Introduction
Colorectal cancer (CRC) is a common malignant
disease which is a profound public health burden (1). The risk of CRC incidence and mortality
in China is enhanced by tobacco smoking, alcohol consumption,
obesity, physical inactivity, low fruit and vegetable consumption
and the high intake of red and processed meat (2). A recent epidemiological study reported
that the incidence of CRC has increased over the past few decades
(3). Despite improvements in the
available treatments, the 5-year survival rate in China remains
low, which is attributed to the majority of patients being
diagnosed at a late disease stage (4). Numerous cases of CRC present with
metastasis of the peripheral lymph nodes and organs (5).
Epithelial-mesenchymal transition (EMT) is
characterized by the loss of epithelial cell characteristics
through the transition to a more malignant, mesenchymal cell
phenotype (6). The loss of cell
polarity combined with the breakdown of tight intercellular
junctions results in a high risk of tumor metastasis (7). Recent evidence has demonstrated that
aberrant EMT activation in CRC is closely associated with
carcinogenesis and tumor progression (8,9).
Following the onset of EMT, E-cadherin is translated into
N-cadherin, which is expressed in interstitial cells. Consequently
the deletion and downregulation of E-cadherin is considered to be
an important marker of EMT (10).
Vimentin is also an important mesenchymal cell marker of EMT, which
serves an important role in maintaining interstitial cell
characteristics. Studies have confirmed that the increased
expression of Vimentin is also associated with tumor invasion and
metastasis (10).
In a previous study, tumor necrosis factor
receptor-associated factor 6 (TRAF6) was identified as a prognostic
factor in CRC (11). The prominent
expression of TRAF6 has also been observed in other human
malignancies, such as lung and gastric cancer, nasopharyngeal
carcinoma and breast cancer (12–15).
TRAF6 belongs to the TRAF family, and acts as an adaptor in the
signaling of channels induced by the TNFR. An increasing number of
studies have indicated that TRAF6 promotes oncogenesis by
attenuating cell apoptosis and accelerating proliferation and
invasion in tumor lesions (14,16).
Notably, TRAF6 is a target gene of miR-124 and the latter was
reported to be involved in EMT in multiple malignant diseases
(17,18). However, the role and relationship
between miR-124 and TRAF6 in CRC remains unclear.
In the present study miR-124 was downregulated in
CRC tissues, and a decrease in the level of miR-124 was associated
with the specific adverse features of CRC, indicating an increased
risk of poor overall survival. The data also demonstrated that
TRAF6 was a target gene for miR-124 and is potentially involved in
EMT. Taken together these results suggest that miR-124 may exhibit
a tumor-suppressive role by regulating TRAF6 expression in CRC.
Materials and methods
Patients and follow up
A total of 80 patients with a median age of 64.5
years (range, 40–72) who underwent surgery at the first
presentation of CRC at the Central Hospital of Wuhan between
January 2008 and December 2012 were selected for inclusion in the
present study. None of the patients had received additional
treatment prior to surgical intervention. Patients' clinical data
are presented in Table I. CRC was
classified according to the American Joint Committee on Cancer
(19) staging system. The endpoint
of this research was described as overall survival. All patients
provided written informed consent to participate prior to surgery.
Ethical approval was given by the medical ethics committee of the
Central Hospital of Wuhan Tongji Medical College, Huazhong
University of Science and Technology. All procedures performed in
this study involving human participants were conducted in
accordance with Chinese ethical standards and the 2008 Declaration
of Helsinki.
 | Table I.Association between miR-124
expression level and the clinicopathological parameters of patients
with colorectal cancer. |
Table I.
Association between miR-124
expression level and the clinicopathological parameters of patients
with colorectal cancer.
|
| miR-124
expression |
|
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Patients, n=80 | Low | High | P-value |
|---|
| Age, years |
|
|
| 0.794 |
|
>65 | 40 | 30 | 10 |
|
|
≤65 | 40 | 31 | 9 |
|
| Sex |
|
|
| 0.657 |
|
Male | 47 | 35 | 12 |
|
|
Female | 33 | 26 | 7 |
|
| Size, cm |
|
|
| 0.273 |
|
>5 | 34 | 28 | 6 |
|
| ≤5 | 46 | 33 | 13 |
|
| Tumor grade |
|
|
| 0.046a |
| Well +
moderate | 57 | 40 | 17 |
|
|
Poor | 23 | 21 | 2 |
|
| Lymph node
status |
|
|
| 0.017a |
|
<1 | 52 | 44 | 8 |
|
| ≥1 | 28 | 17 | 11 |
|
| Distant
metastasis |
|
|
| 0.529 |
|
Yes | 7 | 6 | 1 |
|
| No | 73 | 55 | 18 |
|
| TNM |
|
|
| 0.597 |
| I +
II | 55 | 41 | 14 |
|
| III +
IV | 25 | 20 | 5 |
|
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells and tissues using
TRIzol® reagent (Wuhan Guge Biotechnology Co., Ltd.),
according to the manufacturer's protocol. The Thermo Nano Drop 2000
(Nano Drop Technologies; Thermo Fisher Scientific, Inc.) was used
to determine the concentration and purity of the RNA. The primer
sequences were as follows: Hsa-miR-124-3p forward,
5′-ACACTCCAGCTGGGTAAGGCACGCGGTG-3′, and reverse,
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGGCATTCA-3′. U6 forward,
5′-CTCGCTTCGGCAGCACA-3′, and reverse, 5′-AAACGCTTCACGAATTTGCGT-3′.
RT-qPCR was performed using the iTaq Universal SYBR®
Green Supermix and CFX96 real-time PCR system (Bio-Rad
Laboratories, Inc.). The primers are listed in Table II. RT-qPCR was performed in line
with a commonly used method described previously (20). Primers for U6 and miR-124 were
synthesized and purified by Guangzhou Ribo Bio Co. Ltd. U6 was used
as the endogenous control. Relative fold expressions were
calculated with the comparative quantification cycle
(2−ΔΔCq) method (21).
The expression levels of miR-124 were compared between cancerous
and para-cancerous tissues, with the cancer/para-cancerous ratio as
the ordinate; a ratio >1 indicated a high miR-124 expression
level, and a ratio <1 indicated that miRNA-124 expression was
downregulated.
 | Table II.Primers for gene expression using
reverse transcription-quantitative PCR. |
Table II.
Primers for gene expression using
reverse transcription-quantitative PCR.
| Gene | Primer sequence
(5′-3′) |
|---|
|
hsa-miR-124-3p-RT |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGGCATTCA |
|
hsa-miR-124-3p-S |
ACACTCCAGCTGGGTAAGGCACGCGGTG |
| U6-S |
CTCGCTTCGGCAGCACA |
| U6-A |
AACGCTTCACGAATTTGCGT |
Immunohistochemistry (IHC)
The pathological examination of samples was
performed at the Central Hospital of Wuhan, Tongji Medical College,
Huazhong University of Science and Technology using a two-step
method (22). Tissues were fixed
using 4% paraformaldehyde at room temperature for 24 h.
Paraffin-embedded tissue sections were cut into 5 µm-thick sections
and deparaffinized and rehydrated with xylene and a graded alcohol
series (100, 95, 85 and 75%) at room temperature. Sections were
washed with PBS three times (3 min each time). Antigen retrieval
was performed with 0.01 M citrate buffer at 98°C for 10 min and
cooled to 37°C. The sections were washed three times with PBS for 3
min. Subsequently, 50 µl of 3% hydrogen dioxide solution was added
to each section and incubated at room temperature for 10 min,
followed by washing with PBS. For heat-induced antigen retrieval,
the sections were treated with EDTA buffer, autoclaved and returned
to room temperature. Antibodies against the target proteins were as
follows: TRAF6 (1:50; cat no. ab58369; Abcam), E-cadherin (1:100;
cat. no. MA106302; Thermo Fisher Scientific, Inc.) and Vimentin
(1:100; cat. no. MA106908; Thermo Fisher Scientific, Inc.). IHC
staining was performed according to the following standard
procedure (23) to evaluate TRAF6,
E-cadherin and Vimentin protein expression in CRC tissues.
Cell cultures and transfection
The human colorectal cancer cell line SW480 was
obtained from the China Center for Type Culture Collection. Cells
were cultured in DMEM supplemented with 10% FBS (Sigma-Aldrich,
Merck KGaA), 100 U/ml penicillin (HyClone; GE Health care Life
Sciences), and 100 µg/ml streptomycin (HyClone; GE Health care Life
Sciences). miR-124 mimics and inhibitor were purchased from Gene
Copoeia, Inc. and transfected with a concentration of 50 nM/well
into cells with Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
The cells were harvested 2 days after transfection for further
experimentation.
Western blotting
RIPA lysis buffer (Wuhan Sanying Biotechnology) was
used to extract total cell protein. Proteins (40 µg) were then
separated by 10% SDS-PAGE (Wuhan Guge Biotechnology Co., Ltd.) and
transferred to PVDF membranes. After blocking with 5% non-fat milk
for 1 h at room temperature, the membranes were incubated with the
indicated antibodies overnight at 4°C. The membranes were incubated
with anti-TRAF6 (1:1,000; cat. no. 66498-1-Ig; Protein Tech Group,
Inc.) or GAPDH (1:1,000; cat. no. 10494-1-AP; Protein Tech Group,
Inc.) antibodies overnight at 4°C and subsequently incubated with
matched secondary antibodies (Wuhan Guge Biotechnology Co., Ltd.).
An enhanced chemiluminescence detection system (Wuhan Guge
Biotechnology Co., Ltd.) was used to detect the bands. ImageJ
software (National Institutes of Health Bethesda, MD, USA) was used
to measure the band density. GAPDH was used as a loading
control.
Luciferase reporter assays
The relationship between the expression level of
miR-124 and TRAF6 in SW480 cells was determined by luciferase
reporter assay according to the protocol of a previous study
(21). After analyzing the
biochemical information database, miR-124 was revealed to act on
the 3′-untranslated region (UTR) of TRAF6 and influence its
biological activity. Briefly, the wild-type (WT) or mutant (Mt)
segments of the TRAF6 3 egment were amplified and cloned into
luciferase reporter plasmids (Sino Geno Max Co., Ltd, Beijing,
China) for subsequent experiments. Cells were seeded in 6-well
culture plates at a density of 1×105/ml. Following a
24-h incubation, when the cell confluence had reached 70%, cells
were co-transfected with the reporter plasmids and miR-124 mimics
or inhibitors at a concentration of 4 µg per/well using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). At 48 h post-transfection, the
Dual-Luciferase® Reporter Assay kit (Promega
Corporation) with a Renilla luciferase normalization control was
used to detect the fluorescence intensity of the cells, according
to the manufacturers' protocol. All the procedures were repeated ≥3
times.
Statistical analyses
Non-parametric analysis between groups was performed
using the Mann-Whitney U test, and the Spearman's rank correlation
coefficient was employed to examine the relationship between
miR-124 expression level and clinicopathological parameters.
Categorical variables were analyzed using χ2 tests for
univariate analysis. A paired Student's t-test was performed to
analyze the paired data. ANOVA and the Bonferroni correction
post-hoc test were applied in multiple comparison analysis. The
Kaplan-Meier method was used to draw survival curves, and the
differences were verified using the log-rank test. Whether a factor
was an independent predictor of CRC prognosis was determined by
Cox-multivariate analyses. Statistical analyses were performed
using SPSS software (version 19.0; IBM Corp.). P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-124 expression level is
significantly lower in tumor tissues than in adjacent
para-cancerous tissues
To examine the expression level of miR-124 in
colorectal cancer tissues, RT-qPCR was performed to compare
expression levels in 80 pairs of CRC and adjacent non-cancerous
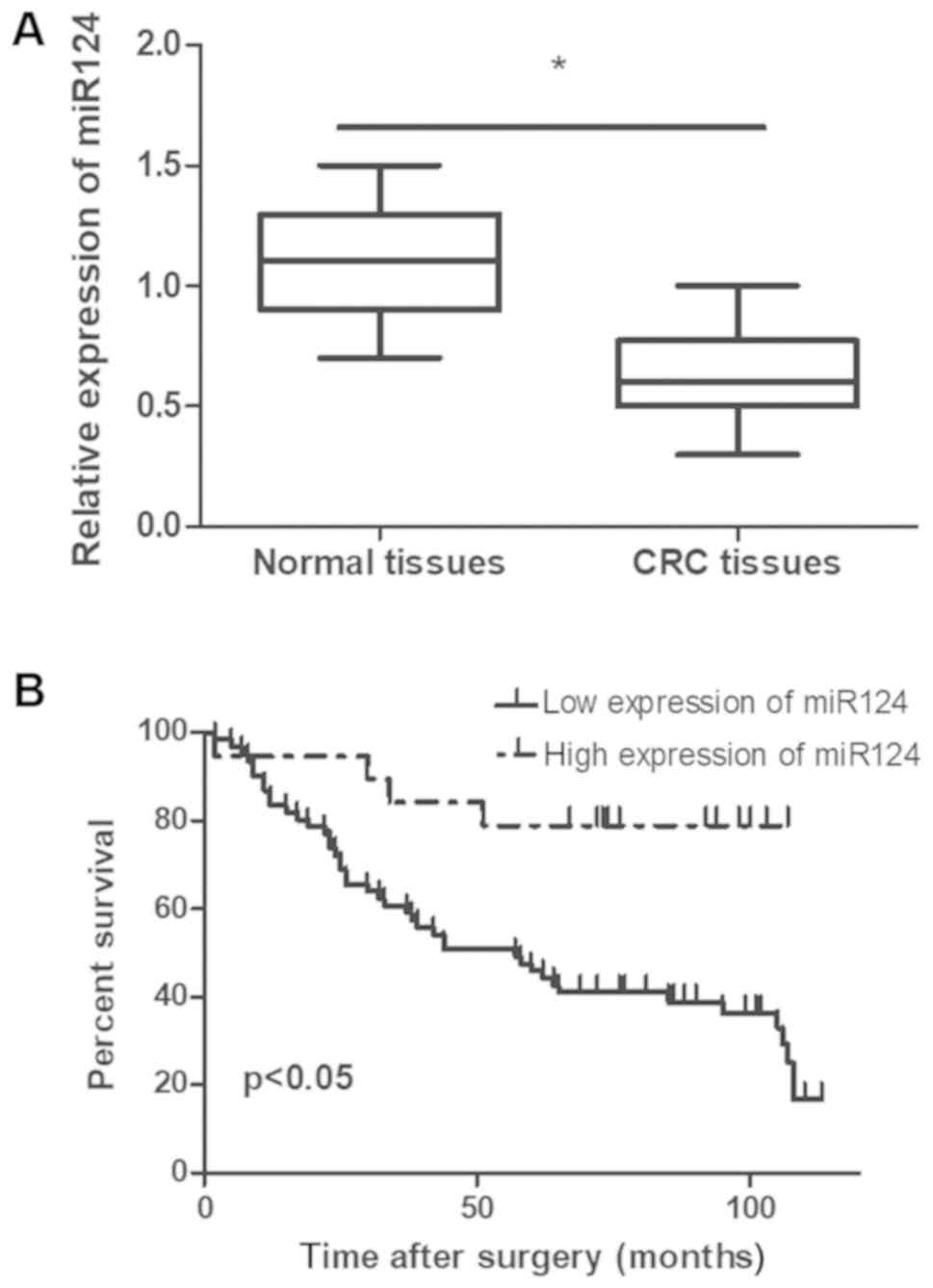
tissues. As illustrated in Fig. 1A,
miR-124 expression level was significantly down regulated in cancer
tissues compared with neighboring para-cancerous tissues
(P<0.05).
Relationship between miR-124
expression and clinical pathological parameters
Recent studies have suggested that clinical features
such as tumor size, pathological grade, TNM stage and lymphatic
metastasis are closely associated with patient prognosis (24,25). On
the basis of the data displayed in Table
I, patients that presented with a low miR-124 expression level
exhibited poor pathological differentiation (P=0.046) and an
increased risk of lymph node metastasis (P=0.017). No significant
associations were identified between miR-124 expression and sex
(P=0657), age (P=0.794), tumor size (P=0.273), distant metastasis
(P=0.597) or advanced TNM stage (P=0.529).
Correlation between miR-124 expression
level and CRC patient prognosis
The time between the date of surgical resection and
mortality or last patient contact was defined as overall survival.
Among the 80 patients with CRC, 47 (58.8%) died during follow-up as
a result of their malignancy. CRC patients with an elevated
expression level of miR-124 had a significantly longer survival
period. Additionally, the prognosis of patients with a low miR-124
expression level was poor compared with patients expressing high
levels of miR-124 (P<0.05; Fig.
1B). Cox regression multivariate analysis revealed that lymph
node status (HR, 0.240; 95% CI, 0.094–0.614; P=0.003), tumor
metastasis (HR, 0.269; 95% CI, 0.093–0.780; P=0.016), histological
grade (HR, 0.474; 95% CI, 0.243–0.927; P=0.029) and miR-124
expression (HR, 6.961; 95% CI, 2.174–22.294; P=0.001) were
independent predictive factors for the overall survival of patients
with CRC (Table III). Furthermore,
survival analyses revealed that patients with a low miR-124
expression level had a significantly poorer 5-year survival
(P<0.05; Fig. 1B). Taken
together, these data suggested that miR-124 may exhibit a
suppressive role in the development of CRC.
 | Table III.Cox multivariate regression analysis
of miR-124 expression, age, sex, depth of invasion, grade of
differentiation, lymph node status and stage in relation to overall
survival in patients with colon cancer. |
Table III.
Cox multivariate regression analysis
of miR-124 expression, age, sex, depth of invasion, grade of
differentiation, lymph node status and stage in relation to overall
survival in patients with colon cancer.
| Variable | Comparison | β | SE | HR | 95%CI | P-value |
|---|
| miR124
expression | High vs. Low | 1.940 | 0.594 | 6.961 | 2.174–22.294 | 0.001 |
| Age | ≤65 vs. >65 | 0.298 | 0.360 | 1.347 | 0.665–2.731 | 0.408 |
| Sex | Male vs.
Female | 0.366 | 0.919 | 1.422 | 0.238–8.743 | 0.690 |
| Size, cm | ≤5 vs. >5 | 0.281 | 0.908 | 0.755 | 0.127–4.477 | 0.757 |
| Tumor
metastasis | Positive vs.
Negative | 1.312 | 0.542 | 0.269 | 0.093–0.780 | 0.016 |
| Histologic
grade | Poor vs. Well,
Moderate | 0.746 | 0.342 | 0.474 | 0.243–0.927 | 0.029 |
| Lymph node
status | Positive vs.
Negative | 1.425 | 0.478 | 0.240 | 0.094–0.614 | 0.003 |
| TNM stage | I+II vs.
III+IV | 0.784 | 0.432 | 0.457 | 0.196–1.064 | 0.069 |
Aberrant expression levels of miR-124
and TRAF6 in CRC
TRAF6 expression confers poor prognosis for patients
with CRC (11), and has been
highlighted as a potential target protein of miR124 in TargetScan
(version 7.1; www.targetscan.org) following bioinformatics analysis.
To determine whether miR-124 functionally modulates TRAF6
expression in CRC, miR-124 and TRAF6 expression levels we measured
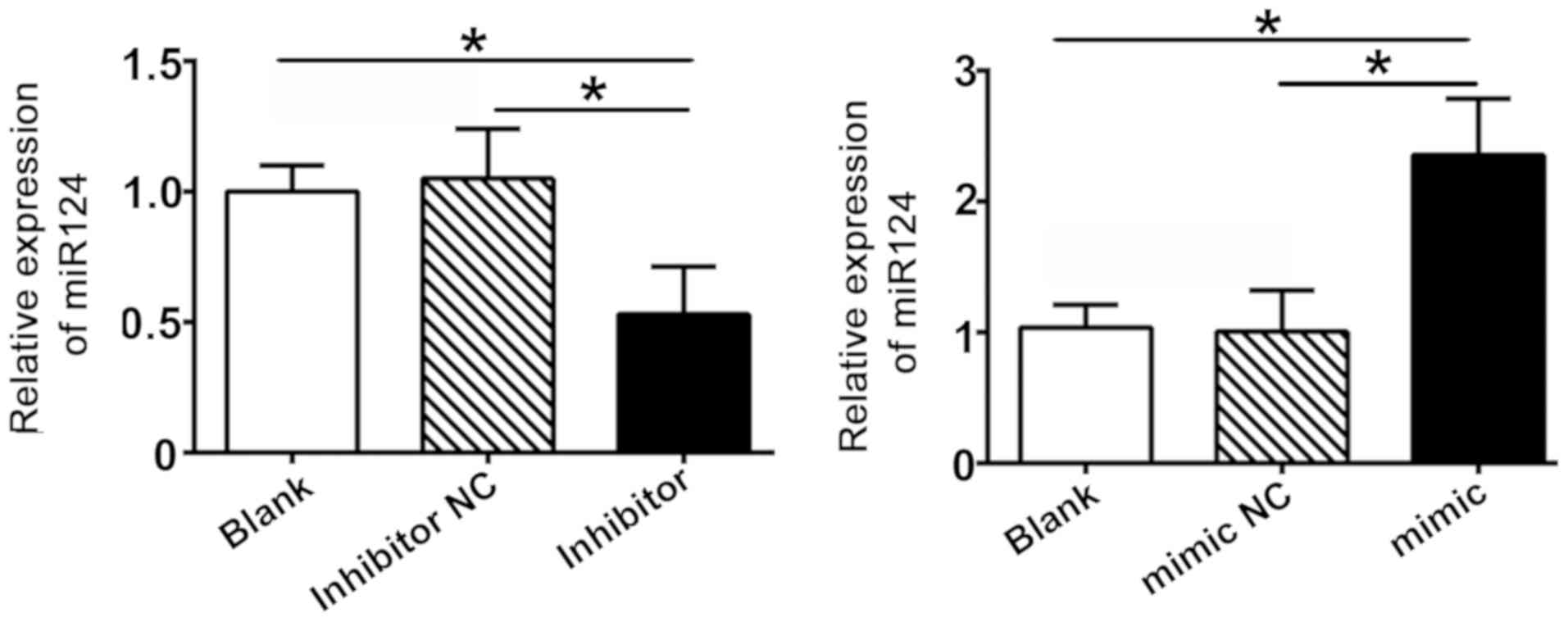
in SW480 cells. Cells transfected with an miR-124 inhibitor
displayed lower miR-124 expression levels compared with the
inhibitor control group, whereas the expression level of miR-124
was markedly upregulated in the miR124 mimic group (Fig. 2). The results revealed that the
transfection of SW480 cells with miR-124 constructs was successful.
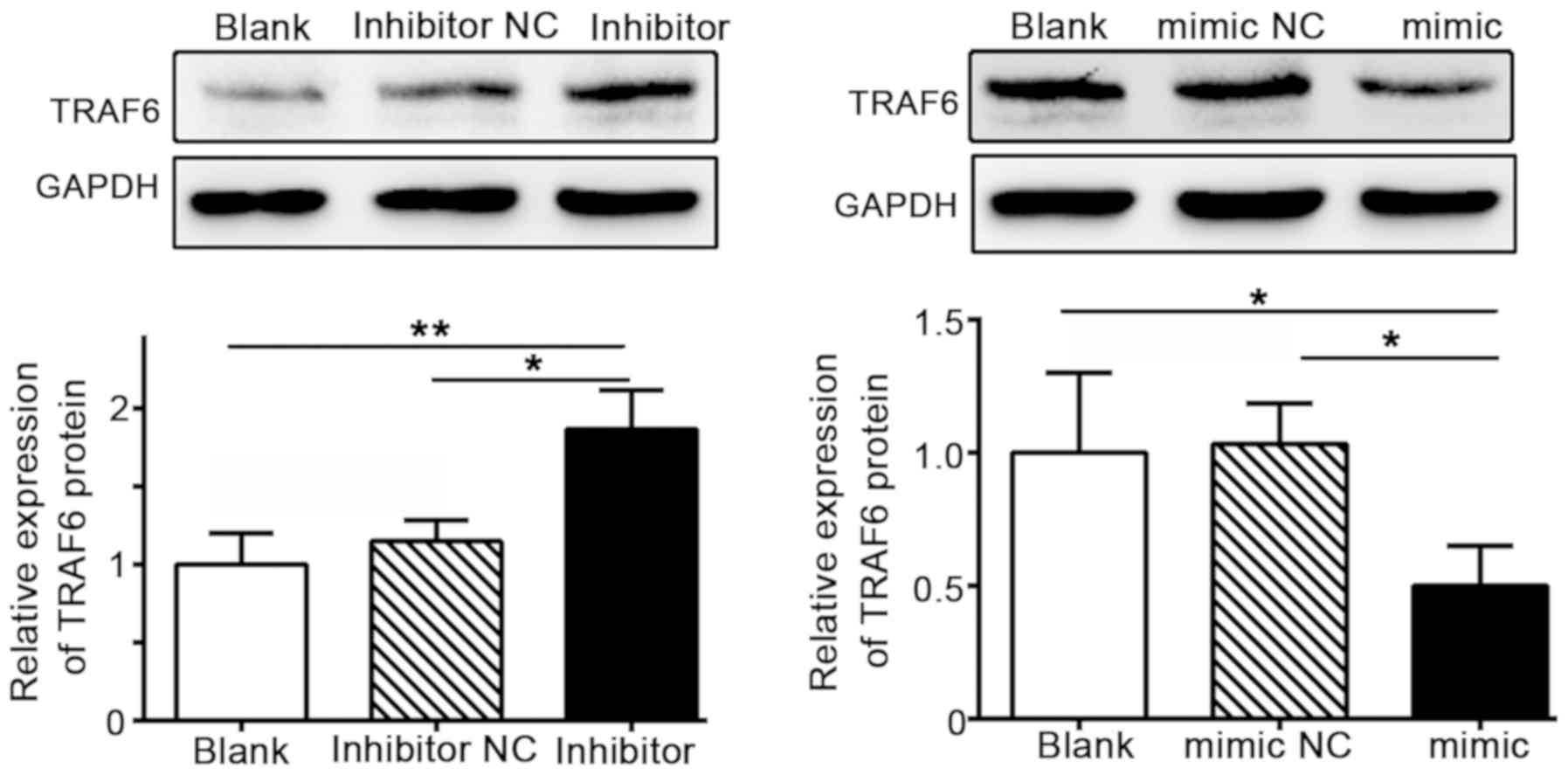
Further evaluation of TRAF6 expression in SW480 cells revealed that
TRAF6 expression levels were markedly increased in cells
transfected with the miR-124 inhibitor, but downregulated in the
miR124 mimic group (Fig. 3). These
findings demonstrated a negative association between the expression
levels of miR-124 and TRAF6.
miR-124 post-transcriptionally
regulates TRAF6 expression
In our previous study, TRAF6 was highly expressed in
CRC tissues, which significantly correlated with Dukes' staging,
degree of cell differentiation and lymphatic metastasis (11). The present study further investigated
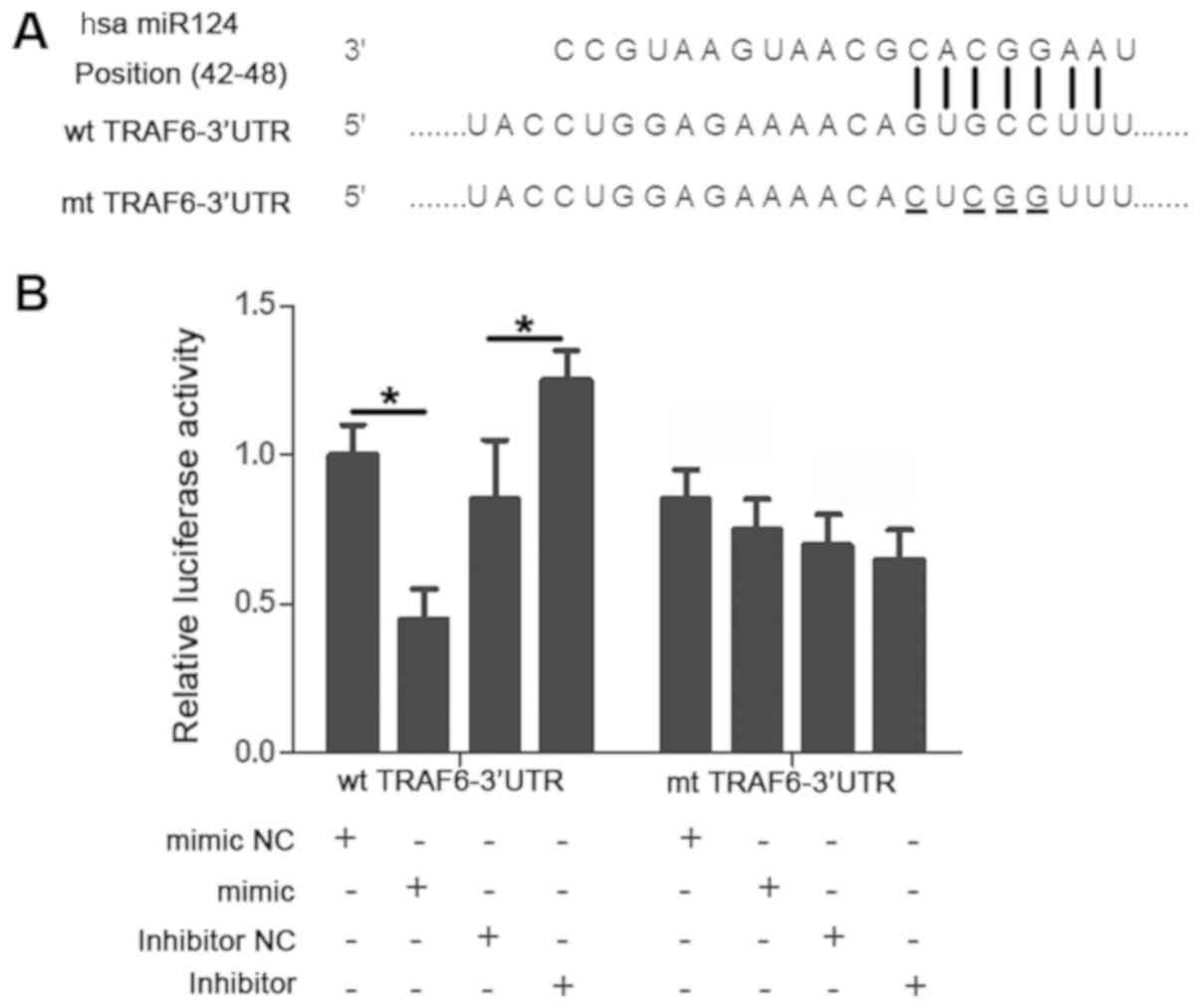
whether TRAF6 was a downstream target of miR-124. As illustrated in
Fig. 4A, a putative binding site for
miR-124 was identified in the 3-UTR of TRAF6. Subsequently, a
luciferase reporter assay was performed to investigate whether
miR-124 bound to this specific site. Upregulating the expression of
miR-124 significantly reduced the luciferase activity of the WT
TRAF6 3′-UTR (P<0.05) whereas downregulating miR124 expression
increased the luciferase activity of the WT 3′-UTR (P<0.05).
However, the altered miR-124 expression level did not substantially
impact the luciferase activity of the MutTRAF63′-UTR (P>0.05;
Fig. 4B).
miR-124 regulates TRAF6 expression and
EMT in CRC tissues
To further confirm the association between miR-124
and TRAF6, and to determine whether these molecules are involved in
EMT and tumor invasion, the association between the EMT-related
biomarkers E-cadherin and Vimentin and TRAF6 in CRC tissues was
analyzed via IHC. As illustrated in Fig.
5, the degree of TRAF6 (Fig. 5A)
and Vimentin staining (Fig. 5E) was
strong in tumors with low miR-124 expression; however, E-cadherin
staining was weak (Fig. 5C). In
addition, weak TRAF6 (Fig. 5B) and
Vimentin (Fig. 5F) staining was
observed in tumors with a high miR-124 expression, whereas
E-cadherin staining was strong (Fig.
5D). Spearman's correlation analysis demonstrated that miR-124
expression was significantly negatively correlated with that of
TRAF6 (r=−0.402; P<0.001) and Vimentin (r=−0.514; P<0.001),
yet positively correlated with E-cadherin expression (r=0.721;
P<0.001) in CRC tissues. These data indicated that miR-124
affects the metastasis of CRC by modulating EMT.
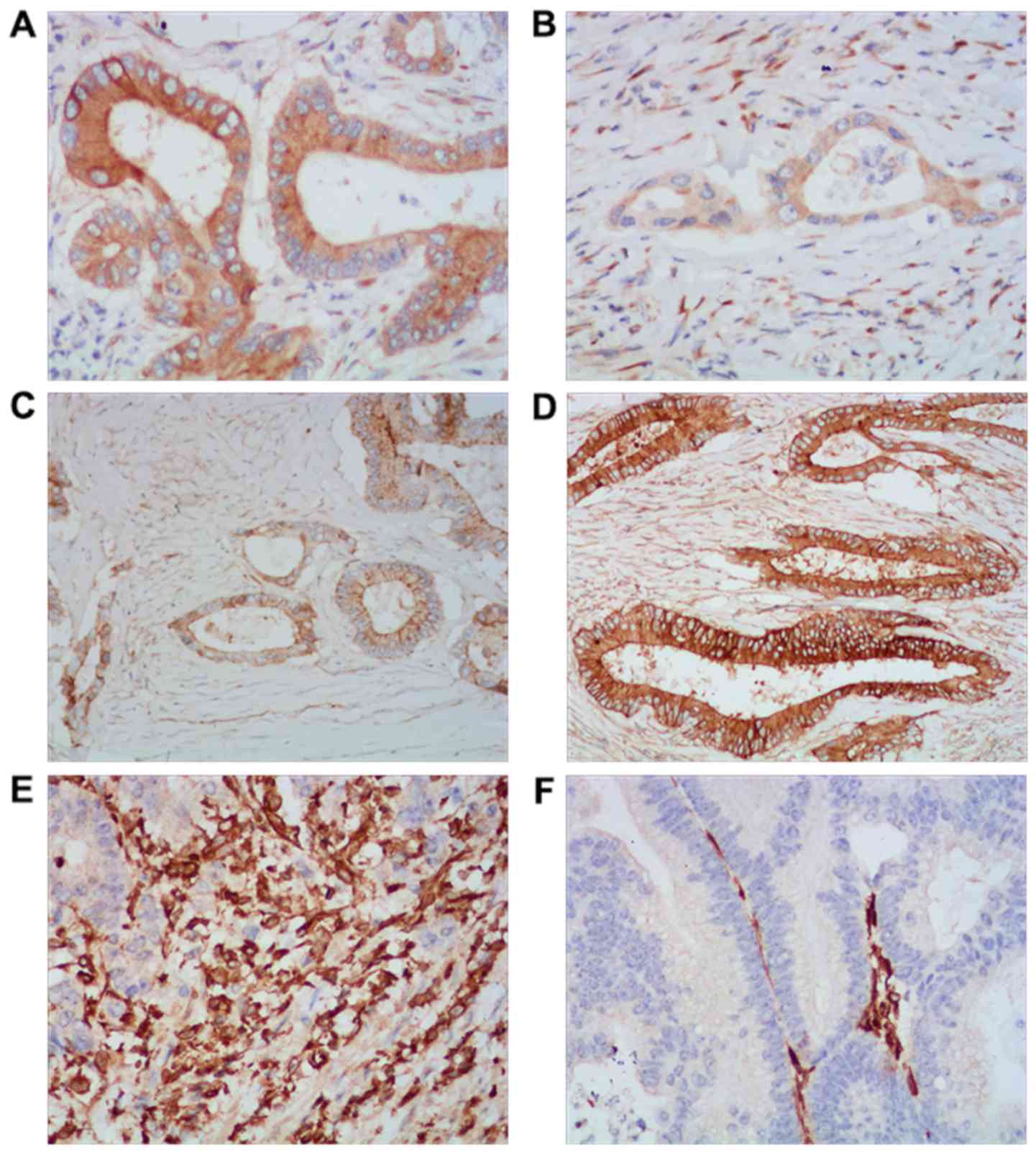 | Figure 5.Expression of TRAF6, E-cadherin, and
Vimentin in CRC tissues. In representative immunohistochemical
staining, miR-124 low-expression tissues exhibited strong (A) TRAF6
and (E) Vimentin staining, and weak (C) E-cadherin staining.
However, miR-124 high-expression tissues presented with weak (B)
TRAF6 and (F) Vimentin staining, and bright staining of (D)
E-cadherin. Magnification, ×400. miR, microRNA; TRAF6, tumor
necrosis factor receptor associated factor 6; CRC, colorectal
cancer. |
Discussion
An increasing number of studies have confirmed that
miRNAs are active participants in the development of CRC (26,27). In
addition, these small non-coding RNAs are regarded as key
regulators of metastasis and EMT in human cancers (28,29).
Thus, determining the expression levels of different miRNAs during
CRC initiation and progression may provide novel insights into the
molecular mechanism of carcinogenesis. Fang et al (30) demonstrated that miRNA 449b inhibits
SW1116 colon cancer stem cell proliferation by downregulating
G1/S-specific cyclin-D1 and transcription factor E2F3
expression. Liu et al (31)
indicated that miR139-3p was an independent prognostic factor of
colon cancer, and He et al (21) revealed that miR-296 attenuated CRC
metastasis and EMT by targeting S100A4. Therefore, miRNAs may
function as prognostic indicators and potential target biomarkers
in the development of novel therapeutics for different types of
cancer.
In the present study, miR-124 was markedly
downregulated in CRC tissues when compared with para-cancerous
tissues. In CRC tissues, the miR-124 expression level was
significantly correlated with histological grade and lymph node
status, which was in agreement with findings from previous studies
(32,33). Therefore, it was hypothesized that
miR-124 was involved in the development and progression of CRC. In
addition, the present study indicated that overall survival time
was decreased in CRC patients with a low miR124 expression level,
compared with those with a higher expression level (P=0.005); this
provides further evidence that reduced miR-124 expression in CRC
may enhance malignant invasion and worsen the prognostic phenotype
of this tumor. In a previous in vitro study, miR-124 was
proposed to inhibit DNA synthesis and proliferation by reducing
ribose-phosphate pyrophosphokinase 1levels in the pentose phosphate
pathway (34). Consistent with these
data, low miR-124 expression level was directly related to poor
prognosis in the present study.
In cancer research, local and/or systemic metastasis
represents poor prognosis in patients with CRC (35). A series of reports (7,36,37)
confirmed that EMT occurs during CRC progression, which provides
cancer cells with invasive and metastatic properties. Therefore,
EMT serves a crucial role in cancer metastasis. In a previous study
(11), TRAF6 was confirmed to be a
weak prognostic marker of CRC and to act on EMT progression.
Therefore, the potential association between miR-124 and TRAF6
expression was investigated in the context to EMT. After analyzing
IHC-stained colorectal tissue samples, it was discovered that
miR-124 expression may be possible negative regulator of EMT in
CRC. Strong TRAF6 and Vimentin staining coupled with weak
E-cadherin staining was observed in tumors with low miR-124
expression levels. Conversely, high miR-124-expressing tumors
presented with positive E-cadherin staining but weak Vimentin and
TRAF6 staining.
TRAF6 has been identified as an oncogene for its
active involvement in malignancy (38,39).
Previous research has confirmed that ectopic TRAF6 expression is
observed in gastrointestinal tumors (40,41). In
the present study, a negative regulatory effect between miR-124
level and TRAF6 expression levels was hypothesized. Strong TRAF6
staining more frequently appeared in CRC tissues with minimal
miR-124 expression than in those with high expression levels, and
vice versa. In addition, miR-124 directly influenced luciferase
reporter activity by interacting with the TRAF6 3′-UTR.
Recently, a study reported that miR-124 inhibited
cell invasion and suppressed gastric cancer invasion and metastasis
by targeting Snail2 (18).
Coincidentally, it was found that high TRAF6 expression levels in
CRC tissues were positively correlated with the expression levels
of EMT biomarkers. The above data illustrated that miR-124 may
serve an important role in EMT in CRC metastasis by regulating the
expression of TRAF6. Therefore, the present study suggests that
miR-124 and TRAF6 are high-risk indicators for poor patient
prognosis, and require further investigation in a larger study
cohort.
In summary, the present study demonstrated that
miR-124 is poor a prognostic factor in patients with CRC; although
miR-124 was shown to influence TRAF6 expression, further evidence
is required to determine whether this is by direct or indirect
regulation.
Acknowledgements
The authors would like to thank Professor Bo Luo and
Hanfeng Zhang from the Pathology Department in Tongji Medical
College, Huazhong University of Science and Technology for their
technical assistance.
Funding
The present study was supported by the Fund of
Health and Family Planning Commission of Wuhan Municipality (grant
no. WX17D05) and the Fund of Scientific Researching of the Central
Hospital of Wuhan (grant no. YB16A02).
Availability of data and materials
The datasets generated and/or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
CW carried out the molecular genetics studies,
participated in sequence alignment and drafted the manuscript. HH
conducted the immunoassays. LL participated in the design of the
study and performed the statistical analysis. ZT conceived the
study, participated in its design and coordination, and helped to
draft the manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
All patients provided written informed consent to
participate prior to surgery. Ethical approval was given by the
medical ethics committee of the Central Hospital of Wuhan Tongji
Medical College, Huazhong University of Science and Technology. All
procedures performed in this study involving human participants
were conducted in accordance with Chinese ethical standards and the
2008 Declaration of Helsinki.
Patient consent for publication
Written informed consent was obtained from patients
for publication of this manuscript and any accompanying images.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
TRAF6
|
tumor necrosis factor
receptor-associated factor 6
|
|
EMT
|
epithelial-mesenchymal transition
|
|
CRC
|
colorectal cancer
|
References
|
1
|
Wang YW, Chen HH, Wu MS and Chiu HM;
Taiwanese Nationwide Colorectal Cancer Screening Program, : Current
status and future challenge of population-based organized
colorectal cancer screening: Lesson from the first decade of
Taiwanese program. J Formos Med Assoc. 117:358–364. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gu MJ, Huang QC, Bao CZ, Li YJ, Li XQ, Ye
D, Ye ZH, Chen K and Wang JB: Attributable causes of colorectal
cancer in China. BMC Cancer. 18:382018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brenner DR, Ruan Y, Shaw E, De P, Heitman
SJ and Hilsden RJ: Increasing colorectal cancer incidence trends
among younger adults in Canada. Prev Med. 105:345–349. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhen YH, Liu XH, Yang Y, Li B, Tang JL,
Zeng QX, Hu J, Zeng XN, Zhang L, Wang ZJ, et al: Phase I/II study
of adjuvant immunotherapy with sentinel lymph node T lymphocytes in
patients with colorectal cancer. Cancer Immunol Immunother.
64:1083–1093. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Deng Y, Chi P, Lan P, Wang L, Chen W, Cui
L, Chen D, Cao J, Wei H, Peng X, et al: Modified FOLFOX6 with or
without radiation versus fluorouracil and leucovorin with radiation
in neoadjuvant treatment of locally advanced rectal cancer: Initial
results of the Chinese FOWARC multicenter, open-label, randomized
three-arm phase III trial. J Clin Oncol. 34:3300–3307. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen Z, He J, Xing X, Li P, Zhang W, Tong
Z, Jing X, Li L, Liu D, Wu Q and Ju H: Mn12Ac inhibits the
migration, invasion and epithelial-mesenchymal transition of lung
cancer cells by downregulating the Wnt/β-catenin and PI3K/AKT
signaling pathways. Oncol Lett. 16:3943–3948. 2018.PubMed/NCBI
|
|
7
|
Mao L, Li Y, Zhao J, Li Q, Yang B, Wang Y,
Zhu Z, Sun H and Zhai Z: Transforming growth factor-β1 contributes
to oxaliplatin resistance in colorectal cancer via epithelial to
mesenchymal transition. Oncol Lett. 14:647–654. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jiang M, Xu B, Li X, Shang Y, Chu Y, Wang
W, Chen D, Wu N, Hu S, Zhang S, et al: O-GlcNAcylation promotes
colorectal cancer metastasis via the miR-101-O-GlcNAc/EZH2
regulatory feedback circuit. Oncogene. 38:301–316. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hashimoto M, Kobayashi T, Tashiro H,
Arihiro K, Kikuchi A and Ohdan H: h-Prune is associated with poor
prognosis and epithelial-mesenchymal transition in patients with
colorectal liver metastases. Int J Cancer. 139:812–823. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chang SN, Lee JM, Oh H, Kim U, Ryu B and
Park JH: Troglitazone inhibits the migration and invasion of PC-3
human prostate cancer cells by upregulating E-cadherin and
glutathione peroxidase 3. Oncol Lett. 16:5482–5488. 2018.PubMed/NCBI
|
|
11
|
Zhang T, Wang H and Han L: Expression and
clinical significance of tumor necrosis factor receptor-associated
factor 6 in patients with colon cancer. Iran Red Crescent Med J.
18:e239312016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lou JS, Yan L, Bi CW, Chan GK, Wu QY, Liu
YL, Huang Y, Yao P, Du CY, Dong TT and Tsim KW: Yu Ping Feng San
reverses cisplatin-induced multi-drug resistance in lung cancer
cells via regulating drug transporters and p62/TRAF6 signalling.
Sci Rep. 6:319262016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Maeda S, Yoshida H, Ogura K, Mitsuno Y,
Hirata Y, Yamaji Y, Akanuma M, Shiratori Y and Omata M: H. Pylori
activates NF-kappaB through a signaling pathway involving IkappaB
kinases, NF-kappaB-inducing kinase, TRAF2, and TRAF6 in gastric
cancer cells. Gastroenterology. 119:97–108. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kong L, Li X, Wang H, He G and Tang A:
Calycosin inhibits nasopharyngeal carcinoma cells by influencing
EWSAT1 expression to regulate the TRAF6-related pathways. Biomed
Pharmacother. 106:342–348. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bilir C, Engin H, Can M, Likhan S,
Demirtas D, Kuzu F and Bayraktaroglu T: Increased serum tumor
necrosis factor receptor-associated factor-6 expression in patients
with non-metastatic triple-negative breast cancer. Oncol Lett.
9:2819–2824. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rezaeian AH, Li CF, Wu CY, Zhang X,
Delacerda J, You MJ, Han F, Cai Z, Jeong YS, Jin G, et al: A
hypoxia-responsive TRAF6-ATM-H2AX signalling axis promotes HIF1α
activation, tumorigenesis and metastasis. Nat Cell Biol. 19:38–51.
2017. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shi B, Wang Y and Yin F:
MALAT1/miR-124/Capn4 axis regulates proliferation, invasion and EMT
in nasopharyngeal carcinoma cells. Cancer Biol Ther. 18:792–800.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li SL, Gao HL, Lv XK, Hei YR, Li PZ, Zhang
JX and Lu N: MicroRNA-124 inhibits cell invasion and
epithelial-mesenchymal transition by directly repressing Snail2 in
gastric cancer. Eur Rev Med Pharmacol Sci. 21:3389–3396.
2017.PubMed/NCBI
|
|
19
|
Liu Q, Luo D, Cai S, Li Q and Li X: P-TNM
staging system for colon cancer: Combination of P-stage and AJCC
TNM staging system for improving prognostic prediction and clinical
management. Cancer Manag Res. 10:2303–2314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen S, Chen H, Gao S, Qiu S, Zhou H, Yu M
and Tu J: Differential expression of plasma microRNA-125b in
hepatitis B virus-related liver diseases and diagnostic potential
for hepatitis B virus-induced hepatocellular carcinoma. Hepatol
Res. 47:312–320. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
He Z, Yu L, Luo S, Li M, Li J, Li Q, Sun Y
and Wang C: miR-296 inhibits the metastasis and
epithelial-mesenchymal transition of colorectal cancer by targeting
S100A4. BMC Cancer. 17:1402017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu W, Zhang Q, Li S, Li L, Ding Z, Qian
Q, Fan L and Jiang C: The relationship between colonic macrophages
and MicroRNA-128 in the pathogenesis of slow transit constipation.
Dig Dis Sci. 60:2304–2315. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li JJ, Luo J, Lu JN, Liang XN, Luo YH, Liu
YR, Yang J, Ding H, Qin GH, Yang LH, et al: Relationship between
TRAF6 and deterioration of HCC: An immunohistochemical and in vitro
study. Cancer Cell Int. 16:762016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hueman MT, Wang H, Yang CQ, Sheng L,
Henson DE, Schwartz AM and Chen D: Creating prognostic systems for
cancer patients: A demonstration using breast cancer. Cancer Med.
7:3611–3621. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang L, Dou X, Liu T, Lu W, Ma Y and Yang
Y: Tumor size and lymph node metastasis are prognostic markers of
small cell lung cancer in a Chinese population. Medicine
(Baltimore). 97:e117122018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Moody L, He H, Pan YX and Chen H: Methods
and novel technology for microRNA quantification in colorectal
cancer screening. Clin Epigenetics. 9:1192017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pecqueux M, Liebetrau I, Werft W,
Dienemann H, Muley T, Pfannschmidt J, Müssle B, Rahbari N, Schölch
S, Büchler MW, et al: A Comprehensive MicroRNA expression profile
of liver and lung metastases of colorectal cancer with their
corresponding host tissue and its prognostic impact on survival.
Int J Mol Sci. 17(pii): E17552016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang J and Ma L: MicroRNA control of
epithelial-mesenchymal transition and metastasis. Cancer Metastasis
Rev. 31:653–662. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hur K, Toiyama Y, Takahashi M, Balaguer F,
Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR and Goel A:
MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT)
in human colorectal cancer metastasis. Gut. 62:1315–1326. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fang Y, Gu X, Li Z, Xiang J and Chen Z:
miR-449b inhibits the proliferation of SW1116 colon cancer stem
cells through downregulation of CCND1 and E2F3 expression. Oncol
Rep. 30:399–406. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu X, Duan B, Dong Y, He C, Zhou H, Sheng
H, Gao H and Zhang X: MicroRNA-139-3p indicates a poor prognosis of
colon cancer. Int J Clin Exp Pathol. 7:8046–8052. 2014.PubMed/NCBI
|
|
32
|
Zhang J, Lu Y, Yue X, Li H, Luo X, Wang Y,
Wang K and Wan J: MiR-124 suppresses growth of human colorectal
cancer by inhibiting STAT3. PLoS One. 8:e703002013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang MJ, Li Y, Wang R, Wang C, Yu YY, Yang
L, Zhang Y, Zhou B, Zhou ZG and Sun XF: Downregulation of
microRNA-124 is an independent prognostic factor in patients with
colorectal cancer. Int J Colorectal Dis. 28:183–189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Qiu Z, Guo W, Wang Q, Chen Z, Huang S,
Zhao F, Yao M, Zhao Y and He X: MicroRNA-124 reduces the pentose
phosphate pathway and proliferation by targeting PRPS1 and RPIA
mRNAs in human colorectal cancer cells. Gastroenterology.
149:1587–1598.e11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ueno H, Hase K, Hashiguchi Y, Shimazaki H,
Yoshii S, Kudo SE, Tanaka M, Akagi Y, Suto T, Nagata S, et al:
Novel risk factors for lymph node metastasis in early invasive
colorectal cancer: A multi-institution pathology review. J
Gastroenterol. 49:1314–1323. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Han L, Jiang Y, Han D and Tan W: Hsp27
regulates epithelial mesenchymal transition, metastasis and
proliferation in colorectal carcinoma. Oncol Lett. 16:5309–5316.
2018.PubMed/NCBI
|
|
37
|
Guo Q, Zhao Y, Chen J, Hu J, Wang S, Zhang
D and Sun Y: BRAF-activated long non-coding RNA contributes to
colorectal cancer migration by inducing epithelial-mesenchymal
transition. Oncol Lett. 8:869–875. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang WL, Wang J, Chan CH, Lee SW, Campos
AD, Lamothe B, Hur L, Grabiner BC, Lin X, Darnay BG and Lin HK: The
E3 ligase TRAF6 regulates Akt ubiquitination and activation.
Science. 325:1134–1138. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang X, Wu L, Xiao T, Tang L, Jia X, Guo
Y, Zhang J, Li J, He Y, Su J, et al: TRAF6 regulates EGF-induced
cell transformation and cSCC malignant phenotype through
CD147/EGFR. Oncogenesis. 7:172018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Han F, Zhang L, Qiu W and Yi X: TRAF6
promotes the invasion and metastasis and predicts a poor prognosis
in gastric cancer. Pathol Res Pract. 212:31–37. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sun H, Li X, Fan L, Wu G, Li M and Fang J:
TRAF6 is upregulated in colon cancer and promotes proliferation of
colon cancer cells. Int J Biochem Cell Biol. 53:195–201. 2014.
View Article : Google Scholar : PubMed/NCBI
|