influenced by CSCs, due to their ability to initiate novel tumors.
Therefore, eradicating CSCs may affect stable, long-lasting
remission and potentially treat cancer (10–12).
A frequently used biomarker for glioma CSC is
prominin-1 [also known as cluster of differentiation 133 (CD133)],
a cell surface pentaspan transmembrane glycoprotein, originally
identified from murine neuroepithelial cells and located in plasma
membrane protrusions (13). A number
of studies have indicated that CD133+ cells exhibit a
greater tumor-forming ability in immunocompromised rodent brains
in vivo compared with CD133− cells. These
CD133+ cells form tumors with as little as 100 cells,
which is consistent with their high self-renewal ability (14,15). As a
subpopulation that is highly resistant to ionizing radiation in
human glioma xenografts, CD133+ cells can more
effectively repair DNA damage compared with CD133− tumor
cells (16). In addition to relative
radioresistance, CD133+ cells have high expression
levels of multiple drug resistance genes compared with the
differentiated bulk tumor, and significant resistance to the
chemotherapy drugs temozolomide, etoposide, carboplatin and
paclitaxel (17–19).
The protein expression of the transcription factor
sex-determining region Y-box 2 (SOX2) putatively contributes to
cellular invasion in tumors of neural and neural crest origin,
including glioma (20). Previous
studies have suggested that the overexpression or gene
amplification of SOX2 is associated with the development of cancer.
The Sox2 gene in mice has a single exon, but no introns, and
is located on chromosome 3q26.3-q27. Sox2 encodes a
317-amino acid protein, which contains a high-mobility group
DNA-binding domain (21). SOX2
positively contributes to the stemness of cells, which is the
ability of cells to self-renew and differentiate into cancer cells,
and to the multiple processes of cancer cells (22,23).
Silencing Sox2 in freshly derived glioblastoma
tumor-initiating cells prevented their proliferation and inhibited
tumorigenicity in immunodeficient mice (24).
While there is considerable literature concerning
CSCs, the number and distribution of CSCs in human GBM remains
unknown. Even within the same grade of glioma, the reported number
of CSCs can vary between <1 and >80% of the total cell
population (17,25). In the present study, it was
hypothesized that the discrepancies in results among studies may be
due to differences in sampling sites. In particular, the present
study investigated whether CSCs can be found in areas of the brain
that appear normal.
Therefore, the present study investigated the
distribution of CSCs within delimited locations in the glioma and
surrounding normal tissues. To the best of our knowledge, this is
the first study to quantify systematically the number of CSCs in
locations ranging from the tumor center to areas of the brain that
appear normal.
Patients and methods
Patients and tissue samples
Two patients (57-year old male and 65-year old male)
with GBM donated their whole brain to the Department of
Neurosurgery, University of Southern California (Los Angeles, CA,
USA) for scientific research. The Ethics Committee of the
University of Southern California approved the present study and
the patients provided prior written informed consent for the use of
their brain tissue upon succumbing to the disease. The areas of the
brains were labeled according to location, separated into several
blocks each, and stored at −80°C. Areas of the brain tissue were
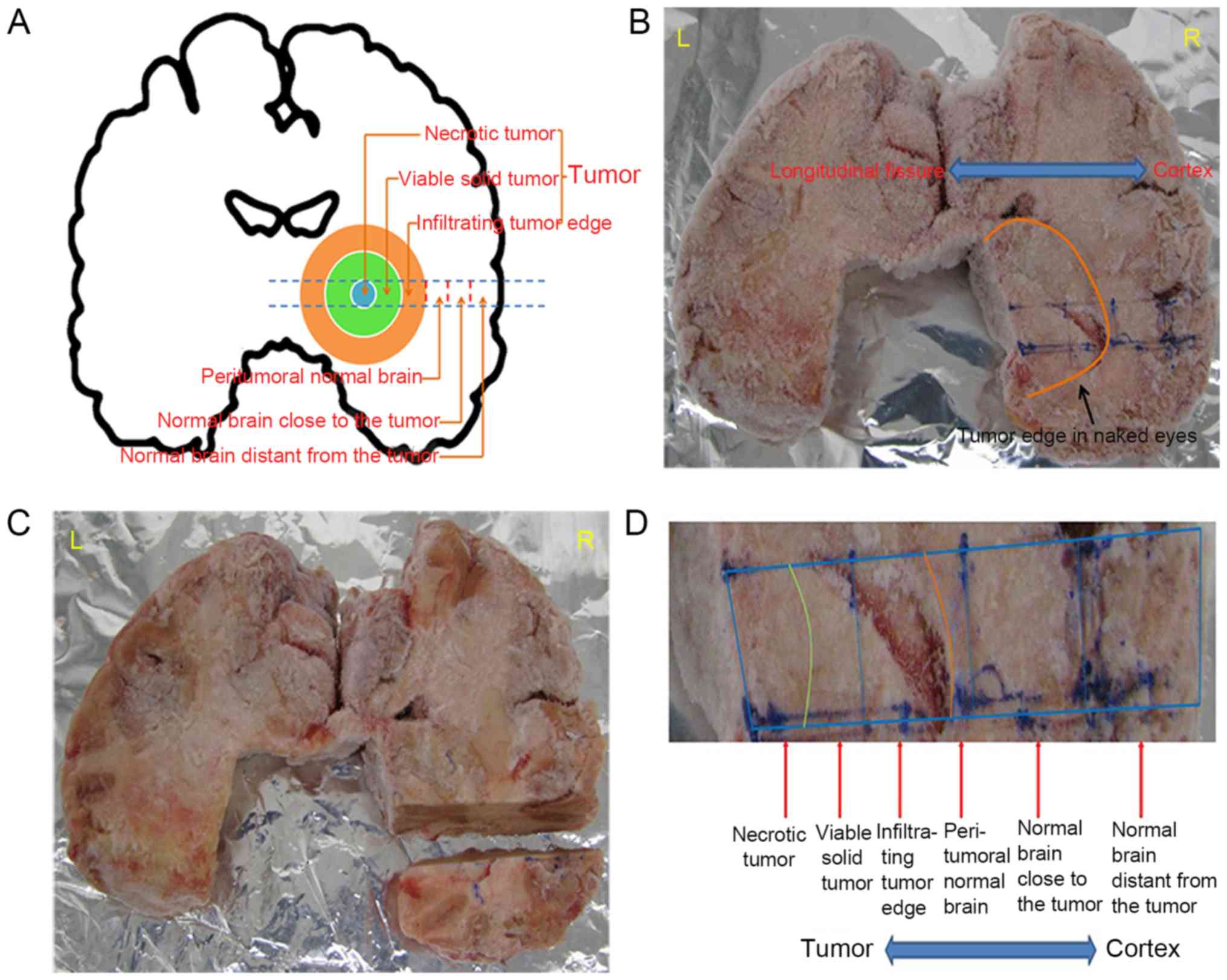
categorized based on their distance from the necrotic center of the
tumor, as follows: Necrotic tumor, viable solid tumor, infiltrating
tumor edge, peritumoral normal brain, normal brain close to the
tumor, and normal brain distant from the tumor (Fig. 1A). The whole brain was snap-frozen,
and the different regions were identified (Fig. 1B). The designated area of the brain
was then cut away (Fig. 1C), each
individual region cut into pieces (Fig.
1D), and each piece sectioned into 10-µm slices on a
cryostat.
Immunohistochemical staining
Cryostat sections (10 µm thick) were fixed in 100%
Acetone for 10 min at room temperature (RT). The slides were
immersed and rehydrated 1X PBS three times, 5 min/each, and
quenched in endogenous peroxidase in 3% hydrogen peroxide in 1× PBS
solution for 10 min at RT, and rinsed once. Subsequently, 10%
skimmed milk in 1× PBS was applied and the slides were incubated
for 30 min at RT to block non-specific binding. Slides were
serially incubated with anti-CD133 mouse monoclonal antibody (cat.
no. MAB4310; 1:100; EMD Millipore, Billerica, MA, USA) or anti-SOX2
mouse monoclonal antibody (cat. no. SAB5300177; 1:200;
Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) at 4°C overnight.
Subsequently the slides were incubated with biotinylated anti-mouse
secondary antibody (cat. no. BP-9200; 1:200), anti-ABC (cat. no.
PK-7200) and anti-AEC (cat. no. SK-4200) (all from Vector
Laboratories, Inc., Burlingame, CA, USA) at RT for 30 min,
according to the manufacturer's protocols. Slides were
counterstained with hematoxylin for 1–2 min at RT. Similarly,
isotype staining with mouse IgG1 negative control antibody (cat.
no. CBL610; 1:100; EMD Millipore) was used as the negative control.
For a control without antibody, no primary antibody was added and
the sections were incubated in 2% skimmed milk only.
Cell quantification
Photomicrographs of all slides were captured with an
EVOS bright-field microscope (magnification, ×20). To quantify the
number of positive cells per region, 5 random images of each region
were taken, and the number of stained cells per total population in
each image was counted by eye. The mean percentage of positive
cells in the 5 images was calculated and reported as the mean ±
standard error of the mean.
Statistical analysis
The percentages of CD133+ and
SOX2+ cells for all 6 regions were compared using
one-way analysis of variance and Student-Newam-Keuls method. The
statistical tests were performed using SPSS 19.0 software (IBM
Corp., Armonk, NY, USA). Differences between means were used to
determine statistical significance. P<0.05 was considered to
indicate a statistically significant difference.
Results
The different areas of the 2 brains were analyzed
for the presence of CSCs by immunostaining for CD133 and SOX2
expression, and the number of CD133+ and
SOX2+ cells was quantified. The 6 areas of the brains
analyzed included the necrotic tumor, the viable solid tumor, the
infiltrating tumor edge, the peritumoral normal brain, the normal
brain close to the tumor and the normal brain distant from the
tumor (Fig. 1).
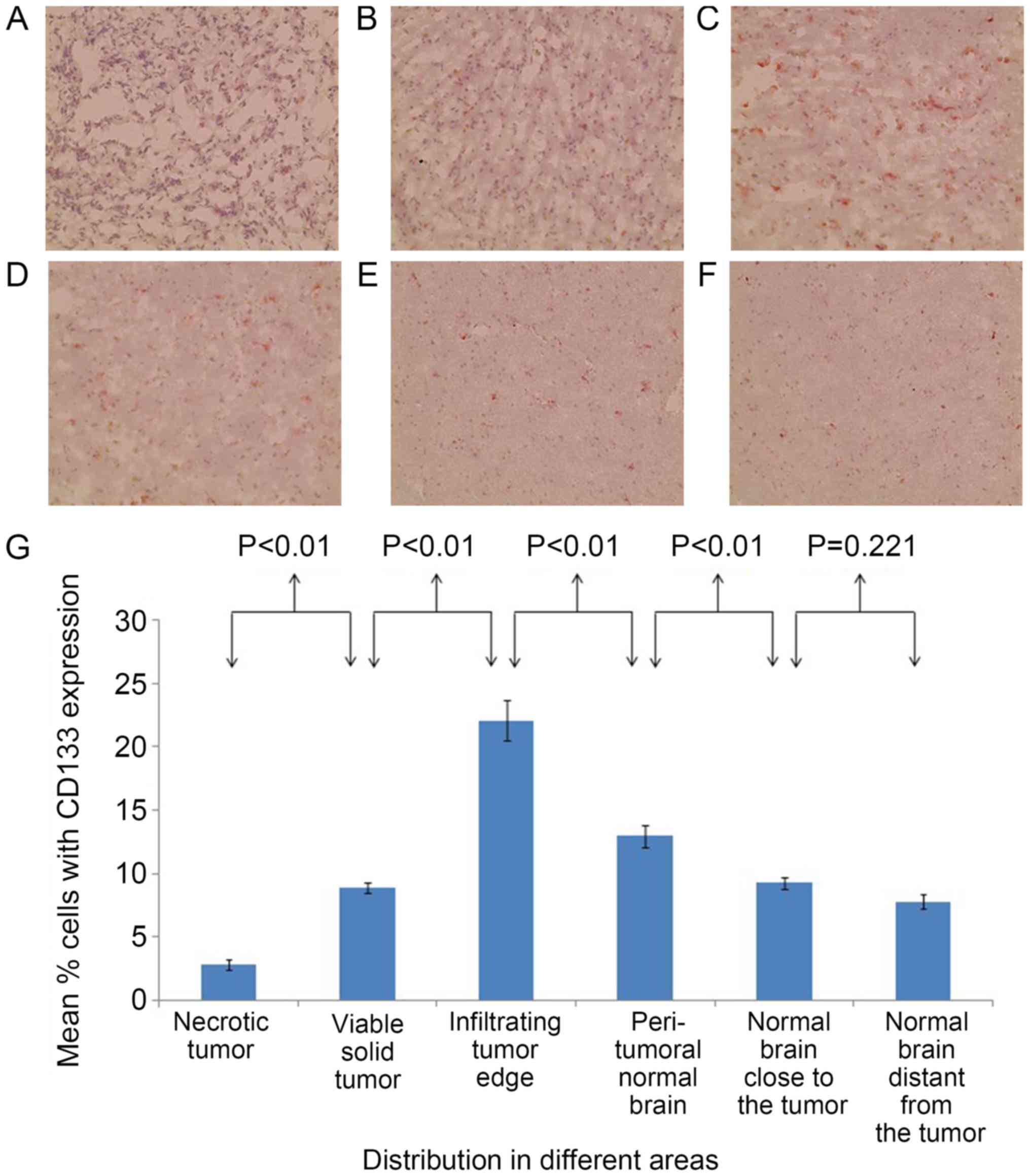
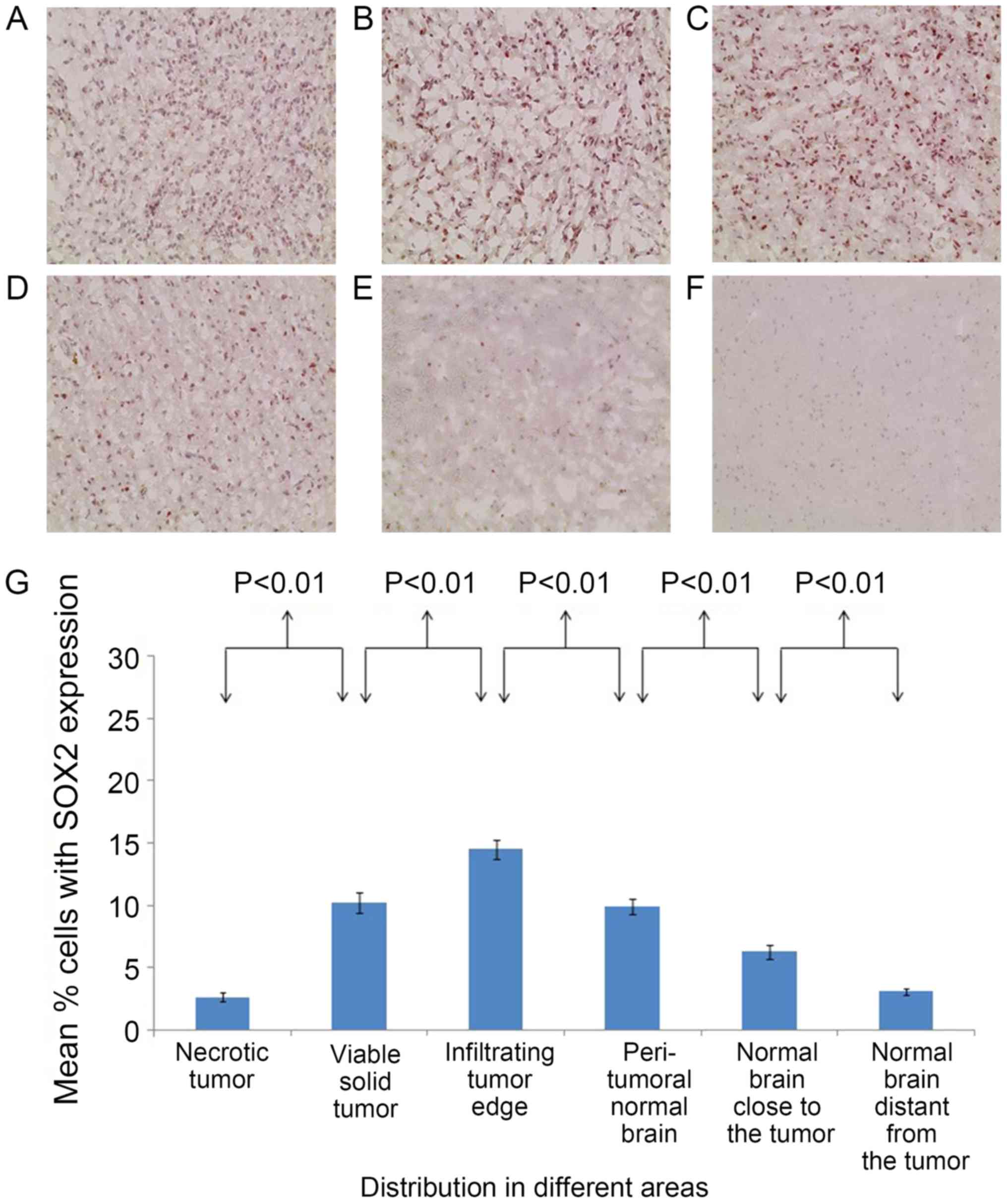
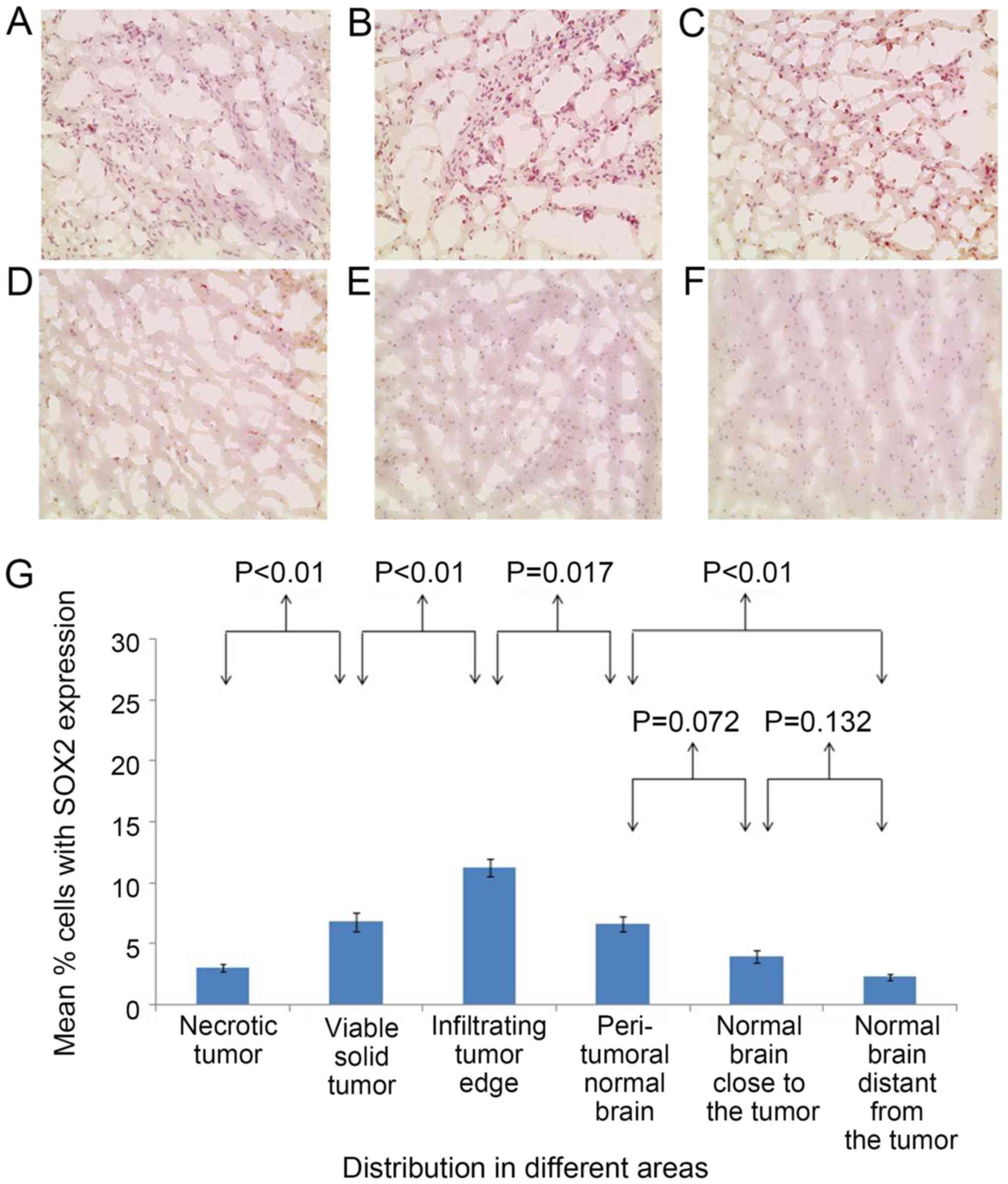
In the brain of patient 1, the percentages of
CD133+ cells (Fig. 2) and
SOX2+ cells (Fig. 3) were
significantly different between all adjacent areas (P<0.01),
with the exception of the percentages of CD133+ cells
between the normal areas close to and distant from the tumor, which
were statistically similar (P=0.221). The percentages of
CD133+ cells (Fig. 2) and
SOX2+ cells (Fig. 3) were
significantly higher at the infiltrating tumor edge compared with
those at any of the other areas (P<0.05).
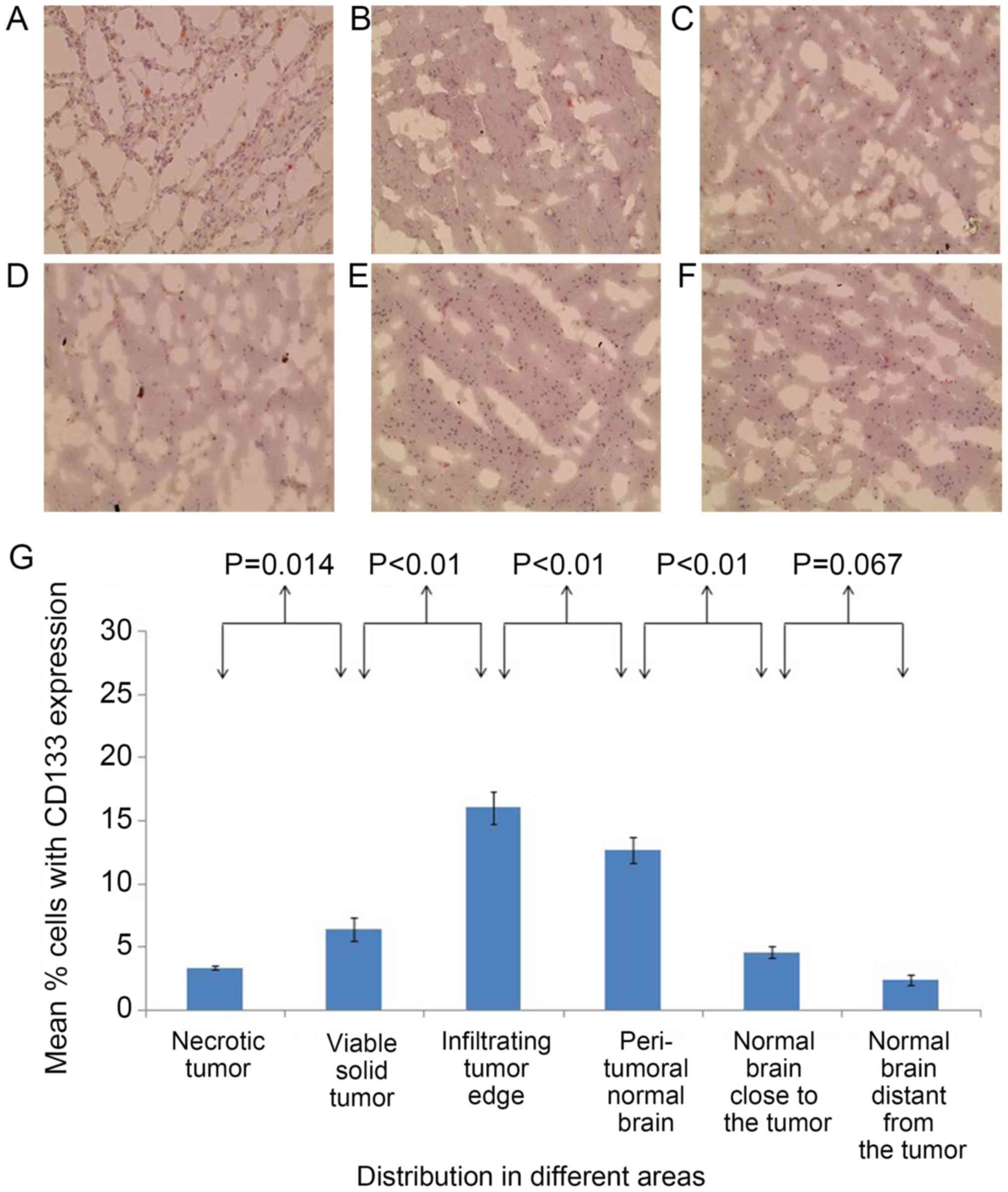
The brain of patient 2 presented with an almost
identical distribution of staining results for CSCs as that of
patient 1. In the second brain, the percentages of
CD133+ cells (Fig. 4) and
SOX2+ cells (Fig. 5) were
significantly different between the majority of adjacent areas.
However, the percentages of SOX2+ cells of the
peritumoral normal brain and the normal brain close to the tumor
were statistically similar, as well as the percentages of
CD133+ cells and SOX2+ cells in the areas
close to and distant from the tumor. The percentage of
SOX2+ cells in the peritumoral normal brain was
significantly higher compared with that of the normal brain distant
from the tumor (P<0.01). The percentages of CD133+
cells (Fig. 4) and SOX2+
cells (Fig. 5) were significantly
higher at the infiltrating tumor edge compared with those at any of
the other areas (P<0.05).
The negative controls, without primary antibody or
with isotype control, exhibited no significant staining (Fig. 6). In the brains of patient 1 and
patient 2, the percentages of CD133+ cells and
SOX2+ cells were highest at the tumor edge, relative to
all the other areas. In particular, in the infiltrating tumor edge
in patient 1, the percentages of CD133+ cells and
SOX2+ cells were 22.07±1.62 and 14.5±0.78%,
respectively. For patient 2, the percentages of CD133+
cells and SOX2+ cells in the infiltrating tumor edge
were 16.03±1.29 and 11.24±0.76%, respectively.
Discussion
The importance of CSCs in recurrent gliomas, as well
as other solid malignancies, has been studied extensively in
previous literature (8,26–30).
Therefore, the relevance of identifying the distribution of CSCs
within the different regions of the glioma is crucial. The present
study indicated, according to the results of the positive staining
for CD133 and SOX2 cells, that CSCs are most prevalent at the tumor
edge. This suggests that the edge of the tumor is the moving front
for tumor progression and invasion, and should be targeted for
therapeutic intervention. Surgery and radiotherapy for glioma and
even other solid tumors should be focused particularly on the area
around the tumor edge. The present study also revealed the presence
of CD133+ and SOX2+ cells in normal brain
areas distant from the tumor, which indicates that these CSCs may
indicate sites of future progression. Therefore, therapeutic
strategies that specifically target CSCs are particularly
important.
The findings of the present study suggest that other
studies have differed with regard to the concentrations of
CD133+ and SOX2+ cells found, even for
gliomas of the same grade, due to different tumor regions being
sampled (31,32). Annovazzi et al (32) examined SOX2 expression in surgical
samples from 133 brain gliomas of different grades of malignancy
and in cell lines from 16 glioblastomas. This revealed a positive
correlation between SOX2 expression and malignancy grade in gliomas
and identified that the expression of SOX2 was different at
different locations of the brain. In medulloblastomas, SOX2
staining was either positive or negative, according to the
occurrence of areas with neuronal differentiation. However, in that
study, samples of peritumoral nervous tissue removed from around
vascular malformations and other normal brain tissue were obtained
from two patients who died following heart attacks. Therefore, the
location of the biopsy is crucial.
Although CD133 and SOX2 are the most common markers
for CSCs, a number of other immunocytochemical markers have also
been used, including nestin and Musashi-1 (33–35). All
these markers identify subpopulations of CSCs. In addition,
different CD133 and SOX2 antibody clones may recognize CD133 and
SOX2 splice variants with epitopes that differ in glycosylation
status (36). Therefore, future
studies are required for the examination of specific markers.
Furthermore, the current study only investigated two specimens as
very few patients wish to donate their whole brains for research.
However, if possible similar research should be performed with a
higher number of specimens to further confirm the conclusion of the
present study.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and
Technology Department of Sichuan Province (grant nos. 2013SZZ002
and 2018JY0404), the Health and Family Planning Commission of
Sichuan Province (grant no. 16PJ557), the government of Luzhou
(grant nos. 14ZC0071-LH09 and 2016LZXNYD-G03), the Southwest
Medical University (grant no. 2013ZRQN068) and the Project Program
of Neurosurgical Clinical Research Center of Sichuan Province
(grant no. 17082).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding authors on reasonable
request.
Authors' contributions
Conception and design: LC and TCC. Acquisition of
data: LP. Analysis and interpretation of data: LP and JF. Drafting
the article: LP and JF. Statistical analysis: LP. Sample collection
and handling: WW and FMH. Critical revision of the article: WW and
FMH. Reviewing of the submitted version of the manuscript: All
authors. Approval of the final version of the manuscript on behalf
of all authors: LC and TCC. Study supervision: TCC.
Ethics approval and consent to
participate
The Ethics Committee of the University of Southern
California (Los Angeles, USA) approved the present study and the
patients provided prior written informed consent for the use of
their brain tissues following mortality.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hassan A, Mosley J, Singh S and Zinn PO: A
comprehensive review of genomics and noncoding RNA in gliomas. Top
Magn Reson Imaging. 26:3–14. 2017.PubMed/NCBI
|
|
2
|
Goodenberger ML and Jenkins RB: Genetics
of adult glioma. Cancer Genet. 205:613–621. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 world health organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 13l:803–820. 2016. View Article : Google Scholar
|
|
4
|
Brada M, Hoang-Xuan K, Rampling R,
Dietrich PY, Dirix LY, Macdonald D, Heimans JJ, Zonnenberg BA,
Bravo-Marques JM, Henriksson R, et al: Multicenter phase II trial
of temozolomide in patients with glioblastoma multiforme at first
relapse. Ann Oncol. 12:259–266. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jordan CT, Guzman ML and Noble M: Cancer
stem cells. N Engl J Med. 355:1253–1261. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Flores DG, Ledur PF, Abujamra AL, Brunetto
AL, Schwartsmann G, Lenz G and Roesler R: Cancer stem cells and the
biology of brain tumors. Curr Stem Cell Res Ther. 4:306–313. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Doherty MR, Smigiel JM, Junk DJ and
Jackson MW: Cancer stem cell plasticity drives therapeutic
resistance. Cancers (Basel). 8:E82016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Batlle E and Clevers H: Cancer stem cells
revisited. Nat Med. 23:1124–1134. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu P and Fan Z: Cancer stem cells and
tumorigenesis. Biophys Rep. 4:178–188. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang PY, Yang YJ, Xue Y, Fu J, Zhang CX,
Wang Y, Yang Y and Shi H: Cancer stem cells: Targeting tumors at
the source. Eur Rev Med Pharmacol Sci. 19:1821–1828.
2015.PubMed/NCBI
|
|
11
|
Pan Q, Li Q, Liu S, Ning N, Zhang X, Xu Y,
Chang AE and Wicha MS: Concise review: Targeting cancer stem cells
using immunologic approaches. Stem Cells. 33:2085–2092. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schmohl JU and Vallera DA: CD133,
selectively targeting the root of cancer. Toxins (Basel).
8:E1652016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jang JW, Song Y, Kim SH, Kim J and Seo HR:
Potential mechanisms of CD133 in cancer stem cells. Life Sci.
184:25–29. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Singh SK, Hawkins C, Clarke ID, Squire JA,
Bayani J, Hide T, Henkelman RM, Cusimano MD and Dirks PB:
Identification of human brain tumour initiating cells. Nature.
432:396–401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Beier D, Hau P, Proescholdt M, Lohmeier A,
Wischhusen J, Oefner PJ, Aigner L, Brawanski A, Bogdahn U and Beier
CP: CD133+ and CD133- glioblastoma-derived cancer stem cells show
differential growth characteristics and molecular profiles. Cancer
Res. 67:4010–4015. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bao S, Wu Q, McLendon RE, Hao Y, Shi Q,
Hjelmeland AB, Dewhirst MW, Bigner DD and Rich JN: Glioma stem
cells promote radioresistance by preferential activation of the DNA
damage response. Nature. 444:756–760. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu G, Yuan X, Zeng Z, Tunici P, Ng H,
Abdulkadir IR, Lu L, Irvin D, Black KL and Yu JS: Analysis of gene
expression and chemoresistance of CD133+ cancer stem cells in
glioblastoma. Mol Cancer. 5:672006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tamura K, Aoyagi M, Ando N, Ogishima T,
Wakimoto H, Yamamoto M and Ohno K: Expansion of CD133-positive
glioma cells in recurrent de novo glioblastomas after radiotherapy
and chemotherapy. J Neurosurg. 119:1145–1155. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Niero EL, Rocha-Sales B, Lauand C, Cortez
BA, de Souza MM, Rezende-Teixeira P, Urabayashi MS, Martens AA,
Neves JH and Machado-Santelli GM: The multiple facets of drug
resistance: One history, different approaches. J Exp Clin Cancer
Res. 33:372014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ikushima H, Todo T, Ino Y, Takahashi M,
Miyazawa K and Miyazono K: AutocrineTGF-beta signaling maintains
tumorigenicity of glioma-initiating cells through Sry-related
HMG-box factors. Cell Stem Cell. 5:504–514. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Scaffidi P and Bianchi ME: Spatially
precise DNA bending is an essential activity of the sox2
transcription factor. J Biol Chem. 276:47296–47302. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu K, Lin B, Zhao M, Yang X, Chen M, Gao
A, Liu F, Que J and Lan X: The multiple roles for Sox2 in stem cell
maintenance and tumorigenesis. Cell Signal. 25:1264–1271. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kuo HY, Hsu HT, Chen YC, Chang YW, Liu FT
and Wu CW: Galectin-3 modulates the EGFR signalling-mediated
regulation of Sox2 expression via c-Myc in lung cancer.
Glycobiology. 26:155–165. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gangemi RM, Griffero F, Marubbi D, Perera
M, Capra MC, Malatesta P, Ravetti GL, Zona GL, Daga A and Corte G:
SOX2 silencing in glioblastoma tumor-initiating cells causes stop
of proliferation and loss of tumorigenicity. Stem Cells. 27:40–48.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu W, Shen G, Shi Z, Shen F, Zheng X, Wen
L and Yang X: Brain tumour stem cells and neural stem cells: Still
explored by the same approach? J Int Med Res. 36:890–895. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ajani JA, Song S, Hochster HS and
Steinberg IB: Cancer stem cells: The promise and the potential.
Semin Oncol. 1 Suppl 42:S3–S17. 2015. View Article : Google Scholar
|
|
27
|
Islam F, Qiao B, Smith RA, Gopalan V and
Lam AK: Cancer stem cell: Fundamental experimental pathological
concepts and updates. Exp Mol Pathol. 98:184–191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sharma A and Shiras A: Cancer stem
cell-vascular endothelial cell interactions in glioblastoma.
Biochem Biophys Res Commun. 473:688–692. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pearson AT, Jackson TL and Nor JE:
Modeling head and neck cancer stem cell-mediated tumorigenesis.
Cell Mol Life Sci. 73:3279–3289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Eun K, Ham SW and Kim H: Cancer stem cell
heterogeneity: Origin and new perspectives on CSC targeting. BMB
Rep. 50:117–125. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cheng JX, Liu BL and Zhang X: How powerful
is CD133 as a cancer stem cell marker in brain tumors? Cancer Treat
Rev. 35:403–408. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Annovazzi L, Mellai M, Caldera V, Valente
G and Schiffer D: SOX2 expression and amplification in gliomas and
glioma cell lines. Cancer Genomics Proteomics. 8:139–147.
2011.PubMed/NCBI
|
|
33
|
Dahlrot RH, Hermansen SK, Hansen S and
Kristensen BW: What is the clinical value of cancer stem cell
markers in gliomas? Int J Clin Exp Pathol. 6:334–348.
2013.PubMed/NCBI
|
|
34
|
Dahlrot RH, Hansen S, Jensen SS, Schroder
HD, Hjelmborg J and Kristensen BW: Clinical value of CD133 and
nestin in patients with glioma: A population-based study. Int J
Clin Exp Pathol. 7:3739–3751. 2014.PubMed/NCBI
|
|
35
|
Dahlrot RH: The prognostic value of
clinical factors and cancer stem cell-related markers in gliomas.
Dan Med J. 61:B49442014.PubMed/NCBI
|
|
36
|
Hermansen SK, Christensen KG, Jensen SS
and Kristensen BW: Inconsistent immunohistochemical expression
patterns of four different CD133 antibody clones in glioblastoma. J
Histochem Cytochem. 59:391–407. 2011. View Article : Google Scholar : PubMed/NCBI
|