Introduction
Colorectal cancer (CRC) is the third most common
cancer worldwide. Its prognosis is poor and the total 5-year
survival rate is 65% (1). CRC is
considered to evolve from conventional adenoma, which gradually
accumulates genetic or epigenetic changes, which eventually lead to
cancer. The most common and characteristic genetic alterations
occur in the APC regulator of WNT signaling pathway, tumor protein
p53, KRAS proto-oncogene GTPase and mismatch repair genes (2).
RAN binding protein 9 (RANBP9) is a 90-kDa protein,
which was initially screened and identified by Nishimoto et
al in a yeast two-hybrid experiment (3). RANBP9 is conserved in various
organisms, including humans, rhesus monkeys, mice and frogs
(4,5). RANBP9 is widely expressed in mammalian
tissues and is distributed in the nucleus and cytoplasm (5); however, its biological functions remain
unclear. RANBP9 overexpression can reduce dendritic arbor and spine
density, and can accelerate loss of dendritic spines in an
Alzheimer's disease mouse model (6,7).
Furthermore, RANBP9 has been demonstrated to be involved in the
nucleation of the central microtubule, affecting cell division and
differentiation (8). Additionally,
RANBP9 has been suggested as a platform for the interaction of cell
signaling molecules, including cell surface receptors, nuclear
receptors, transcription factors and cytoplasmic kinases (4,9). Similar
to the majority of RAN binding proteins, RANBP9 is functionally
associated with the β-importin receptor family, which is
responsible for transporting proteins into the nucleus (8). Additionally, RANBP9 has been associated
with osteosarcoma, lung, gastric and breast cancer (10–14);
however, its systematic effects in cancer remain to be
investigated.
In the present study, overexpression of RANBP9 in
CRC was identified. Additionally, its suppression by short hairpin
RNA (shRNA) promoted cell proliferation and transition from S
phase. Furthermore, cyclin A2 expression was demonstrated to be
associated with RANBP9 knockdown. In conclusion, the findings of
the present study suggested that RANBP9 may be a potent
anti-oncogene in CRC.
Materials and methods
Clinical specimens and
immunohistochemistry (IHC) scoring
A total of 75 consecutive specimens (tumors and
paired normal tissues) from 53 (70.7%) male patients and 22 (29.3%)
female patients (median, 65 years; range, 32–81) with CRC who
underwent radical colectomy were collected at the Department of
General Surgery, Jinshan Hospital, Fudan University (Shanghai,
China) between January and June 2012. IHC was performed and
investigated as described previously (15). In January 2018, 12 fresh specimens
(tumors and paired normal tissues) from patients with CRC were
randomly collected from the same hospital for detection of RANBP9
expression using western blotting (WB). The age range was 36–79
years (median, 56 years), including 9 (75.0%) male patients and 3
(25.0%) female patients. Ethical approval was obtained from the
Clinical Research Ethics Committee of Jinshan Hospital, Fudan
University. Written informed consent for the acquisition and use of
tissue samples was obtained from all patients.
Cell culture
The CRC cell lines HCT116, HT29, SW480, SW620, RKO,
Lovo, Caco2 and DLD1 were obtained from Nanjing KeyGen Biotech Co.,
Ltd. (Nanjing, China). HCT116 and HT29 cells were maintained in
McCoy's 5A medium (Nanjing KeyGen Biotech Co., Ltd.), while the
other cell lines were cultured in Dulbecco's modified Eagle's
medium (HyClone; GE Healthcare Life Sciences, Logan, UT, USA). The
media were supplemented with 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) and cultured in a
humidified atmosphere containing 5% CO2/95% air at
37°C.
Plasmids and cell transfection
RANBP9-shRNA (NM_005493.2) targeting sequence
(5′-GGAATTGGATCCTGCGCAT-3′) was designed and constructed. Non-sense
sequence (5′-TTCTCCGAACGTGTCACGT-3′) was used as a control-shRNA.
shRNA sequences were then cloned into GV112 plasmids (Shanghai
GeneChem Co., Ltd., Shanghai, China), which were packed into a
lentivirus using 293T cells, and the lentivirus was harvested and
purified by Shanghai GeneChem Co., Ltd. HCT116 and HT29 cells at a
density of 104 cells per well in 6-well plates were
infected with the lentiviruses at 20 multiplicity of infection. The
stably infected cells were enriched by puromycin (2 µg/ml;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The time interval
between infection and subsequent experimentation was >1 month.
To induce overexpression of RANBP9, HCT116 and HT29 cells
(1×106) were transfected with 2 µg/ml
GV141-RANBP9 or empty vectors (Shanghai GeneChem Co., Ltd.)
using FuGENE HD transfection reagent (Promega Corporation, Madison,
WI, USA). After 72 h of transfection, WB analysis was conducted to
verify RANBP9 expression with a RANBP9 antibody. In the shRNA
experiment, HCT116 or HT29 cells were infected with the empty
vector in the blank control group.
Cell Counting kit-8 (CCK-8) assay
HCT116 and HT29 cells (4,000 cells/well) were seeded
into 96-well plates. Cell viability was measured using a CCK-8
assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) at
several time points over 3 days. Briefly, cells were incubated with
10 µl CCK-8 for 1 h at 37°C. Subsequently, the optical density was
detected at 450 nm using a multifunctional plate reader (BioTek
Instruments, Inc., Winooski, VT, USA) according to the
manufacturer's protocol.
Anchorage-independent colony formation
assay
Complete medium with 0.5% agarose was layered in a
6-well plate and placed at room temperature to concretion. HCT116
and HT29 cells (200/well) were inoculated into the plate. Matrigel
(250 µg/ml; BD Biosciences, San Jose, CA, USA) was mixed with the
cell solutions for 1 min in advance. When the number of cells in
the majority of the single colonies were >50, the cells were
stained with 0.005% crystal violet (Biosharp, Hefei, Anhui, China)
for 1 h at room temperature. Subsequently, the number of visible
colonies was counted.
Flow cytometry (FCM)
HCT116 and HT29 cells were harvested and prepared as
cell suspensions. Adherent cells were digested with EDTA-free
trypsin and washed twice with ice-cold PBS. The cells were
subsequently fixed with 70% ethanol at −20°C for 2 h, and stained
with PI/RNase staining solution (BD Biosciences) for 1 h at 4°C.
Following incubation with PI for 15 min at room temperature in the
dark, the cells were immediately quantified using FCM (Beckman
Coulter, Inc., Brea, CA, USA) and the results were analyzed using
ModFit LT™ 4.1 software (Verity Software House, Inc., Topsham, ME,
USA).
WB
HCT116 and HT29 cells were homogenized in a lysate
buffer (Beyotime Institute of Biotechnology, Shanghai, China), and
protein concentrations were determined with a bicinchoninic acid
kit (Beyotime). Protein extracts (30 µg) were separated by 10%
SDS-PAGE and transferred to polyvinylidene difluoride membranes
(EMD Millipore, Billerica, MA, USA). The blots were blocked with 5%
non-fat milk for 1 h at room temperature, followed by incubation
with primary antibodies overnight at 4°C. The primary antibodies
used were: Anti-GAPDH (1:2,000 dilution; cat. no. 10494-1-AP; Wuhan
Sanying Biotechnology, Wuhan, China), anti-RANBP9 (1:1,000
dilution; cat. no. 17755-1-AP; Wuhan Sanying Biotechnology),
anti-cyclin A2 (1:1,000 dilution; cat. no. 18202-1-AP; Wuhan
Sanying Biotechnology) and anti-cyclin B1 (1:1,000 dilution; cat.
no. 55004-1-AP; Wuhan Sanying Biotechnology). A horseradish
peroxidase-conjugated anti-rabbit antibody as the secondary
antibody (1:10,000; cat. no. KGAA35; Nanjing KeyGen Biotech Co.,
Ltd.) was incubated for 1 h at room temperature. The immunoreactive
bands were visualized by the Tanon-4500 Gel Imaging system (Tanon
Science and Technology Co., Ltd., Shanghai, China) using an
enhanced chemiluminescence kit (Thermo Fisher Scientific,
Inc.).
In vivo tumorigenicity assay
A total of 12 specific pathogen-free male BALB/c
mice (4 weeks old; 14–18 g) were purchased from Shanghai SIPPR-Bk
Lab Animal Co., Ltd. (Shanghai, China). All animals were housed
under a temperature of 22±1°C and a relative humidity of 50±1% in a
12-h light/dark cycle with free access to food and water. All
studies were approved by the Shanghai Public Health Clinical Center
Laboratory Animal Welfare and Ethics Committee (IRB no.
2018-A001-01; Shanghai, China). All mice were handled according to
the National Institutes of Health Guidelines for the Care and Use
of Laboratory Animals (16). Three
animals per group (4 groups: The control and RANBP9-shRNA
group for HCT116 cells or HT29 cells) were used in each experiment.
Mice tissues were homogenized in a lysate buffer (Beyotime) and
underwent WB. HCT116 or HT29 cells (expressing control-shRNA or
RANBP9-shRNA) in 100 µl PBS at 1×108 cells/ml
were inoculated into the armpit of the mice. The tumor volumes were
measured weekly and calculated using the following formula: π/6 ×
length × width2.
Statistical analysis
RANBP9 expression in human tissues was analyzed by
Wilcoxon test. All data are expressed as the means ± standard
deviation of three independent experiments. Results were analyzed
using a Student's t-test, or one-way analysis of variance followed
by a least significant difference post hoc test, using the SPSS
version 23 software (IBM Corp., Armonk, NY, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
RANBP9 is overexpressed in human
CRC
RANBP9 expression was detected in the
paraffin-embedded CRC tissue samples using IHC (Fig. 1A-G) and in fresh tissue samples using
WB (Fig. 1H). The results revealed
that RANBP9 was overexpressed in CRC tissues compared with in
adjacent normal tissues (two-sided Wilcoxon test, P<0.0001;
Table I). RANBP9 was detected in the
nucleus and cytoplasm. These results indicated that high levels of
RANBP9 were present in CRC. In the majority of fresh tissues,
RANBP9 expression was higher in the tumor tissue than in the
adjacent normal tissue. Additionally, the protein expression levels
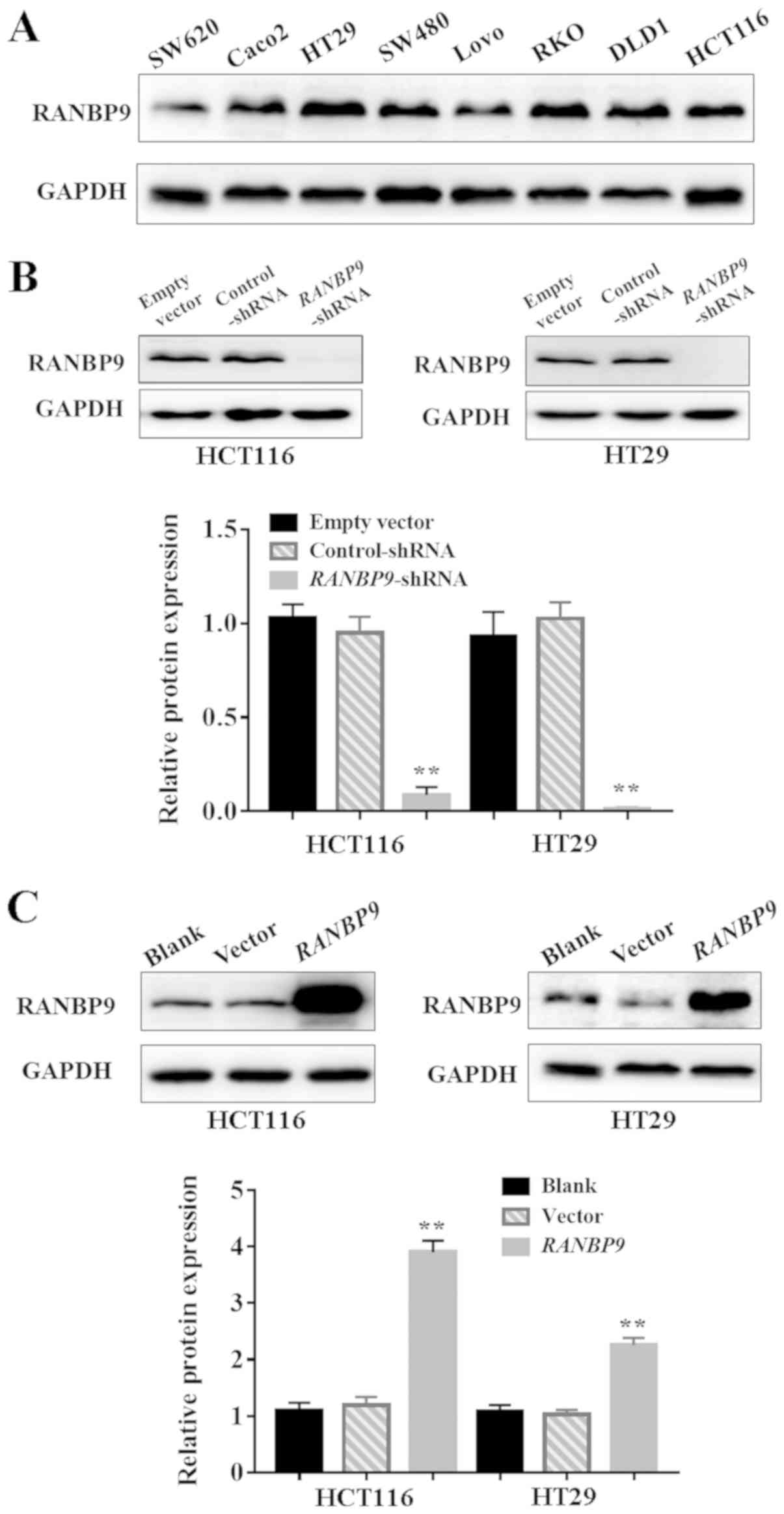
of RANBP9 in CRC cell lines were confirmed by WB (Fig. 2A). HCT116 and HT29 cells were
selected for subsequent experiments as they are highly invasive CRC
cell lines and exhibited high expression levels of RANBP9.
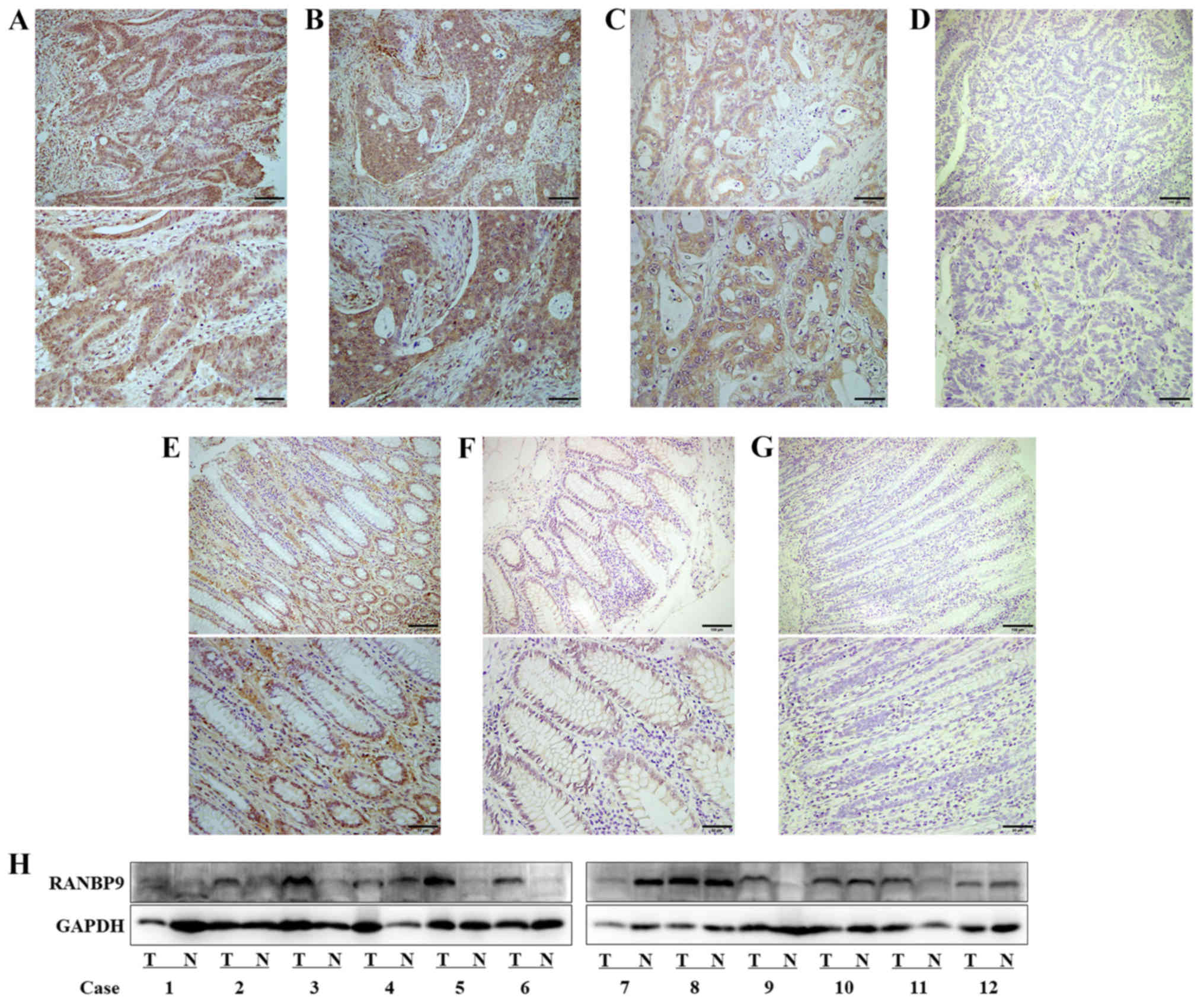 | Figure 1.Expression of RANBP9 in
paraffin-embedded and fresh colorectal cancer tissue samples. In
tumor samples, (A) high-intensity RANBP9 expression was scored as
‘+++’, (B) moderate-intensity RANBP9 expression as ‘++’, (C)
low-intensity RANBP9 expression as ‘+’ and (D) the near absence of
RANBP9 staining was scored as ‘-’. (E-G) Moderate, low and negative
expression levels of RANBP9 in normal mucosa, respectively. (H)
Expression levels of RANBP9 in 12 colorectal cancer samples as
demonstrated by western blotting. Scale bars: Upper rows, 100 µm;
lower rows, 50 µm. N, normal mucosa; RANBP9, RAN binding protein 9;
T, tumor. |
 | Table I.Expression of RANBP9 in 75 colorectal
cancer samples. |
Table I.
Expression of RANBP9 in 75 colorectal
cancer samples.
|
| RANBP9
immunostaining |
|
|
|---|
|
|
|
|
|
|---|
| Sample type | +++ | ++ | + | − | Positive rate
(%) |
P-valuea |
|---|
| Cancer | 14 | 24 | 31 | 6 | 92.0 | <0.0001 |
| Normal mucosa | 0 | 10 | 28 | 37 | 50.7 |
|
RANBP9 knockdown promotes
proliferation of CRC cells
RANBP9 overexpression in CRC suggested that it may
be associated with the biological behavior of CRC.
RANBP9-shRNA and control-shRNA were successfully infected
into HCT116 and HT29 cells using a lentiviral vector (Fig. 2B). HCT116 and HT29 cells were
successfully transfected with RANBP9 plasmid or empty vector
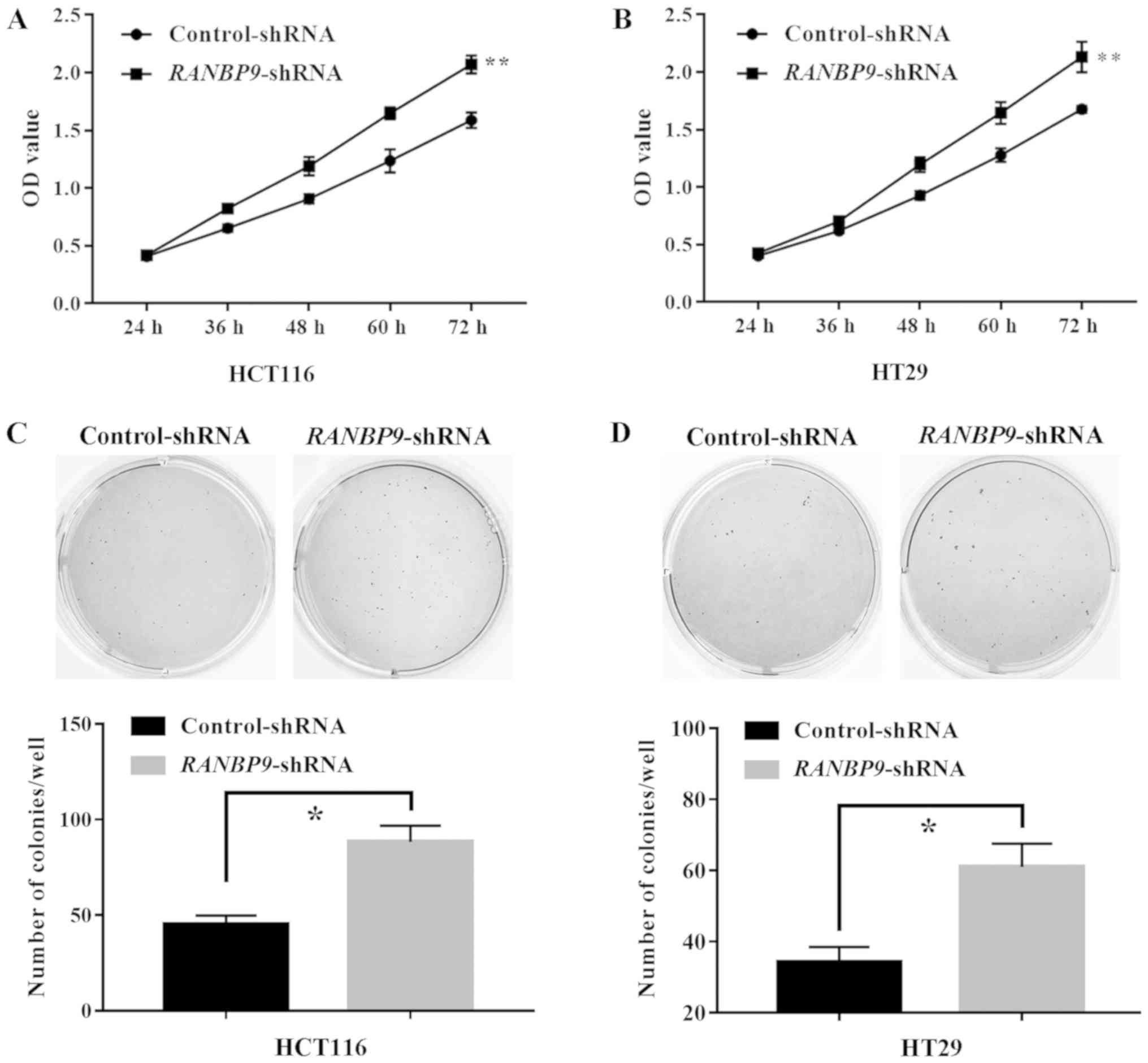
(Fig. 2C). A CCK-8 assay revealed
that the number of viable cells at the last time point (72 h) in
the RANBP9-shRNA group was significantly higher compared
with in the control group (P<0.01; Fig. 3A and B). In addition, a colony
formation assay was performed to determine tumorigenesis in
vitro. Compared with in the control group, the number of HCT116
and HT29 cell colonies in the RANBP9-shRNA group was
significantly higher (P<0.05; Fig. 3C
and D). These results suggested that RANBP9-shRNA
exhibited a positive effect on CRC cell proliferation.
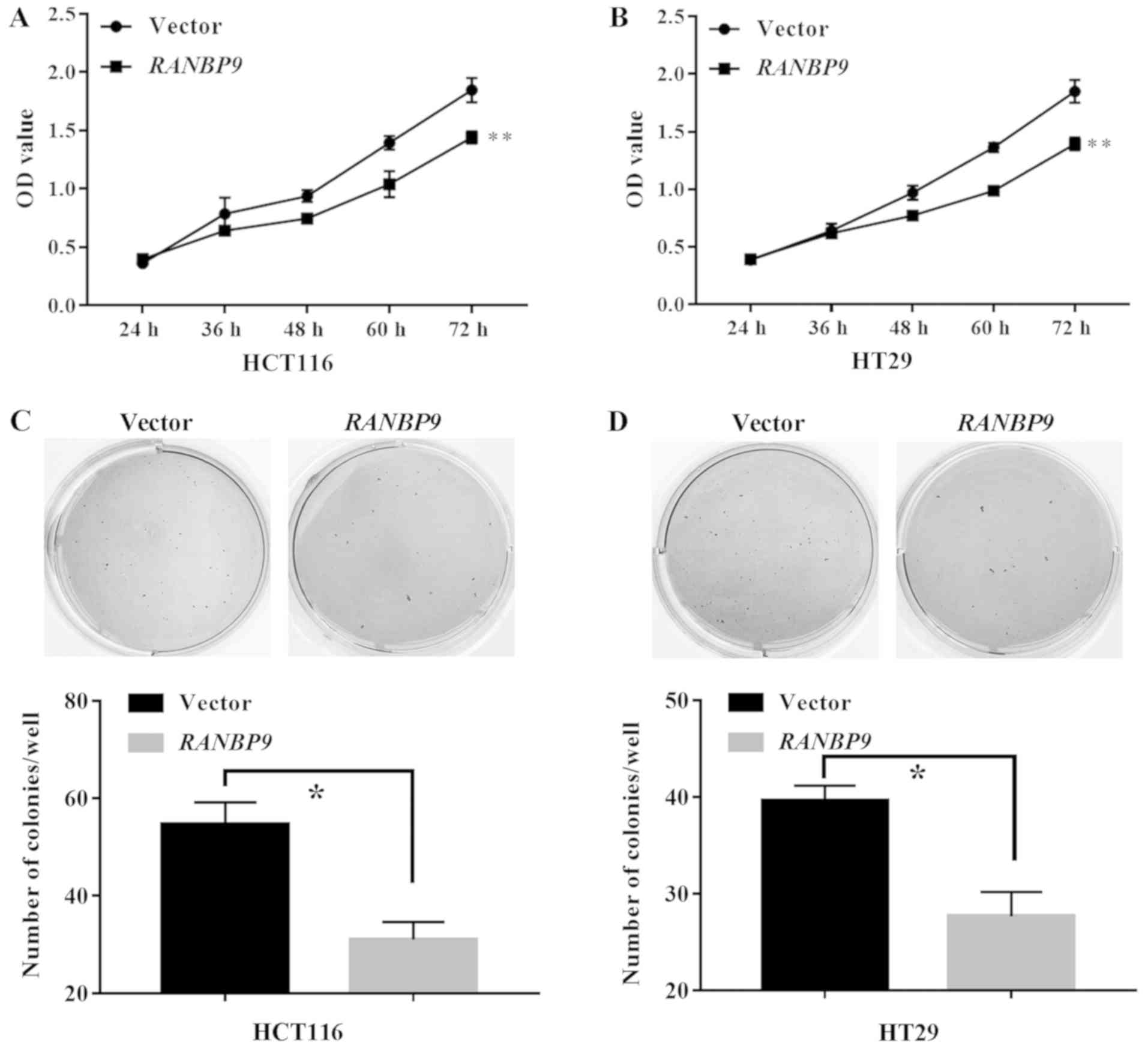
RANBP9 overexpression hinders the
proliferation of CRC cells
A CCK-8 assay revealed that the number of viable
cells at the final timepoint (72 h) in the RANBP9 group was
significantly lower compared with in the empty vector group
(P<0.01; Fig. 4A and B).
Similarly, in a colony formation assay, the number of HCT116 and
HT29 cell colonies in the RANBP9 group was significantly
lower compared with in the empty vector group (P<0.05; Fig. 4C and D). These data suggested an
inhibitory role of RANBP9 in CRC cell proliferation.
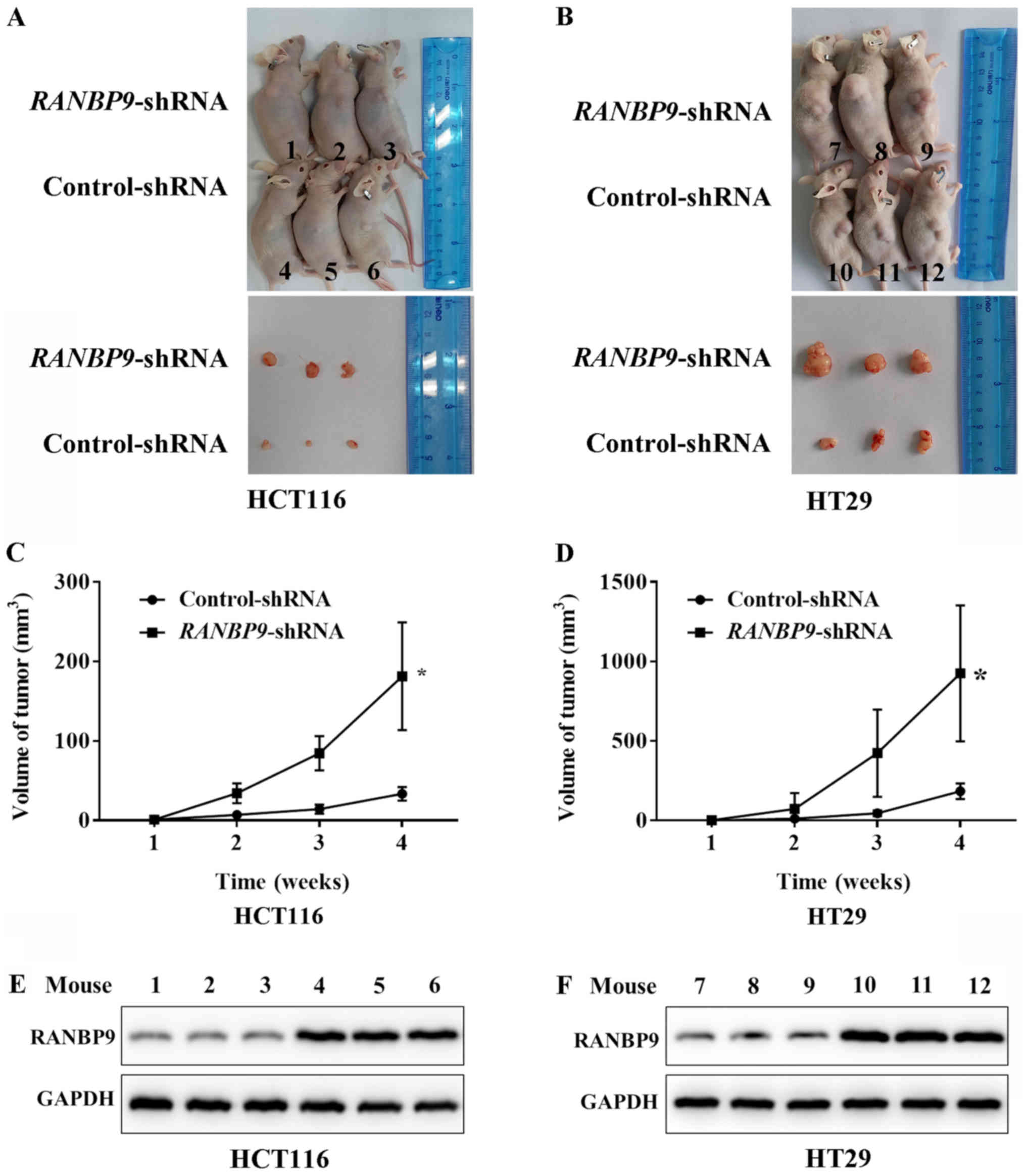
RANBP9-shRNA promotes colon
tumorigenesis in vivo
To examine the effects of RANBP9 knockdown on tumor
growth in vivo, HCT116 and HT29 cells that stably expressed
RANBP9-shRNA or control-shRNA were inoculated into the
armpit of BALB/c mice. All animals developed tumors within 1 week
following inoculation. At 4 weeks post-inoculation, tumor sizes of
the RANBP9-shRNA animals were larger than those of the
control-shRNA group (Fig. 5A and B).
The mean tumor volume after 4 weeks was 181.3±67.7 mm3
in mice inoculated with HCT116 cells that expressed
RANBP9-shRNA compared with 33.7±8.5 mm3 in the
control-shRNA group (Fig. 5C). The
mean tumor volume of mice inoculated with HT29 cells transfected
with RANBP9-shRNA was 925.3±427.3 mm3 compared
with a volume of 184.3±48.5 mm3 in the control group
(Fig. 5D). These data demonstrated
the role of RANBP9-shRNA in promoting the growth of CRC
cells in vivo. The efficiency of RANBP9-knockdown in
lentivirus-infected HCT116 and HT29 cells was ensured according to
the detection of RANBP9 expression level in transplanted tumors
using WB (Fig. 5E and F).
RANBP9-shRNA facilitates the
transition from S phase in CRC cells
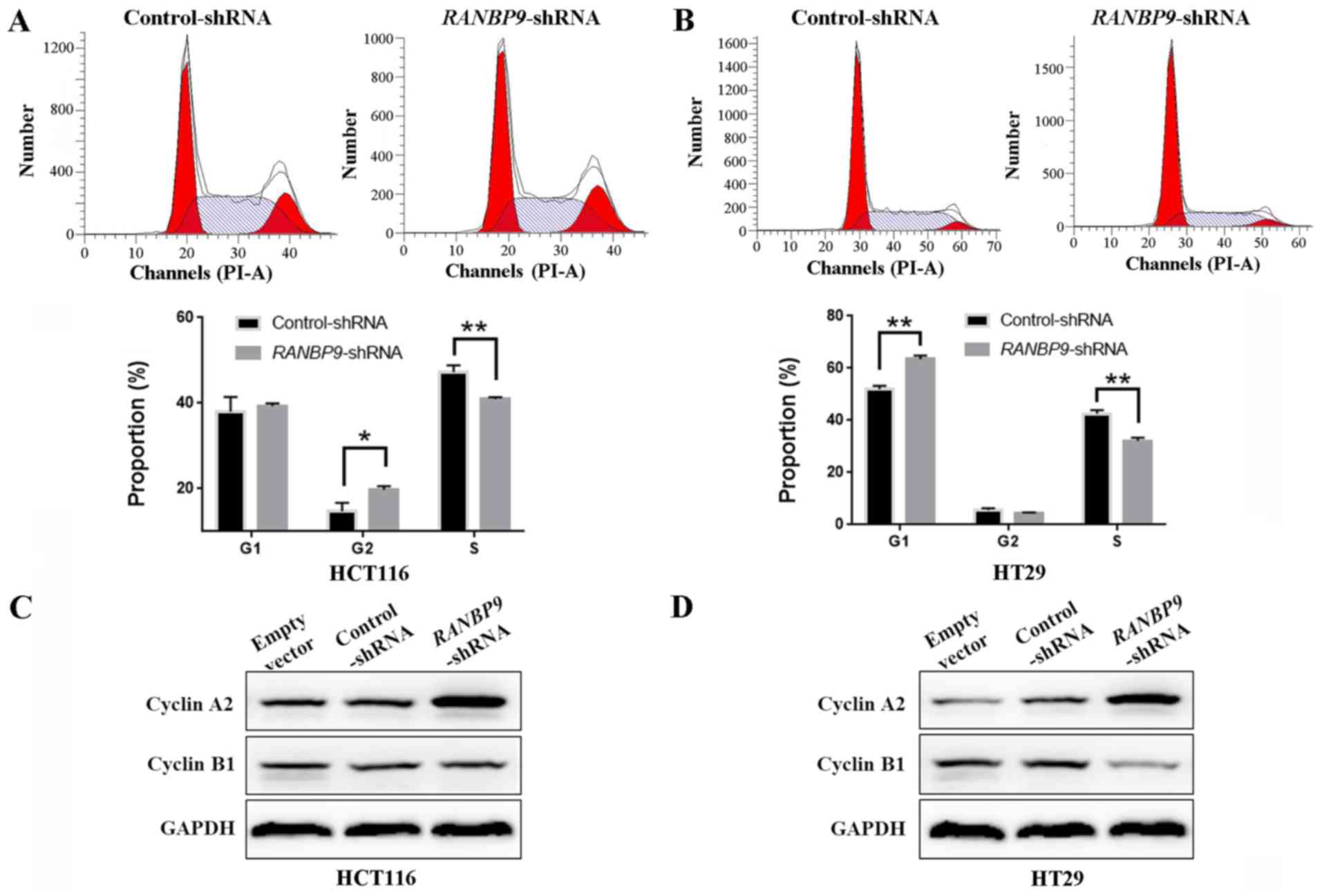
Cell cycle profiling by FCM indicated that knockdown
of RANBP9 expression resulted in a decreased number of cells in S
phase in HCT116 and HT29 cells (Fig. 6A
and B). However, no consistency was observed in the
distribution between G1 and G2 stages for the
two cell lines. These results suggested that RANBP9-shRNA
promoted the proliferation of CRC cells indicated in the CCK-8
assay and the colony formation assay, by facilitating the
transition from S phase. Additionally, cell cycle-associated
protein expression levels were examined by WB following knockdown
of RANBP9. RANBP9 knockdown yielded a variably increased protein
level of cyclin A2 in the two cell lines (Fig. 6C and D). In addition, HT29 cells
exhibited a lower expression of cyclin B1.
Discussion
In the present study, RANBP9 expression was
significantly increased in tumor tissues compared with in paired
normal mucosa tissues in patients with CRC, which indicated that
RANBP9 may be associated with CRC. To further explore the function
of RANBP9, its expression in several CRC cell lines was examined.
The suppression of RANBP9 promoted proliferation and strengthened
colony formation in CRC cells; accordingly, its overexpression led
to suppressed proliferation and impaired colony formation.
Additionally, shortening of the S phase was identified in two CRC
cell lines. The present study revealed that RANBP9 may be a
potential anti-oncogene in CRC. These results were consistent with
studies on osteosarcoma, lung and gastric cancer (10–13).
The present study demonstrated that RANBP9 exhibited
an inhibitory role in CRC, yet it remains unclear why malignant
cells overexpress RANBP9. Similar phenomena have been observed in
osteosarcoma, lung and gastric cancer (10–13).
Cancer cells are associated with different sets of oncogenes and
tumor suppressor genes that are normally involved in stabilizing
negative feedback loops. These genes do not act individually, but
rather execute their cellular functions in cooperation with other
genes, including oncogene mouse double minute 2 and tumor
suppressor gene p53, which are overexpressed in hepatocellular
cancer (17,18), or Wnt signaling and conductin, which
are upregulated in colorectal and liver tumors (19). Nevertheless, further studies should
be conducted to elucidate the oncogene partner of RANBP9 in
CRC.
The cell cycle is a complex system, which includes
phases that are comprehensively regulated by cyclin-dependent
kinases (CDKs), cyclins, catalytic partners and CDK inhibitors,
directly affecting the proliferation and tissue differentiation of
mammalian cells (20,21). In numerous diseases, the mechanism of
normal cell division is constantly altered, particularly in several
types of cancer. Cyclins and CDKs, including cyclin D, cyclin E,
CDK4 and CDK6, have been classified as proto-oncogenes, while CDK
inhibitors, including p16 and p21, are regarded as tumor
suppressors; all of these are associated with the occurrence or
prognosis of CRC (20,22,23).
Therefore, cell cycle-targeted therapy is a possible way to control
the disease. For example, palbociclib, which is an orally
administered and specific CDK4/6 inhibitor, can prominently inhibit
tumor growth in mice bearing colon cancer (24). Flavopiridol, which inhibits CDK1,
CDK2 and CDK4, has also been widely studied in gastrointestinal
cancer (25,26). There are additional cell cycle
inhibitors in different stages of development; nonetheless, to the
best of our knowledge, there are still no encouraging reports. In
the present study, RANBP9-shRNA promoted S phase transition
in HCT116 and HT29 cells, although there were no consistent results
for the G1 or G2 phases. CDK/Cyclin A2
complexes have been reported to phosphorylate proteins, including
pocket proteins Rb, p107, p130, and proteins involved in DNA
synthesis, thereby driving S phase progression (27,28). In
the present study, it was indicated that RANBP9-knockdown increased
protein level of cyclin A2 in the two cell lines. In addition,
RANBP9-shRNA promotes cell proliferation by CCK8 assay and
colony formation assay. These suggest that RANBP9-shRNA may
facilitate S phase transition rather than induce G1 or
G2/M arrest. Further studies are required in order to
prove that RANBP9 overexpression induces cell cycle arrest at S
phase.
At present, eukaryotic cyclins, including cyclin
A-H, which are synthesized and degraded in different phases of the
cell cycle, are fluctuant and periodic-phase specific (29). Cyclin A serves a role in the later
stages of the S phase; cyclin B is mainly associated with the
G2/M phase; cyclin D and cyclin E serve a key regulatory
role in the G1/S phase; and cyclin F and cyclin G are
associated with the late S and G2/M phase (29,30). The
results of the present study revealed that there may be an
association between cyclin A and RANBP9 knockdown in CRC cells.
However, it may be a phenomenon rather than a direct regulatory
association, which requires further examination.
There are limitations in the present study. Firstly,
the clinical sample size was small and obtained at a single center,
which may lead to selective bias. In addition, RANBP9 expression in
fresh normal intestinal epithelium in certain cases was higher
compared with in matched tumor tissues, which remains unexplained.
RANBP9 has numerous characteristics as a scaffold protein,
including a protein interaction sequence, cytoskeleton binding
domain and multiple classical anchor loci for signal transduction
molecules (4,31); it can interact with SRC
proto-oncogene non-receptor kinase, growth factor receptor bound
protein 2 and hepatocyte growth factor receptor in the
mitogen-activated protein kinase and extracellular signal-regulated
kinase signaling pathways (4,32–34).
The downstream mechanism of RANBP9 in CRC cells should be examined
in subsequent experiments.
In conclusion, RANBP9 is a molecule associated with
the complex cell cycle regulation network. The present study
revealed that RANBP9 contributed to tumor suppression in CRC.
Exploring the underlying mechanisms of RANBP9 activity may lead to
its use in the treatment of patients with CRC.
Acknowledgements
Not applicable.
Funding
The study was funded by the Health and Family
Planning Commission of Jinshan District, Shanghai, China (grant no.
JSKJ-KTMS-2017-01).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
CQ and GW designed the study and analyzed the data.
CQ and QZ performed the experiments. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
All patients were required to provide written
informed consent prior to their inclusion. The study was approved
by the Ethics Committee of Jinshan Hospital, Fudan University. All
animal studies were approved by the Shanghai Public Health Clinical
Center Laboratory Animal Welfare and Ethics Committee (IRB no.
2018-A001-01).
Patient consent for publication
All patients provided written informed consent for
the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fearon ER and Vogelstein B: A genetic
model for colorectal tumorigenesis. Cell. 61:759–767. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yokoyama N, Hayashi N, Seki T, Panté N,
Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T and Aebi U: A giant
nucleopore protein that binds Ran/TC4. Nature. 376:184–188. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Murrin LC and Talbot JN: RanBPM, a
scaffolding protein in the immune and nervous systems. J
Neuroimmune Pharmacol. 2:290–295. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Salemi LM, Loureiro SO and Schild-Poulter
C: Characterization of RanBPM molecular determinants that control
its subcellular localization. PLoS One. 10:e01176552015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang H, Lewsadder M, Dorn E, Xu S and
Lakshmana MK: RanBP9 overexpression reduces dendritic arbor and
spine density. Neuroscience. 265:253–262. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang R, Palavicini JP, Wang H, Maiti P,
Bianchi E, Xu S, Lloyd BN, Dawson-Scully K, Kang DE and Lakshmana
MK: RanBP9 overexpression accelerates loss of dendritic spines in a
mouse model of Alzheimer's disease. Neurobiol Dis. 69:169–179.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Valiyaveettil M, Bentley AA, Gursahaney P,
Hussien R, Chakravarti R, Kureishy N, Prag S and Adams JC: Novel
role of the muskelin-RanBP9 complex as a nucleocytoplasmic mediator
of cell morphology regulation. J Cell Biol. 182:727–739. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rex EB, Rankin ML, Yang Y, Lu Q, Gerfen
CR, Jose PA and Sibley DR: Identification of RanBP 9/10 as
interacting partners for protein kinase C (PKC) gamma/delta and the
D1 dopamine receptor: Regulation of PKC-mediated receptor
phosphorylation. Mol Pharmacol. 78:69–80. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao Z, Cheng S, Zabkiewicz C, Chen J,
Zhang L, Ye L and Jiang WG: Reduced expression of ranBPM is
associated with poorer survival from lung cancer and increased
proliferation and invasion of lung cancer cells in vitro.
Anticancer Res. 37:4389–4397. 2017.PubMed/NCBI
|
|
11
|
Shao S, Sun PH, Satherley LK, Gao X, Ji
KE, Feng YI, Jia Y, Ji J, Jiang WG and Ye L: Reduced ranBPM
expression is associated with distant metastasis in gastric cancer
and chemoresistance. Anticancer Res. 36:1295–1303. 2016.PubMed/NCBI
|
|
12
|
Dai H, Lv YF, Yan GN, Meng G, Zhang X and
Guo QN: RanBP9/TSSC3 complex cooperates to suppress anoikis
resistance and metastasis via inhibiting Src-mediated Akt signaling
in osteosarcoma. Cell Death Dis. 7:e25722016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu LL, Wang CH, Yang HP and Shu WH:
Expression of cartilage antitumor component RanBP9 in osteosarcoma.
J Biol Regul Homeost Agents. 30:103–110. 2016.PubMed/NCBI
|
|
14
|
Emberley ED, Gietz RD, Campbell JD,
HayGlass KT, Murphy LC and Watson PH: RanBPM interacts with
psoriasin in vitro and their expression correlates with specific
clinical features in vivo in breast cancer. BMC Cancer. 2:282002.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qin C, Ren L, Ji M, Lv S, Wei Y, Zhu D,
Lin Q, Xu P, Chang W and Xu J: CDKL1 promotes tumor proliferation
and invasion in colorectal cancer. Onco Targets Ther. 10:1613–1624.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang H, Yang R, Zhong L, Zhu XY, Ma PP,
Yang XQ, Jiang KL and Liu BZ: Location of NLS-RARalpha protein in
NB4 cell and nude mice. Oncol Lett. 13:2045–2052. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Meng X, Franklin DA, Dong J and Zhang Y:
MDM2-p53 pathway in hepatocellular carcinoma. Cancer Res.
74:7161–7167. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang MF, Zhang ZY, Fu J, Yang YF and Yun
JP: Correlation between expression of p53, p21/WAF1, and MDM2
proteins and their prognostic significance in primary
hepatocellular carcinoma. J Transl Med. 7:1102009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lustig B, Jerchow B, Sachs M, Weiler S,
Pietsch T, Karsten U, van de Wetering M, Clevers H, Schlag PM,
Birchmeier W and Behrens J: Negative feedback loop of Wnt signaling
through upregulation of conductin/axin2 in colorectal and liver
tumors. Mol Cell Biol. 22:1184–1193. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Malumbres M: Cyclin-dependent kinases.
Genome Biol. 15:1222014. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Morgan DO: Cyclin-dependent kinases:
Engines, clocks, and microprocessors. Annu Rev Cell Dev Biol.
13:261–291. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ogino S, Nosho K, Irahara N, Shima K, Baba
Y, Toyoda S, Chen L, Giovannucci EL, Meyerhardt JA and Fuchs CS: A
cohort study of cyclin D1 expression and prognosis in 602 colon
cancer cases. Clin Cancer Res. 15:4431–4438. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao P, Hu YC and Talbot IC: Expressing
patterns of p16 and CDK4 correlated to prognosis in colorectal
carcinoma. World J Gastroenterol. 9:2202–2206. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fry DW, Harvey PJ, Keller PR, Elliott WL,
Meade M, Trachet E, Albassam M, Zheng X, Leopold WR, Pryer NK and
Toogood PL: Specific inhibition of cyclin-dependent kinase 4/6 by
PD 0332991 and associated antitumor activity in human tumor
xenografts. Mol Cancer Ther. 3:1427–1438. 2004.PubMed/NCBI
|
|
25
|
Aklilu M, Kindler HL, Donehower RC, Mani S
and Vokes EE: Phase II study of flavopiridol in patients with
advanced colorectal cancer. Ann Oncol. 14:1270–1273. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Motwani M, Rizzo C, Sirotnak F, She Y and
Schwartz GK: Flavopiridol enhances the effect of docetaxel in vitro
and in vivo in human gastric cancer cells. Mol Cancer Ther.
2:549–555. 2003.PubMed/NCBI
|
|
27
|
Yam CH, Fung TK and Poon RY: Cyclin A in
cell cycle control and cancer. Cell Mol Life Sci. 59:1317–1326.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zindy F, Lamas E, Chenivesse X, Sobczak J,
Wang J, Fesquet D, Henglein B and Bréchot C: Cyclin A is required
in S phase in normal epithelial cells. Biochem Biophys Res Commun.
182:1144–1154. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bloom J and Cross FR: Multiple levels of
cyclin specificity in cell-cycle control. Nat Rev Mol Cell Biol.
8:149–160. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Malumbres M and Barbacid M: Cell cycle,
CDKs and cancer: A changing paradigm. Nat Rev Cancer. 9:153–166.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lakshmana MK, Chung JY, Wickramarachchi S,
Tak E, Bianchi E, Koo EH and Kang DE: A fragment of the scaffolding
protein RanBP9 is increased in Alzheimer's disease brains and
strongly potentiates amyloid-beta peptide generation. FASEB J.
24:119–127. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Brannetti B and Helmer-Citterich M: iSPOT:
A web tool to infer the interaction specificity of families of
protein modules. Nucleic Acids Res. 31:3709–3711. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang D, Li Z, Messing EM and Wu G: The
SPRY domain-containing SOCS box protein 1 (SSB-1) interacts with
MET and enhances the hepatocyte growth factor-induced
Erk-Elk-1-serum response element pathway. J Biol Chem.
280:16393–16401. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Woo JA, Roh SE, Lakshmana MK and Kang DE:
Pivotal role of RanBP9 in integrin-dependent focal adhesion
signaling and assembly. FASEB J. 26:1672–1681. 2012. View Article : Google Scholar : PubMed/NCBI
|