Introduction
Glioma, which has a poor prognosis, is the most
common malignant primary brain tumor; it often displays unique
biological features, including strong propensity for proliferation
and metastasis (1). Despite the use
of the most effective treatments available, including surgery and
chemoradiotherapy, the median survival time of patients with
glioblastoma has no optimistic estimate of being extended (1). Glioma often develops resistance to a
number of chemotherapy drugs, such as nitrosourea. There are
several approaches for glioma to obtain resistance, including
reduced drug absorption, increased drug discarding, enhanced
ability to decompose anticancer drugs, increased proliferation and
reduced apoptosis by stimulation of cytokine excretion to alter the
microenvironment (2). Drug
resistance genes, including O6-methylguanine-DNA methyltransferase
(MGMT), topoisomerase II (TOPO II) and multiple drug
resistance 1, are involved in these activities (3). Temozolomide (TMZ) has been used
extensively for the treatment of glioblastomas (GBMs) over the past
two decades; it is an alkylating agent that causes DNA mismatches
resulting in cell apoptosis in GBMs (4,5).
Nevertheless, TMZ does not increase life span significantly,
possibly due to high-grade glioma developing resistance to the
majority of chemotherapy drugs, including TMZ. The underlying
mechanisms require investigation and solutions to the problem of
drug resistance are being sought.
MicroRNAs (miRNAs) are involved in gene expression
regulation by binding to mRNA 3′-untranslated region to suppress
translation or induce cleavage of the target mRNA directly to
regulate protein expression (6).
They serve important roles in all physiological functions; for
example, cellular differentiation, proliferation, cell cycle
control, cell death and organ development (7). miRNAs are also involved in a variety of
human diseases, including inflammation and cancer (8). Our previous study demonstrated that
miRNA-9 (miR-9) inhibited vasculogenic mimicry and modulated
migration by suppressing stathmin expression (9), and the silencing of stathmin was able
to enhance chemotherapeutic sensitivity of glioma to TMZ (10). However, the effects of miR-9 on
chemotherapeutic sensitivity of glioma to TMZ and the underlying
mechanisms are unknown.
In the present study U251 glioma cells were
transfected with lentiviral (LV) vectors carrying a miR-9 mimic or
inhibitor and subsequently treated with gradient concentrations of
TMZ. Cell viability, cell apoptosis, cell cycle and proteins
responsible for apoptosis and drug resistance were examined. The
role of nuclear factor κB (NF-κB) in the regulation of sensitivity
to chemotherapy was also investigated. The aim of the present study
was to evaluate the effects of miR-9 on chemotherapy of glioma to
provide a target for glioma chemotherapy.
Materials and methods
Cell culture and miR-9
interference
The malignant glioma cell line U251 was obtained
from the American Type Culture Collection (Manassas, VA, USA) and
was cultured in complete Dulbecco's modified Eagle's medium (DMEM)
at 37° with 5% CO2. The Hsa-miR-9 sequence was obtained
from the miRbase database (www.miRbase.org). The miR-9 mimic sequence
(5′-TCTTTGGTTATCTAGCTGTATGA-3′) and inhibitor sequence
(5′-TCATACAGCTAGATAACCAAAGA-3′) were synthesized, inserted into
green fluorescent protein (GFP)-containing lentivirus vectors
(GV280 for inhibitor and GV369 for mimic and negative control), and
then embedded in lentiviral particles by Shanghai GeneChem Co.,
Ltd. (Shanghai, China). The cells (5×104 per well of a
6-well plate) were transfected with lentivirus particles using
Lentifectin™ transfection reagent (ABM Inc., Richmond,
BC, Canada) as per the protocol provided by the lentivirus
manufacturer (virus titer, 5×108 transducing units/ml;
multiplicity of infection, 5). Next, the cells were incubated in
the cell incubator. miR-9 transfection efficiency was examined by
fluorescence microscopy 72 h following transfection and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) 24 h
following transfection. The cells transfected with lentivirus
particles carrying empty vectors were used as negative controls
(NC) and the cells treated with transfection reagent only were
regarded as the mock group. For the following steps, cells were
incubated for 120 h after transfection.
TMZ treatment
TMZ (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
was dissolved in dimethyl sulfoxide (DMSO) to make a 10 mM stock
solution that was serially diluted in DMEM to the following
concentrations: 31.25, 62.5, 125, 250, 500 and 1,000 µM. All groups
had the same final concentration of DMSO (1% v/v). A further
control with DMSO only was named the 0 µM control. U251 cells were
plated and treated with the all concentrations of TMZ to assess
cell viability by MTT assay, or for apoptosis and cell cycle
assays, with 100 µM TMZ, at which concentration cell viability is
inhibited.
MTT assay
Glioma cells were plated in 96-well plates (6,000
cells/well) and transfected and/or treated with TMZ as
aforementioned. Transfected and non-transfected cells were
incubated for 24, 48 or 72 h. Then cell viability was analyzed by
MTT colorimetric assay. MTT (500 mg) was dissolved in 100 ml PBS to
make 5 mg/ml stock solution. This MTT solution (20 µl) was added to
each well and incubated for 4 h, then the crystal was dissolved in
150 µl DMSO after the culture medium was discarded. The absorbance
at a wave length of 490 nm was detected using a microplate reader
(M2009PR; Tecan infinite; Thermo Fisher Scientific, Inc., Waltham,
MA, USA). Experiments were performed three times.
Apoptosis assay
Transfected cells were treated with 100 µM TMZ for
24 h, washed with Hank's D solution, harvested and counted. The
eBioscience™ Annexin V Apoptosis Detection Kit APC (cat. no.
88-8007-72; Invitrogen; Thermo Fisher Scientific, Inc.) was used to
measure apoptosis according to the manufacturer's protocol. A total
of 1×105 cells were resuspended in 100 µl binding
buffer, and 10 µl of Annexin V and 5 µl of propidium iodide (PI)
were added. The cells were then incubated in the dark for 15 min at
room temperature, and subsequently analyzed using an Epics Altra II
cytometer (Beckman Coulter Inc., Brea, CA, USA). The data were
analyzed by Kaluza analysis software (version 1.3; Beckman Coulter
Inc.) and the apoptotic rate (%) was determined by adding the cell
population positive for PI and annexin V (late apoptosis) and the
population positive for annexin V only (early apoptosis). The
experiment was repeated three times.
NF-κB signaling pathway
interference
Bay117082 (Sigma-Aldrich; Merck KGaA,), an inhibitor
of the NF-κB signaling pathway, was dissolved in DMSO to make a 10
mM stock solution. U251 cells were plated at a concentration of
5×104 cells/ml (100 µl for 96-well plates, and 1 ml for
6-well plates) and treated with 10 µM Bay117082 for 24 h at 37°C in
a cell incubator with 5% CO2.
Western blot analysis
Total protein was extracted from cells in one well
of a 6-well plate (~1×106 cells) following 24-h
treatment with 100 µM TMZ as aforementioned and the concentrations
were measured using a spectrophotometer (Bio-Rad Laboratories,
Inc., Hercules, CA, USA) with a bicinchoninic acid protein assay
kit (Beyotime Institute of Biotechnology, Shanghai, China). Protein
at a concentration of >5 µg/µl (50 µg in total) was separated by
10% or 12% SDS-PAGE and was transferred onto polyvinylidene
difluoride membranes (EMD Millipore, Billerica, MA, USA), followed
by blocking with skimmed milk dissolved in TBS and 0.05% Tween-20
(TBST) for 1 h at room temperature. The membranes were incubated
with primary antibodies (Table I) at
4°C overnight, washed three times with TBST and were incubated in
horseradish peroxidase-conjugated secondary antibodies at 1:5,000
dilution (sc-2004 and sc2005; Santa Cruz Biotech, Santa Cruz, CA,
USA) for 1 h at room temperature after washing with TBST three
times. Following washing, the protein bands were detected with ECL
substrates or DAB Detection System (both OriGene Technologies,
Inc., Beijing, China). GAPDH was used as a loading control, and all
experiment were repeated three times. Quantitative analysis was
performed using the Quantity One Software (version 4.6.2; Bio-Rad
Laboratories, Inc.).
 | Table I.Primary antibodies used in western
blot analysis. |
Table I.
Primary antibodies used in western
blot analysis.
| Protein | Supplier | Cat. no. | Origin | Dilution | Molecular weight |
|---|
| Bax | Abcam | Ab32503 | Rabbit | 1:1,000 | 21 kDa |
| Caspase-3 | Abcam | Ab4051 | Rabbit | 1:500 | 32 kDa |
| Bcl-2 | Abcam | Ab692 | Mouse | 1:500 | 26 kDa |
| GAPDH | Santa Cruz | Sc-32233 | Mouse | 1:2,000 | 36 kDa |
| TOPO II | Abcam | Ab52934 | Rabbit | 1:1,000 | 174 kDa |
| MMP-2 | Cell Signal
Tech | 13132 | Rabbit | 1:1,000 | 72 kDa |
| MMP-9 | Cell Signal
Tech | 13667 | Rabbit | 1:1,000 | 92 kDa |
| NF-κB | Abcam | Ab32536 | Rabbit | 1:10,000 | 65 kDa |
| IκB | Santa Cruz | Sc52900 | Mouse | 1:1,000 | 36 kDa |
| Cyclin D | Santa Cruz | Sc450 | Mouse | 1:1,000 | 35 kDa |
| N-Cad | Cell Signal
Tech | 13116 | Rabbit | 1:1,000 | 140 kDa |
| R-Cad | Abcam | Ab109242 | Rabbit | 1:10,000 | 150 kDa |
Cell cycle assay
Transfected cells were treated with 100 µM TMZ for
24 h in a cell incubator at 37°C and subsequently harvested, washed
with ice-cold PBS and fixed with 70% ethanol at 4°C overnight. The
ethanol was removed by centrifugation at 300 × g for 10 min at room
temperature and ~1×106 cells were resuspended in PBS
containing PI (50 µg/ml) and RNase A (50 µg/ml; both Sigma-Aldrich;
Merck KGaA) for 30 min in the dark prior to analysis by flow
cytometry (FACScalibur; BD Biosciences, San Jose, CA, USA). The
data were analyzed by ModFit LT™ software (Verity
Software House, Inc., Topsham, ME, USA) and the percentage of cells
at G0/G1, S or G2/M phase was calculated. DMSO-treated cells were
used as untreated controls. Experiments were repeated three
times.
RT-qPCR
Cells of one well of 6-well plate (~1×106
cells) were lysed with TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) following 24-h treatment with 100
µM TMZ as aforementioned and total mRNA was extracted. The mRNA was
reverse-transcribed into cDNA in a reverse-transcription reaction
with the following reagents: M-MLV reverse transcriptase (M1705);
dNTPs (U1240; both Promega Corporation, Madison, WI, USA); and
Oligo dT primer (Sangon Biotech Co., Ltd., Shanghai, China). For
PCR analysis, cDNA was diluted to a final concentration of 10
ng/µl. qPCR was performed with a Universal Master Mix (Nantong
Chem-Base Co. Ltd, Nantong, China). cDNA (50 ng) was used to
determine the relative amounts of mRNA by qPCR using a MAX3000
Sequence-Detection System (Nantong Chem-Base Co. Ltd,) using
specific primers with SYBR Green dye. The thermocycling conditions
were as follows: An initial step of 95°C for 30 sec, followed by 40
cycles of 95°C for 5 sec, 60°C for 30 sec, 95°C for 15 sec, 55°C
for 30 sec and 95°C for 15 sec. The primers used for PCR were as
follows: miR-9 forward, 5′-GTGCAGGGTCCGAGGT-3′ and reverse,
5′-GCGCTCTTTGGTTATCTAGC-3′. U6 was amplified as reference for miR-9
using the following primers: U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. The experiment was
performed three times, and the 2−ΔΔCq method was used
for determining relative expression levels of mRNA or microRNA
(11).
Statistical analysis
The results were presented as mean ± standard
deviation. Statistical analyses were performed using SPSS 16.0
statistical software (SPSS, Inc., Chicago, IL, USA). One-way or
two-way analysis of variance (ANOVA) were used to compare the
differences between the groups, and Dunnett's or Tukey post hoc
test was used for multiple comparisons. P<0.05 was considered to
indicate a statistically significant difference. The graphs were
drawn using GraphPad Prism 7 (GraphPad Software, Inc., La Jolla,
CA, USA).
Results
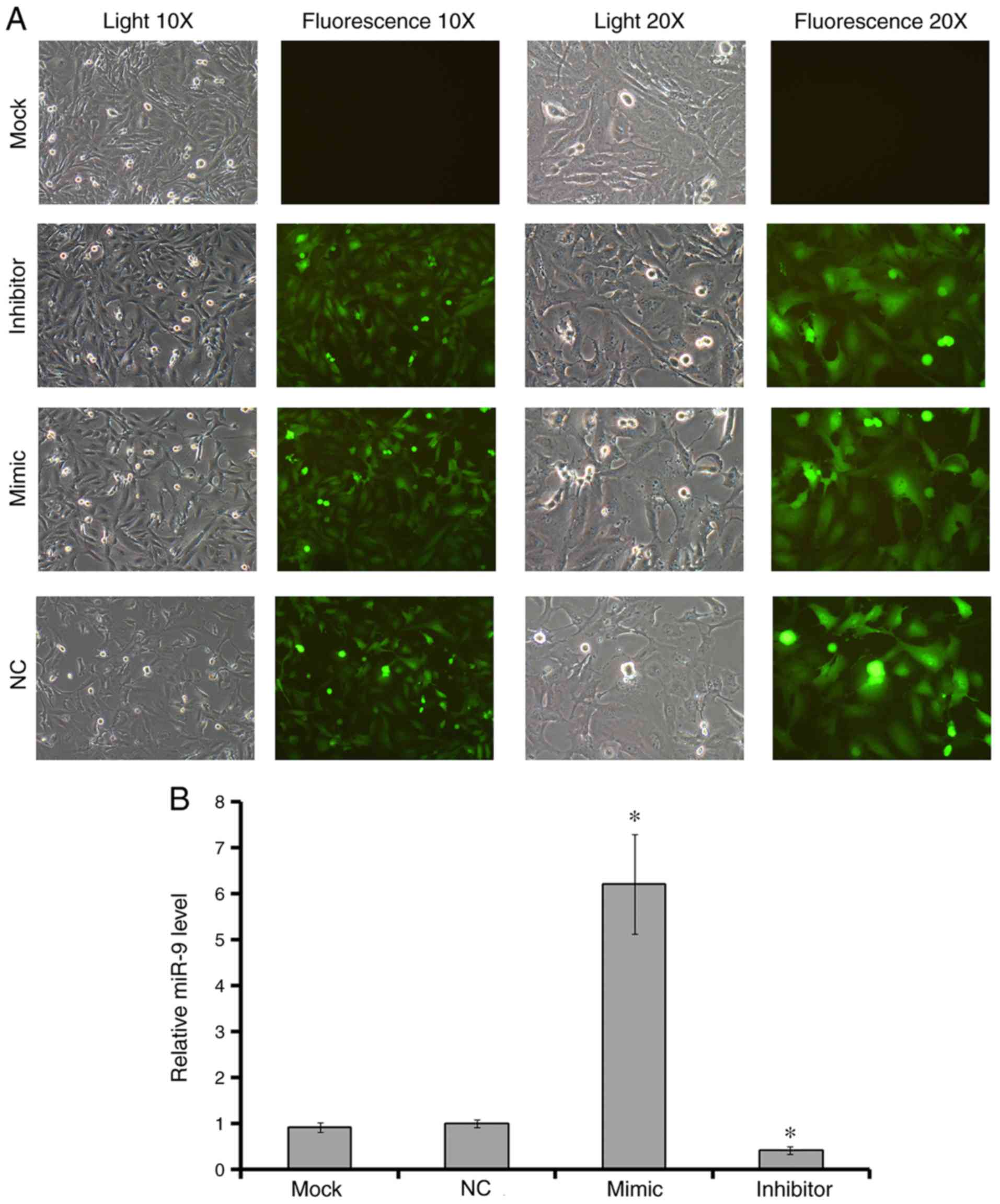
Verification of transfection
U251 cells were transfected with LV carrying miR-9
mimic or inhibitor. An empty virus was used as positive
transfection control group, and cells incubated with transfection
reagent only were used as an untreated control (mock). As the
results demonstrated, green fluorescence was detected in >95%
cells of the transfected groups (Fig.
1A). The levels of miR-9 expression in test groups were
evaluated by RT-qPCR. The results revealed that there were
significant differences between the mimic and the control groups,
as well as the inhibitor and the control groups (Fig. 1B).
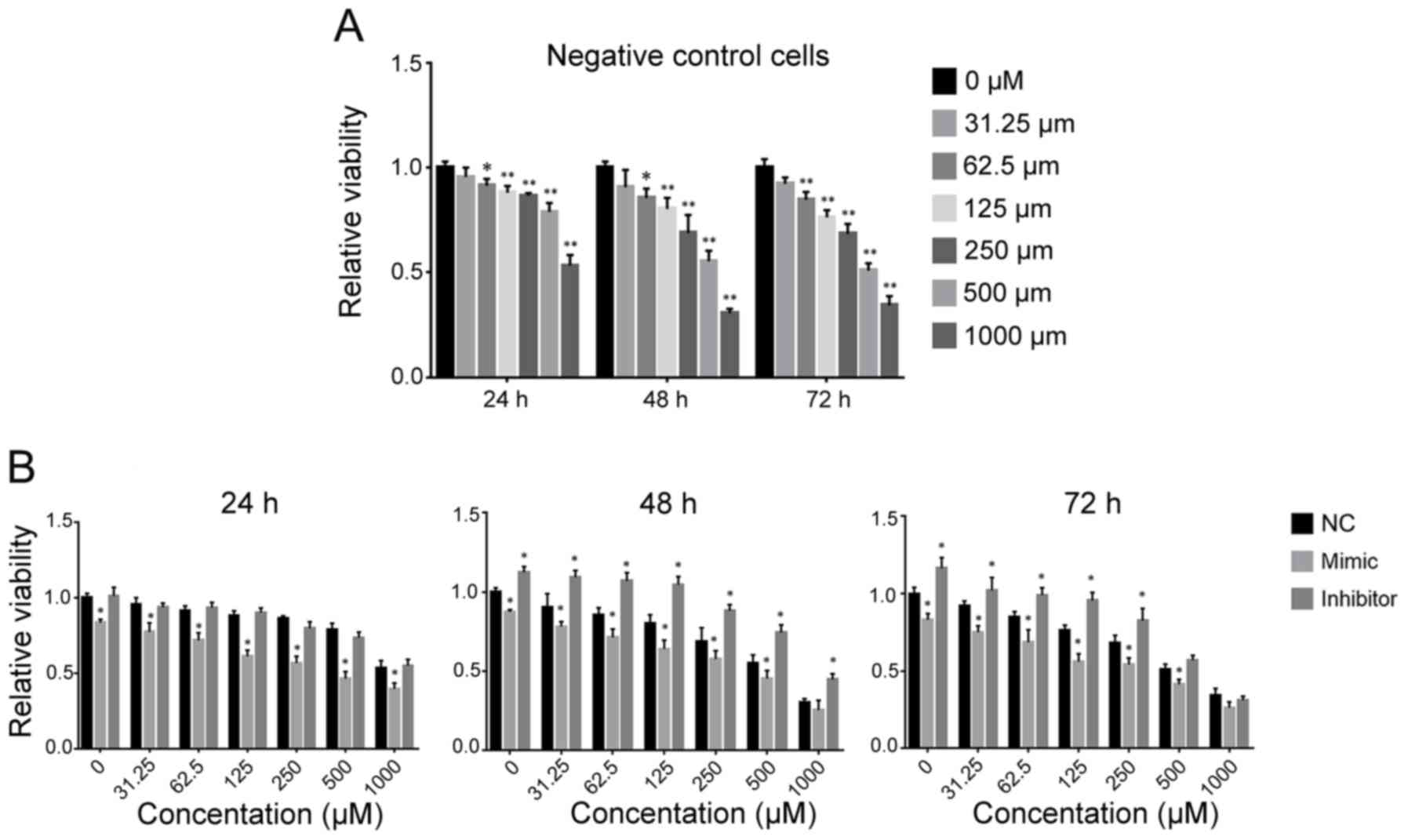
miR-9 enhances TMZ-induced inhibition
of cell viability
When U251 cells were treated with TMZ (NC group),
the cell viability decreased with increasing TMZ concentration.
When the concentration of TMZ was >62.5 µM, cell viability was
significantly decreased compared with the 0 µM group (Fig. 2A; 24 h, P=0.0469; 48 h, P=0.0322; 72
h, P=0.0013).
When miR-9 was overexpressed, the viability of
glioma cells was significantly lower compared with the negative
control group (NC) 24, 48 or 72 h following TMZ treatment (Fig. 2B). Conversely, when the miR-9
inhibitor was transfected, cell viability was significantly higher
compared with the NC group 48 or 72 h following TMZ treatment
(Fig. 2B). However, when TMZ
concentration was >500 µM, there was no significant difference
in cell viability compared to the NC group (500 µM at 72 h,
inhibitor group; 1,000 µM at 72 h, inhibitor group and mimic group;
1,000 µM at 48 h, mimic group).
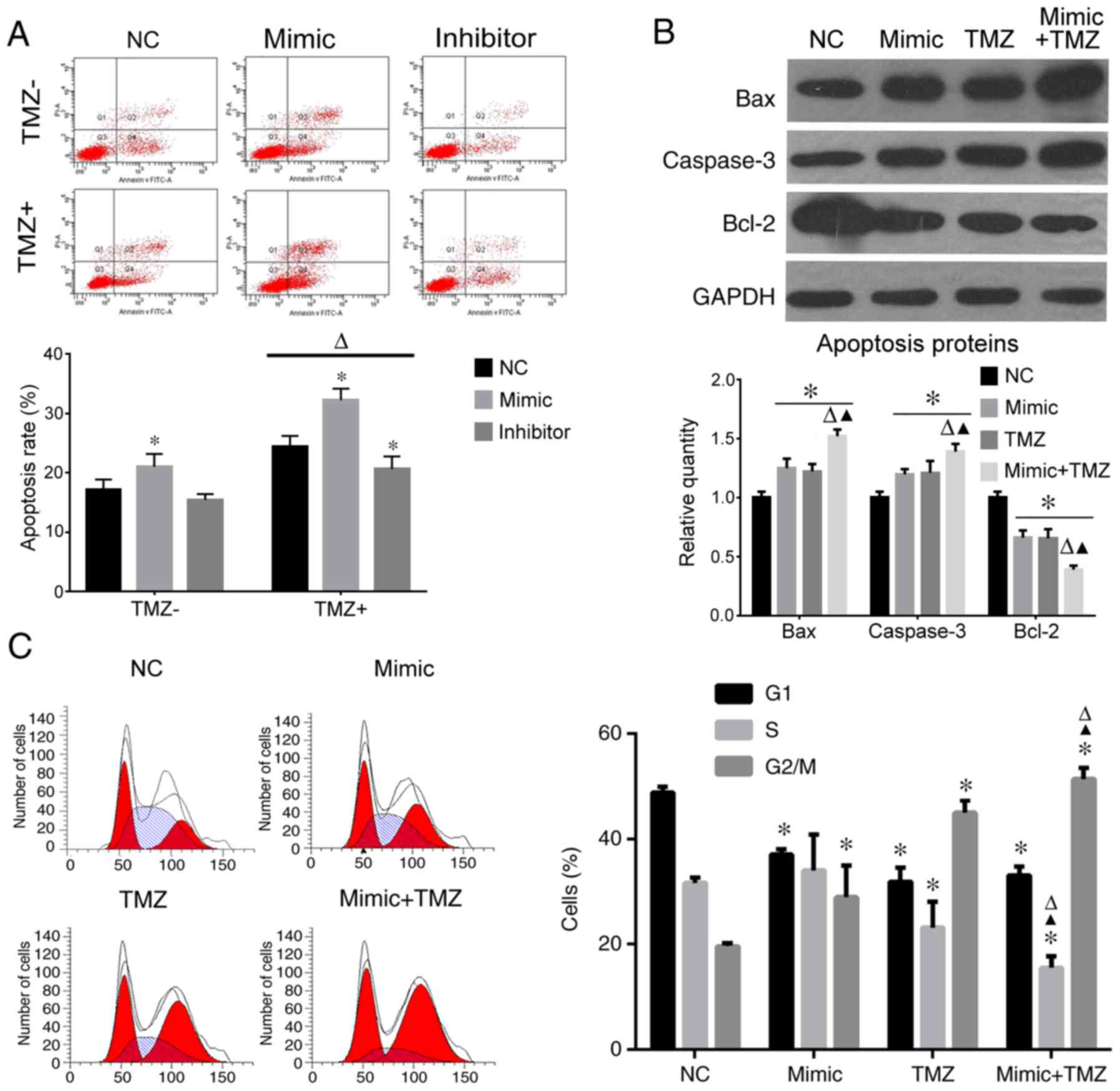
miR-9 overexpression increases the
apoptotic rate and aggravates G2/M stage arrest induced by TMZ
Cell cycle and apoptotic rate were also analyzed.
When treated with TMZ, the apoptotic rate of U251 cells increased
significantly compared with the groups without TMZ (Fig. 3A). In miR-9 mimic-transfected cells,
the apoptotic rate increased, whereas in the inhibitor-transfected
cells, the apoptotic rate reduced (Fig.
3A). In cells co-treated with TMZ and miR-9 mimic the apoptotic
rate increased significantly compared with miR-9 alone.
When cells were treated with miR-9 or TMZ, caspase-3
and Bax expression increased while Bcl-2 decreased compared to the
NC group. When co-treated with miR-9 and TMZ, Bax and caspase-3
protein expression levels markedly increased compared with miR-9-
or TMZ-only treated cells; Bcl-2 expression further decreased
compared to the miR-9 or TMZ groups (Fig. 3B). Furthermore, TMZ or miR-9
significantly induced G2/M stage arrest compared with the NC group,
and when the treatments were combined, the rate of G2/M stage
increased significantly compared to the miR-9 or TMZ groups
(Fig. 3C).
miR-9 overexpression inhibits TOPO II
expression via the NF-κB signaling pathway
Proteins responsible for drug resistance, including
TOPO II, MGMT and p170, were analyzed. The expression of TOPO II
was not significantly changed in inhibitor group (P=0.9977;
Fig. 4A and B) and was notably
downregulated in the mimic group compared to the NC group
(P=0.0433; Fig. 4A and B).
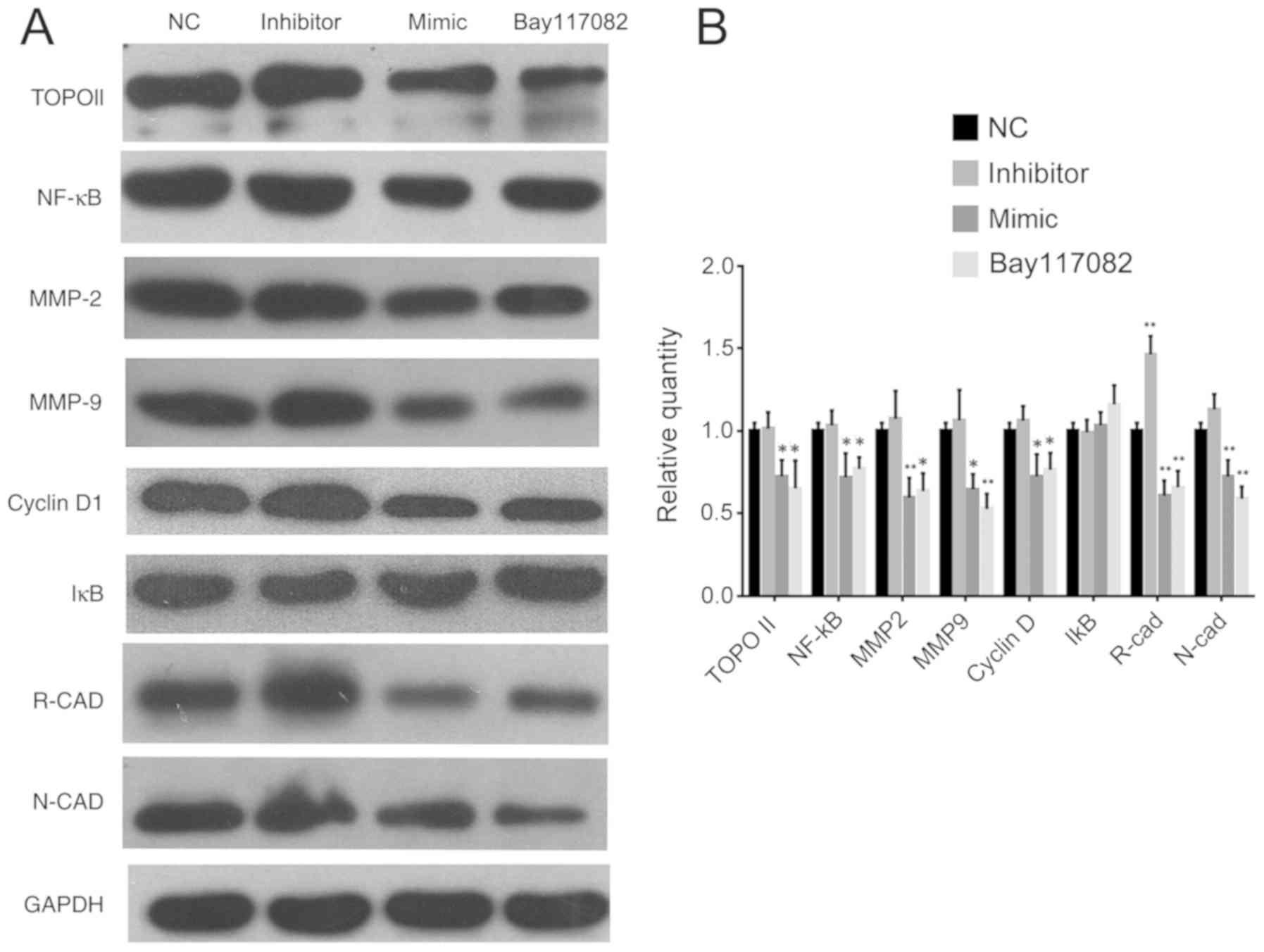 | Figure 4.Western blot analysis of NF-κB and
proteins associated with invasion. Experiments were repeated three
times. (A) western blot images and (B) densitometric analysis of
proteins; *P<0.05 vs. NC, **P<0.01 vs. NC. TOPO II,
topoisomerase II; NF-κB, nuclear factor κB; MMP, metalloproteinase;
IκB, inhibitor of κB; R-CAD, retinal cadherin; N-CAD, neural
cadherin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; NC,
negative control; Bay117082, inhibitor of the NF-κB signaling
pathway. Data for each protein were analyzed with one-way ANOVA and
Dunnett's post hoc test. |
The role of the NF-κB pathway in the modulation of
chemotherapy sensitivity was investigated using an inhibitor of
NF-κB activation, Bay117082. When miR-9 mimic or Bay117082 was
applied, NF-κB expression was suppressed, as well as the downstream
genes matrix metalloproteinase (MMP)-2, MMP-9 and cyclin D1
(Fig. 4). Proteins associated with
drug resistance and invasion, including TOPO II, retinal
(R)-cadherin and neural (N)-cadherin were also suppressed as NF-κB
signal pathway was blocked. When the inhibitor was transfected, the
expression of all proteins were not significantly altered except
for R-cad, which was significantly upregulated (Fig. 4). IκB was slightly upregulated when
Bay117082 was applied, but the change was not significant
(P=0.1005; Fig. 4), and it was not
altered in the mimic or inhibitor groups.
Discussion
miR-9 is a tissue-specific miRNA of the central
nervous system (CNS) that is expressed in embryonic stages and
serves vital roles during CNS development; it promotes neural stem
cell differentiation and prevents neurons from mutating into
gliocytes (12). In cerebral
development, miR-9 promotes the formation of cortex construction by
modulating the expression of forkhead box G1 (FoxG1)
(13). Furthermore, miR-9 also
serves important roles in spinal cord and peripheral neural system
development (14). Function
impairment of miR-9 results in severe malformation or deficiencies
of the nervous system (15). miR-9
also participates in tumor development, proliferation, metastasis
and invasion. For the tissue specificity of cancers, as well as the
different target genes modulated, miR-9 acts as a tumor suppressor
or a tumor-promoting factor depending on the situation. For
example, miR-9 promotes breast cancer metastasis and invasion by
inhibiting epithelial-cadherin expression (16) and enhances the invasion of
endometrial cancer by suppressing the expression of FoxO1
(17). However, miR-9 acts as tumor
suppressor in certain other cancers. For example, overexpression of
miR-9 in gastric adenoma, which expresses low levels of miR-9,
stimulated the expression of phosphatase and tensin homolog and
induced apoptosis, inhibiting proliferation, differentiation and
invasion (18). In ovarian cancer,
miR-9 inhibited tumor progression by suppressing the expression of
fibroblast growth factor, BCL-2 and B-Raf proto-oncogene,
serine/threonine kinase (19).
As miR-9 is specifically expressed in primary tumors
of the nervous system, it may be regarded as a marker for
differentiation between primary and metastatic tumors, tumor
progression and prognosis for patients with glioma (20). The functions of miR-9 in glioma may
be complicated and need further investigation, as its target genes
include both tumor suppressors and tumor promoters (21,22). Our
previous study demonstrated that miR-9 inhibited glioma cell
proliferation, metastasis and vasculogenic mimicry both in
vitro and in vivo through the suppression of stathmin
expression (9). When
stathmin-targeted small interfering (si)RNA was applied,
chemotherapeutic sensitivity of glioma cells to TMZ was enhanced
(10). miR-9 overexpression induced
G2/M stage arrest; the same effect was observed when TMZ and
stathmin-targeted siRNA were applied (23). Therefore, stathmin is involved in TMZ
chemotherapeutic sensitivity. In the present study, miR-9 applied
together with TMZ induced more substantial apoptosis and inhibition
of viability by suppressing TOPO II expression. However, as miR-9
is an upstream modulating factor, it may enhance chemotherapy
sensitivity through the regulation of a drug resistance gene,
TOPOII, as the current study showed.
TOPO II, which is located on chromosome 17,
is involved in DNA repair and replication, as well as chromosome
segregation and replication (24).
TOPO II is an essential nuclear enzyme that induces repair and
replication of DNA by protecting the double helix, insuring
stability and genomic integrity when DNA faces physical or chemical
damage (24). As opposed to MGMT and
excision repair associated protein ERCC that are abundant in normal
tissue, TOPO II levels are higher in glioma tissue, correlating
with an increased glioma grade (3).
TOPO II may stimulate and promote tumor growth and metastasis and
inhibit apoptosis, and they are involved in maintaining the glioma
stem cell character (25). Silencing
of TOPO II expression resulted in increased apoptosis and cell
cycle arrest at G0/G1 (26). TOPO II
is essential for TMZ resistance, as glioma cells resistant to TMZ
have higher TOPO II expression (27). Downregulation of TOPO II expression
by overexpression of leucine-rich repeats and immunoglobulin-like
domains protein-1 in U251 cells resulted in hypersensitivity to TMZ
(28). TOPO II may be a good target
for glioma chemotherapy, as the strategy to interfere and generate
enzyme-mediated DNA damage is proven to be effective for cancer
chemotherapy (24). Results from the
present study revealed that miR-9 suppressed TOPO II expression and
induced apoptosis and sensitization to TMZ in glioma cells.
NF-κB serves an essential role in cancer development
and is a direct target gene of miR-9 (16,18). The
role of NF-κB in the regulation of chemotherapy sensitivity
mediated by miR-9 has been studied. MMPs and cadherins, important
for glioma metastasis, also serve vital roles in drug resistance
(3). The present results
demonstrated that when miR-9 was overexpressed or a signaling
pathway inhibitor was applied, NF-κB expression was suppressed, and
the expression levels of proteins regulated by NF-κB were also
lower, including MMP-2, MMP-9, N-cadherin and R-cadherin. However,
the expression of inhibitor of κB (IκB) had no significant change,
which may be due to miR-9 potentially inhibiting NF-κB
independently of IκB, and Bay110782 inhibiting IκB phosphorylation
only. NF-κB also has strong interactions with TOPO II, as a
previous study reported that some chemotherapy drugs suppressed
TOPO II activity through the downregulation of NF-κB activity to
achieve cell toxicity (29). TOPO II
is essential for NF-κB activation in mitoxantrone-induced apoptosis
(30), whereas miR-106a silencing
modulated TOPO II and glutathione S-transferase π expression by
inhibition of NF-κB activation and AKT expression (31). Results from the present study
revealed that when the NF-κB signaling pathway was blocked, TOPO II
expression was downregulated. These results provided further
evidence for TMZ sensitization by miR-9, and the NF-κB signaling
pathway serves an important role in that regulation.
Further elucidation of the mechanisms of tumor
chemotherapy resistance may provide more precise and effective
anticancer therapies. Results from the present study revealed that
miR-9 enhanced chemotherapeutic sensitivity of glioma to TMZ by
suppressing TOPO II via the NF-κB signaling pathway, which
suggested that miR-9 may be used as a novel therapeutic target for
glioma treatment.
Acknowledgements
The authors would like to thank Miss Shaohong Fang
and Mr Jiangtian Tian from the Key Laboratory of Myocardial
Ischemia Mechanism and Treatment Ministry (Harbin, China) for
laboratory support.
Funding
The present study was supported by grants from
Heilongjiang Education Funds (no. 12511304), Heilongjiang Health
and the Family Planning Commission Project (no. 2014-377) and
Heilongjiang Postdoctoral scientific research development fund (no.
LBH-Q17125).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QL performed cell incubation and transfection. YC
performed the western blot and PCR. LM performed the cell cycle and
cell apoptosis assay. YS designed the experiment, and was a major
contributor in writing the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Komori T: The 2016 WHO Classification of
tumors of central nervous system: The major point of revision.
Neurol Med Chir. 57:301–311. 2017. View Article : Google Scholar
|
|
2
|
Chung AS, Wu X, Zhuang G, Ngu H, Kasman I,
Zhang J, Vernes JM, Jiang Z, Meng YG, Peale FV, et al: An
interleukin-17-mediated paracrine network promotes tumor resistance
to anti-angiogenic therapy. Nat Med. 19:1114–1123. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhao Y, Xue Y, Zhang Q, Wang K, Yin J and
Lou M: Transcriptional expression of glioma chemotherapy drugs
associated marker molecules in gliomas and normal brain tissues.
Cancer Biomark. 13:59–66. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hermisson M, Klumpp A, Wick W, Wischhusen
J, Nagel G, Roos W, Kaina B and Weller M: O6-methylguanine DNA
methyltransferase and p53 status predict temozolomide sensitivity
in human malignant glioma cells. J Neurochem. 96:766–776. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Roos WP, Batista LF, Naumann SC, Wick W,
Weller M, Menck CF and Kaina B: Apoptosis in malignant glioma cells
triggered by the temozolomide-induced DNA lesion O6-methylguanine.
Oncogene. 26:186–197. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Omura N, Li CP, Li A, Hong SM, Walter K,
Jimeno A, Hidalgo M and Goggins M: Genome-wide profiling of
methylated promoters in pancreatic adenocarcinoma. Cancer Biol
Ther. 7:1146–1156. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hildebrandt MA, Gu J, Lin J, Ye Y, Tan W,
Tamboli P, Wood CG and Wu X: Hsa-miR-9 methylation status is
associated with cancer development and metastatic recurrence in
patients with clear cell renal cell carcinoma. Oncogene.
29:5724–5728. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song Y, Mu L, Han X, Li Q, Dong B, Li H
and Liu X: MicroRNA-9 inhibits vasculogenic mimicry of glioma cell
lines by suppressing Stathmin expression. J Neurooncol.
115:381–390. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song Y, Mu L, Han X, Liu X and Fu S: siRNA
targeting stathmin inhibits invasion and enhances chemotherapy
sensitivity of stem cells derived from glioma cell lines. Acta
Biochim Biophys Sin (Shanghai). 46:1034–1040. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Krichevsky AM, Sonntag KC, Isacson O and
Kosik KS: Specific microRNAs modulate embryonic stem cell-derived
neurogenesis. Stem Cells. 24:857–864. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shibata M, Kurokawa D, Nakao H, Ohmura T
and Aizawa S: MicroRNA-9 modulates cajal-retzius cell
differentiation by suppressing Foxg1 expression in mouse medial
pallium. J Neurosci. 41:10415–10421. 2008. View Article : Google Scholar
|
|
14
|
Otaegi G, Pollock A, Hong J and Sun T:
MicroRNA miR-9 modifies motor neuron columns by a tuning regulation
of FoxP1 levels in developing spinal cords. J Neurosci. 31:809–818.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bonev B, Pisco A and Papalopulu N:
MicroRNA-9 reveals regional diversity of neural progenitors along
the anterior-posterior axis. Dev Cell. 20:19–32. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang J, Gu Z, Ni P, Qiao Y, Chen C, Liu X,
Lin J, Chen N and Fan Q: NF-kappaB P50/P65 hetero-dimer mediates
differential regulation of CD166/ALCAM expression via interaction
with micoRNA-9 after serum deprivation, providing evidence for a
novel negative auto-regulatory loop. Nucleic Acids Res.
39:6440–6455. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Myatt S, Wang J, Monteiro L, Christian M,
Ho KK, Fusi L, Dina RE, Brosens JJ, Chaem-Maghami S and Lam EW:
Definition of microRNAs that repression expression of the tumor
suppressor gene FOXO1 in endometrial cancer. Cancer Res.
70:367–377. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wan H, Guo L, Liu T, Liu M, Liu X and Tang
H: Regulation of the transcription factor NF-κB1 by microRNA-9 in
human gastric adenocarcinoma. Mole Cancer. 9:162010. View Article : Google Scholar
|
|
19
|
Laios A, O'Toole S, Flavin R, Martin C,
Kelly L, Ring M, Finn SP, Barrett C, Loda M, Gleeson N, et al:
Potential role of miR-9 and miR-223 in recurrent ovarian cancer.
Mole Cancer. 7:352008. View Article : Google Scholar
|
|
20
|
Nass D, Rosenwald S, Meiri E, Gilad S,
Tabibian-Keissar H, Schlosberg A, Kuker H, Sion-Vardy N, Tobar A,
Kharenko O, et al: MiR-92b and miR-9/9* are specifically expressed
in brain primary tumors and can be used to differentiate primary
from metastatic brain tumors. Brain Pathol. 19:375–383. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schraivogel D, Weimann L, Beier D,
Tabatabai G, Eichner A, Zhu JY, Anton M, Sixt M, Weller M, Beier
CP, et al: CAMTA1 is a novel tumour suppressor regulated by
miR-9/9* in glioblastoma stem cells. EMBOJ. 30:4309–4322. 2011.
View Article : Google Scholar
|
|
22
|
Ben-Hamo R and Efroni S: Gene expression
and network-based analysis reveals a novel role for hsa-miR-9 and
drug control over the p38 network in glioblastoma multiforme
progression. Genome Med. 3:772011. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Newlands ES, Stevens MF, Wedge SR,
Wheelhouse RT and Brock C: Temozolomide: A review of its discovery,
chemical properties, pre-clinical development and clinical trials.
Cancer Treat Rev. 23:35–61. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nitiss JL: Targeting DNA topoisomerase II
in cancer chemotherapy. Nat Rev Cancer. 9:338–350. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Galli R, Binda E, Orfanelli U, Cipelletti
B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F and Vescovi
A: Isolation and characterization of tumorigenic, stem-like neural
precursors from human glioblastoma. Cancer Res. 64:7011–7021. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hong Y, Sang M, Shang C, Xue YX and Liu
YH: Quantitative analysis of topoisomerase II alpha and evaluation
of its effects on cell proliferation and apoptosis in glioblastoma
cancer stem cells. Neurosci Lett. 518:138–143. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Arivazhagan A, Kumar DM, Sagar V, Patric
IR, Sridevi S, Thota B, Srividya MR, Prasanna K, Thennarasu K,
Mondal N, et al: Higher topoisomerase 2 alpha gene transcript
levels predict better prognosis in GBM patients receiving
temozolomide chemotherapy: Identification of temozolomide as a
TOP2A inhibitor. J Neurooncol. 107:289–297. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qi XC, Xie DJ, Yang QF, Wang YR, Zhu YX,
Qian C and Yang SX: LRG1 dictates the chemo-sensitivity of
temzolomide in U251 glioblastoma cells via downregulation of
EGFR/topoisomerase-2/bcl-2. Biochem Biophys Res Commun.
437:565–572. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wong HY, Tsai KD, Liu YH, Yang SM, Chen
TW, Cherng J, Chou KS, Chang CM, Yao BT and Cherng JM: Cinnamomum
verum component 2-Methoxycinnamaldehyde: A novel anticancer agent
with both anti-topoisomerase I and II activities in human lung
adenocarcinoma A549 cells in vitro and in vivo. Phytother Res.
30:331–340. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Boland MP, Fitzgerald KA and O'Neill LA:
Topoisomerase II required for mitoxantrone to signal nuclear factor
κB activation in HL60 cells. J Biol Chem. 275:25231–25238. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Q, Wang Z, Chu L, Li X, Kan P, Xin X,
Zhu Y and Yang P: The effects and molecular mechanisms of MiR-106a
in multidrug resistance reversal in human glioma U87/DDP and U251/G
cell lines. PLoS One. 10:e01254732015. View Article : Google Scholar : PubMed/NCBI
|