Introduction
In the whole world, esophageal cancer is the sixth
cause of death among various cancer types. Esophageal cancers are
mainly classified into two histological types, esophageal squamous
cell carcinoma (ESCC) and adenocarcinoma (1). Then, ESCC is thought to be the main
histological type which accounts for more than 90% in Asian
countries including Japan, Korea and China (2), and it is known that ESCC is a highly
malignant malignancy among many cancer types (3–5).
Esophagectomy remains the mainstay potential curative treatment for
ESCC (6). However, esophagectomy is
still a highly invasive surgical procedure with high morbidity and
mortality (7). Although remarkable
advances have been made in chemotherapy and chemotherapy with
radiotherapy as cancer therapies, These therapeutic effects can be
said to be extremely limited as curative treatment (8,9).
Therefore, understanding the characteristics of ESCC and developing
new therapeutic tools are urgently required.
Both genetic mechanisms and epigenetic alterations
are thought to be closely involved in the development and
progression of ESCC (10). Several
epigenetic abnormalities have been reported, including DNA
methylation, histone modifications and non-coding RNAs (11,12). In
our studies, epigenetic modifications play crucial roles in the
regulation of gene expression in ESCC (12–19). In
particular, the methylation of lysine residues on histone proteins
in the chromatin structure has received attention due to their
potential regulatory ability on DNA-based nuclear processes such as
transcription, replication and repair (20). The methylation of histone lysine
residues was first reported in the 1960s and was considered an
irreversible posttranslational modification (21). In 2004, however, a lysine demethylase
was discovered, and the methylation of histone lysine residues is
now regarded as a dynamic modulation (22).
Abnormalities in histone lysine methylation are
frequently observed in various cancers (23–26).
Lysine-specific histone demethylase 1 (LSD 1), a histone
demethylase, is an amine oxidase that removes monomethyl and
dimethyl moieties from Lys 4 of histone H 3 and produces a
demethylated H3 tail (27).
Identifying the key points of regulation in the histone methylation
network for cancer development and progression can provide
innovative targets for cancer therapies.
In the present study, we focused on the mechanisms
underlying how demethylated Lys4 of H3 influences the gene
expressions in ESCC cells. We investigated microarray and chromatin
immunoprecipitation sequencing (ChIP-seq) in order to explore the
effect of demethylated Lys4 of H3 on the transcriptional state of
ESCC cells and identified genes affecting cancer growth.
Materials and methods
Cell culture and chemicals
The human esophageal cell lines T.Tn and TE2 were
cultured in DMEM (Life Technologies, Grand Island, NY, USA)
supplemented with 10% fetal calf serum. T.Tn cells were acquired
from the Japanese Cancer Research Resources Bank (Tsukuba, Japan),
and TE2 cells were obtained from Tohoku University (Sendai, Japan).
NCL1, an LSD1 inhibitor, was provided by Kyoto Prefectural
University of Medical Science (Graduate School of Medicine) (Kyoto,
Japan) in cooperation. NCL1 showed higher inhibitory activity than
the known LSD1 inhibitor, trans-2-phenylcyclopropylamine. Moreover,
in the presence of NCL1, the methylation activity of H3K4 is
observed and cell proliferation is inhibited in experiments using
cancer cells (28–30). NCL1 was dissolved in dimethyl
sulfoxide and used for in vitro studies.
Messenger RNA preparation and a cDNA
microarray analysis
T.Tn or TE2 cells were seeded into a
225-cm2 flask, incubated for 48 h, treated with or
without an IC80 concentration of LSD1 inhibitor and
harvested at 24 h. Subsequently, the cells were washed with
phosphate-buffered saline (PBS; cat. no: 14190-250, Invitrogen,
Carlsbad, CA, USA) and total RNA was extracted using RNeasy Plus
Mini kit (Qiagen, Inc., Chatsworth, CA, USA). Changes in gene
expression were compared between 5.5 tor of total RNA extracted
from cells cultured by exposure to NCL 1 and 5.5 tor of total RNA
extracted from cells cultured in a control culture using an
Affymetrix Human Exon 1.0ST array (Affymetrix, Santa Clara, CA,
USA). Hybridization signals were detected with a GeneChip scanner
3000 7 G (Affymetrix), and the scanned images were analyzed using
the GeneChip command console software (AGCC). All the processes
were basically carried out according to the previous report
(31). All experiments were done in
duplicate and the averaged data were subjected to statistical
analysis.
ChIP-seq analyses
ChIP-seq analyses were performed using the
SimpleChIP plus enzymatic chromatin IP kit (Magnetic Beads; Cell
Signaling Technology, Danvers, MA, USA). T.Tn or TE2 cells were
cultured for 48 h in a 225-cm2 flask, then incubated
under the condition with or without an IC80
concentration of LSD1 inhibitor and harvested at 24 h. The Cells
were crosslinked with 1% formaldehyde for 10 min at room
temperature, then washed twice with PBS containing 0.5 mM EDTA and
collected. The cell pellet was lysed with 0.3 ml of cell lysis
buffer (50 mM Tris-HCl [pH 8.1], 10 mM EDTA, 1% SDS, and protease
inhibitor) and incubated on ice for 10 min. Lysates of the cells
were sonicated to obtain DNA fragments of 150 to 900 base pair (bp)
in size. About 50 µg of cross-linked sheared chromatin solution was
then used for immunoprecipitation. The solution with the
Anti-Histone H3 (di methyl K4) antibody-ChIP Grade (Abcam, Inc.,
Cambridge, UK; cat. no: ab7766) was incubated overnight at 4°C on a
rotating shaker for immunoprecipitation. Magnetic beads were added
to the solution, incubated at 4°C for 1 h, and then washed with
washing buffer. The cross-linking was reversed by adding NaCl at a
final concentration of 200 mM and heating at 65°C for 30 min. The
DNA fragments were purified using a spin column. A sequencing
library was prepared and massively parallel high throughput
sequencing was performed with the Illumina HiSeq 2000 system
(Illumina, Inc., San Diego, Calif., USA) and a 50-bp reads were
aligned against the reference genome on a Burrows-Wheeler
transform, and a minimum mapping quality filter 20 was applied
(32). Enriched regions for each
condition were detected and analyzed with MACS v1.4.0 (model-based
analysis for ChIP-Seq) (33) and
CEAS v1.0.2 (cis-regulatory element annotation system) (34,35).
Peaks with overlaps in both cell lines were merged into a broad
peak domain using BEDTools (36).
All of the count data from the ChIP-Seq assays were analyzed with
DESeq to normalize the peak signal (37).
The reverse transcription-quantitative
PCR (RT-qPCR) for measuring the LDHB and AEG-1/MTDH mRNA
expression
The mRNA expression of DUSP5, BHLHE40 and MXRA5 were
examined by a RT-qPCR. T.Tn or TE2 cells were seeded into a
225-cm2 flask, incubated for 48 h, treated with or
without an IC80 concentration of LSD1 inhibitor and
harvested at 24 h. Subsequently, the cells were washed with
phosphate-buffered saline (PBS) and total RNA was extracted using
an RNeasy Plus Mini kit (Qiagen, Inc., Chatsworth, CA, USA). The
cDNA templates for the qPCR were synthesized from 1 µg of total RNA
using a High Capacity RNA-to-cDNA kit (Applied Biosystems).The
Actin alpha 1 (ACTA1) gene served as an internal control. The PCR
reaction consisted of Sso Fast Eva Green Supermix (BioRad;
containing dNTPs, Sso7d fusion polymerase, MgCl2, EvaGreen dye,
stabilizers), the primers (each 1 µM) and cDNA. All reactions were
run in duplicate on the MyiQ2 Two-Color Real-Time PCR detection
system (BioRad). The PCR processes were as follows: initial
denaturation at 95°C for 30 sec, followed by 40 cycles of
denaturation at 95°C for 5 sec, annealing at 55°C for 10 sec. The
following primer sequences were used: DUSP5;
5′-CCTGCTAAAACTGGGATGGA-3′ and 5′-ACCTACCCTGAGGTCCGTCT-3′: BHLHE40;
5′-GGCATAGCACGGTAGTGGTT-3′ and 5′-TCAGACCTTGGTTTGGTTCC-3′: MXRA5;
5′-CTGTCCAGTCCTCAGGAAGC-3′ and 5′-TCCTGTGGAAACCTTTGTCC-3′: ACTA1;
5′-CCTTCATCGGTATGGAGTC-3′ and 5′-GTTGGCATACAGGTCCTT-3′.
The comparative quantitative cycle (Cq)
method was applied to quantify the expression levels of mRNAs. The
relative amount of DUSP5, BHLHE40 and MXRA5 to ACTA1mRNA was
calculated using the following equation:
2q−ΔC, where ΔCq=(Cq DUSP5,
BHLHE40 or MXRA5 or AEG-1/MTDH-Cq ACTA1).
Statistical analyses
The Student's t-test was performed to compare the
differences in the mRNA expression levels. P<0.05 was considered
to indicate a statistically significant difference. The SPSS v.16.0
(SPSS, Inc., Chicago, IL, USA) software program were used for the
analyses.
Results
Effects of NCL1 on the expression of
various genes in microarray analyses
We used a cDNA microarray to identify genes induced
by LSD1 exposure in ESCC. We extracted genes with expression levels
more than two-fold or greater compared to control, whether
decreased or increased, as significant. In both T. Tn and TE2 cell
lines, expression of 18 genes was increased, while expression of 9
genes was decreased (Table I).
 | Table I.List of genes up- or downregulated
(>2-fold change) in T.Tn and TE2 cells. |
Table I.
List of genes up- or downregulated
(>2-fold change) in T.Tn and TE2 cells.
| A, Upregulated
(>2-fold changes) |
|---|
|
|---|
|
|
| Maximum fold
change |
|
|---|
|
|
|
|
|
|---|
| Genbank accession
no. | Gene symbol | T.Tn | TE2 | Gene
description |
|---|
| BC005008 | CEACAM6 | 2.84 | 3.62 | carcinoembryonic
antigen-related cell adhesion molecule 6 (non-specific cross
reacting antigen) |
| BC012172 | ACSS2 | 1.10 | 1.24 | acyl-CoA synthetase
short-chain family member 2 |
| BC008723 | ASNS | 1.92 | 1.96 | asparagine
synthetase (glutamine-hydrolyzing) |
| L19501 | CBS | 1.15 | 1.18 |
cystathionine-β-synthase |
| M29540 | CEACAM5 | 2.45 | 2.90 | carcinoembryonic
antigen-related cell adhesion molecule 5 |
| BC019625 | CHAC1 | 1.34 | 2.11 | ChaC, cation
transport regulator homolog 1 (E. coli) |
| U03688 | CYP1B1 | 1.49 | 1.67 | cytochrome P450,
family 1, subfamily B, polypeptide 1 |
| BC007333 | ETV5 | 1.73 | 1.81 | ets variant 5 |
| AF110400 | FGF19 | 1.17 | 1.26 | fibroblast growth
factor 19 |
| EF152283 | GCNT3 | 1.17 | 1.66 | glucosaminyl
(N-acetyl) transferase 3, mucin type |
| AF019770 | GDF15 | 2.01 | 1.75 | growth
differentiation factor 15 |
| BC033089 | LCN2 | 1.62 | 2.78 | lipocalin 2 |
| BC004863 | PSAT1 | 1.34 | 2.87 | phosphoserine
aminotransferase 1 |
| AF539739 | S100P | 1.26 | 2.06 | S100 calcium
binding protein P |
| AF097514 | SCD | 1.25 | 2.38 | stearoyl-CoA
desaturase (∆-9-desaturase) |
| BC000658 | STC2 | 1.95 | 1.59 | stanniocalcin
2 |
| BC011703 | TMPRSS4 | 1.03 | 1.24 | transmembrane
protease, serine 4 |
| AF022375 | VEGFA | 1.62 | 1.50 | vascular
endothelial growth factor A |
|
| B, Downregulated
(>2-fold changes) |
|
|
|
| Maximum fold
change |
|
|
|
|
|
|
| Genbank
accession no. | Gene
symbol | T.Tn | TE2 | Gene
description |
|
| J04948 | ALPPL2 | −1.01 | −1.69 | alkaline
phosphatase, placental-like 2 |
| BC098561 | EFEMP1 | −1.59 | −1.76 | EGF-containing
fibulin-like extracellular matrix protein 1 |
| AB031548 | GPR87 | −1.11 | −1.06 | G protein-coupled
receptor 87 |
| M19154 | TGFB2 | −1.72 | −1.11 | transforming growth
factor, β2 |
| BC142678 | PHLDB2 | −1.14 | −1.04 | pleckstrin
homology-like domain, family B, member 2 |
| BC146868 | COL12A1 | −1.13 | −1.22 | collagen, type XII,
α1 |
| BC017782 | WISP2 | −1.12 | −1.31 | WNT1 inducible
signaling pathway protein 2 |
| AF098807 | LHFP | −1.21 | −1.26 | lipoma HMGIC fusion
partner |
| AK123348 | C3orf57 | −1.14 | −1.72 | chromosome 3 open
reading frame 57 |
ChIP-seq analyses
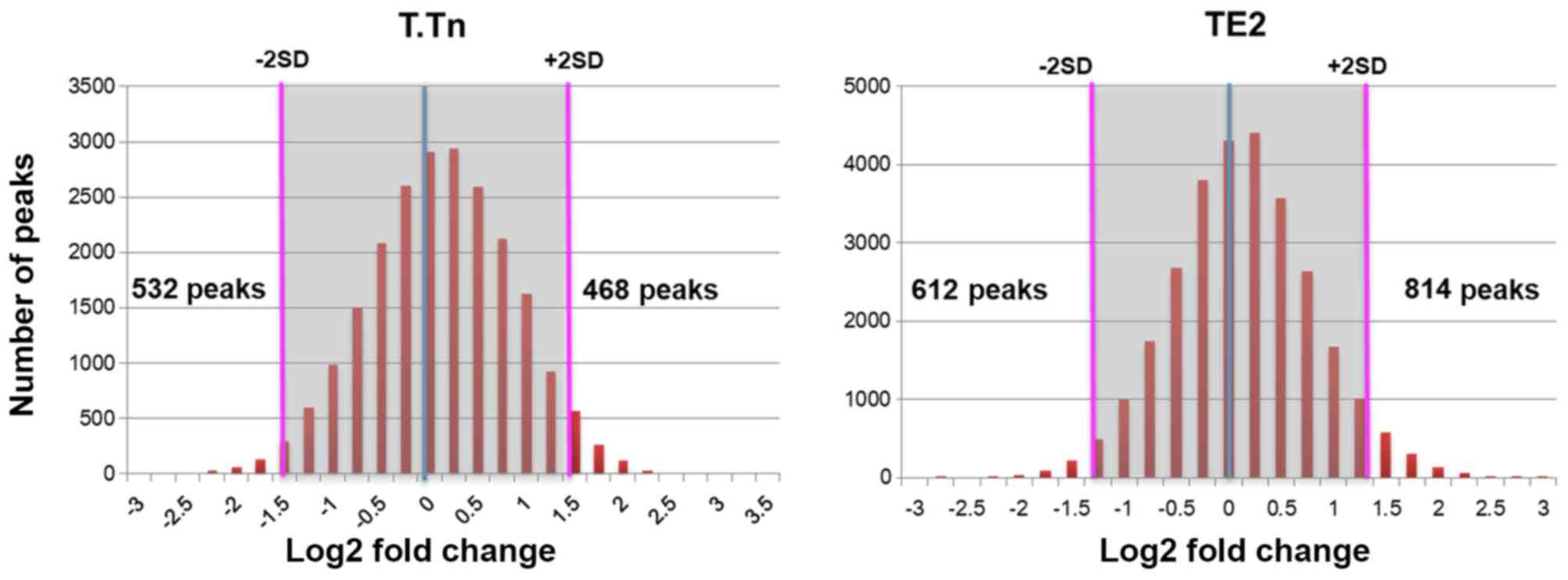
To assess the functional significance of
demethylated Lys4 of H3 in ESCC cells, we also analyzed the
genome-wide modified targets of demethylation Lys4 of H3 using deep
sequencing based on chromatin immunoprecipitation (ChIP-seq). When
we compared the findings with control cells (without LSD1
inhibitor), we identified up-regulated peaks in 468 and 814
demethylated Lys4 of H3-specific modification sites in T.Tn and TE2
cells, respectively (Fig. 1). We
also identified down-regulated peaks in 532 and 612 demethylated
Lys4 of H3-specific modification sites in T.Tn and TE2 cells,
respectively (Fig. 1).
Identifying the relationship between
histone modification states and the gene expression in ESCC
cells
To clarify the gene expression change by the state
of histone modification, the genes with up- or down-regulated
expression were investigated using microarray data, and that the
promoter region of these genes may be the targets of histone
modification. The expression of some of these genes whose promoters
were detected as candidates for targets of demethylated Lys4 of H3
were markedly changed according to the microarray data (Table II). The results showed that 17 genes
were commonly up-regulated, while 16 genes were commonly
down-regulated (Table III). These
identified genes were categorized based on their function,
referring to GENE ONTOLOGY™, and classified into 7
groups: apoptosis, cell cycles, defense and immunity, metabolism,
signal transduction and transcription, structural protein, and
unclassified. The frequencies of these functionally classified
genes in each cluster are shown in Table IV.
 | Table II.List of numbers of genes up- or
down-regulated in the chromatin immunoprecipitation-seq
analysis. |
Table II.
List of numbers of genes up- or
down-regulated in the chromatin immunoprecipitation-seq
analysis.
| Cell line | Upregulated peaks,
n | Downregulated
peaks, n |
|---|
| T.Tn | 468 | 532 |
| TE2 | 814 | 612 |
 | Table III.List of gene symbols commonly up- or
downregulated in both the microarray and ChIP-seq assay. |
Table III.
List of gene symbols commonly up- or
downregulated in both the microarray and ChIP-seq assay.
|
| Microarray |
|---|
|
|
|
|---|
| ChIP-seq | Upregulated | Downregulated |
|---|
| Upregulated | DUSP5, BHLHE40,
TMC5 GNE, PMAIP1, TIMP3 C6orf223, PHLDA1,ERRFI1 MID1IP1, ULBP1,
FGF19 GCNT3, HMOX1, TRIB3 VEGFA, CEACAM6 | DIO2, RBMS3, LHFP
PLK2, CP, TOM1L2 MXRA5, DKK1, EPHA4 EGLN3, CCDC80, MID1 SLC16A7,
RAPGEF4, TOX VCL, MAP7D2, RASAL2 HAS2, TNS3, FLNA NEDD4L, KIAA1217,
PSAPL1 SEMA3A, GPR126, EGFR |
| Downregulated | GDF15, CLGN | RHOB, KLHL13,
ARID5B DIO2, MXRA5, THBS1 ALDH1A1, DKK1, C1orf116 SOX2, CACNG4,
LHFP FGFR2, EPHA4, EFEMP1, PALMD |
 | Table IV.Categorization of genes regulated by
the LSD1 inhibitor based on their functions, referring to GENE
ONTOLOGY™, and classified into 7 groups. |
Table IV.
Categorization of genes regulated by
the LSD1 inhibitor based on their functions, referring to GENE
ONTOLOGY™, and classified into 7 groups.
|
| Microarray and
ChIP-seq analysis |
|---|
|
|
|
|---|
| Function | Upregulated | Downregulated |
|---|
| Apoptosis | PMAIP1,TIMP3,
PHLDA1, FGF19, TRIB3, CEACAM6 | RHOB, FGFR2 |
| Cell cycles | HMOX1, VEGFA | KLHL13, THBS1 |
| Defense and
immunity | GCNT3 | – |
| Metabolism | MID1IP1 | ALDH1A1 |
| Signal transduction
and transcription | DUSP5, BHLHE40,
GNE, ERRFL1, ULBP1 | ARID5B, DKK1, SOX2,
EPH!4, EFEMP1 |
| Structural
protein | TMC5, | DIO2, MXRA5,
C1orf116, CACNG4, LHFP, PALMD |
| Unclassified | C6orf223 | – |
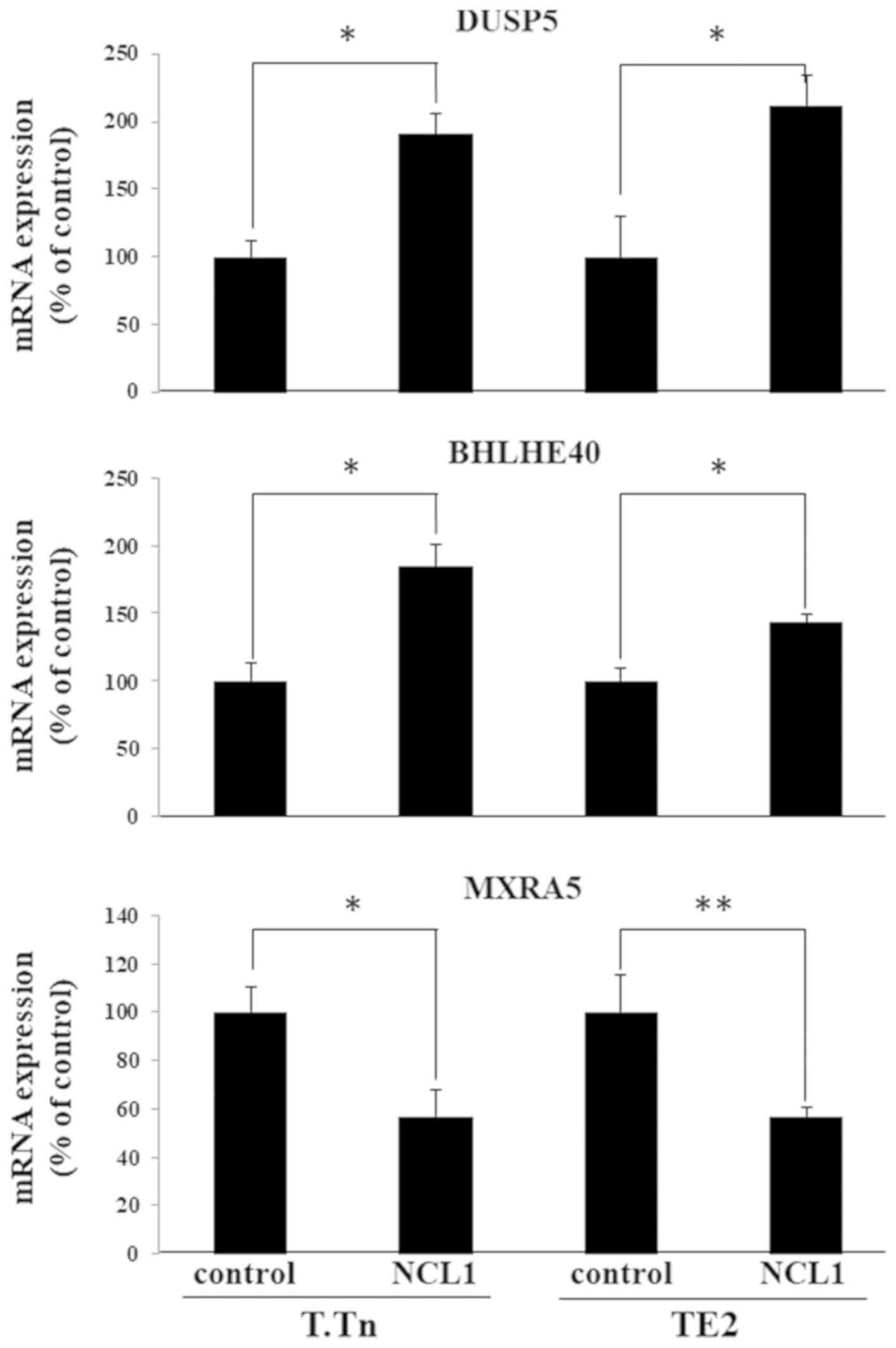
Validation of the gene expression
changes induced by NCL1
Among the mRNAs that showed altered expression
levels in both microRNA and ChIP-seq experiments, changes in the
expression levels of DUSP5, BHLHE40 and MXRA5 were confirmed by a
RT-qPCR (Fig. 2). As expected, the
expression of DUSP5 and BHLHE40 increased and the expression of
MXRA5 decreased.
Discussion
In this study, we tried to clarify the changes in
the gene expression due to histone demethylase LSD1 inhibitor using
a microarray and ChIP-Seq analyses. Some LSD1 inhibitors have shown
potent anti-cancer effects, and their pharmacological mechanisms
have been elucidated (38,39). ORY-1001 is an LSD1 inhibitor that was
shown to selectively inhibit KDM1A in clinical trials and is
currently being assessed for its utility in treating patients with
leukemia and solid tumors (40).
Although clinical trials of LSD1 inhibitors are being conducted
around the world, very few describe the mechanisms in detail
(41,42).
We have already elucidated the anti-tumor effect of
LSD1 inhibitors on ESCC, and this effect was shown to be caused by
changes in the gene expression induced by the agent, with PHLDB2
reported to demonstrate a particularly enormous change in
expression (19). In the present
study, in addition to changes in the gene expression, genome-wide
CHIP-Seq analyses were performed, and the histone methylation that
occurred was evaluated.
DUSP5 is one of the nuclear localization members of
the MKP/DUSP family and it is induced in response to the activation
of ERK, specifically dephosphorylated, and has the function of
anchoring the ERK in the nucleus (43). Furthermore, DUSP5 has been reported
to increase RAF, MEK and ERK activities in the cytoplasm, in
addition to its role in ERK nuclear inactivation. This activity has
been shown to be caused by alleviation of upstream kinase
inhibition and depends on its ability to sequester DUSP5 turnover
rate and inactive ERK in the nucleus (44). Also, the expression of BRAFV 600 E
oncoprotein, which has mutations in BRAF that are not sensitive to
feedback inhibition, changes the function of DUSP 5 to become an
inhibitor of the entire cell of ERK, and that the cell avoids
hyperactivation and aging of ERK. These analysis results explain
that DUSP5 functions as a tumor suppressor or a tumor promoter
(45).
BHLHE 40 is an up-regulated gene and is a basic
helix-loop-helix type transcription factor that has been shown to
be involved in epithelial-mesenchymal transition (EMT). According
to Asanoma et al (46), BHLHE
40 inhibited tumor cell invasion by suppressing the transcription
of the EMT factors SNAI 1, SNAI 2 and TWIST 1. In addition, they
showed an association between the transcription factor SP1 and the
basal transcriptional activity of TWIST1 and BHLHE40 and competes
with SP1 to regulate DNA transcription and control gene
transcription. Therefore, BHLHE 40 is thought to function as a
tumor suppressor.
It is thought that p53 reactivation and mass
apoptosis induction (PRIMA-1), a low-molecular compound, restores
the function of mutant TP53 to the function of wild-type TP53 and
induces p53-mediated apoptosis (47). PRIMA-1 and its methylated form
PRIMA-1 Met (APR-246) are thought to have antitumor effects and its
effects are evident in several types of cancers such as
osteosarcoma, multiple myeloma, lung cancer, breast cancer and
colon cancer (48–52). Furthermore, several clinical trials
using APR-246 have been performed, indicating its tolerability and
clinical effects in hematologic malignancies and prostate cancer
(53). Also in ESCC, Furukawa et
al (47) reported that PRIMA-1
may restore the function of mutant TP 53 in ESCC with a TP 50
missense mutation, due to the enhanced expression of Noxa. Tissue
inhibitor of metalloproteinase-3 (TIMP 3) which is one of the four
members of the protein family is initially classified according to
their function of inhibiting matrix metalloproteinases (MMP)
(54–56). TIMP3 is thought to induce apoptosis
in malignant cells, such as melanoma (57) human colon carcinoma (58), cervical carcinoma cells and breast
cancer cells (59). The death domain
of TIMP3, a region that inhibits the function of MMP, is localized
at its N-terminus (60). TIMP3 has
been reported in colon cancer cells and melanoma cells to increase
susceptibility to apoptosis via stabilization of the TNF-α receptor
on the cell surface (58,61). In ESCC, expression of TIMP-3 protein
is correlated with depth of tumor infiltration, number of lymph
node metastasis and stage of disease as a result of
immunohistochemical analysis using clinical specimens (54). TIMP-3 protein was localizes in a
shallow region of the tumor, and even in the same tumor, its
expression was decreased in the deep part. Furthermore, the
prognosis of cancer patients who lost TIMP-3 expression was
significantly worse than that of TIMP-3-positive cancer
patients.
PHLDA1 is a cell death mediator that induces cells
into apoptosis and exerts antiproliferative activity (62–65). The
overexpression of PHLDA1 inhibits cell proliferation and induces
cell death in various malignant tumors, including breast cancer,
melanoma and cervical carcinoma cells (66–68). Low
expression of PHLDA1 mRNA and protein is strongly related to breast
cancer and melanoma progression (63,64).
Conversely, it has been found that in oral squamous cell carcinoma,
high expression of PHLDA1 is associated with more advanced stage.
Therefore, the function of PHLDA1 is still controversial, but from
these results, it is possible that PHLDA1 functions as a tumor
suppressor in ESCC.
The genes shown to be down-regulated on either the
microarray analysis or ChIP-seq analysis in our study may have a
potential oncogenic function in ESCC.
Rho protein belongs to the Ras superfamily and is a
small molecule that functions as a binary switch in a wide range of
signaling pathways (69,70). Rho proteins are a family of 20
intracellular signaling molecules, including RhoA, RhoB, RhoC,
RhoG, RhoE, Rac1, Rac2, Cdc42Hs and TC10 (71). RhoB has the function of molecular
switch, and it circulates between inactive GDP-bonded type and
active GTP-bound type (72). RhoB
was reported as a molecule that induces Ras-induced fibroblast
transformation. New evidence suggests a potential role of RhoB in
supporting the tumorigenic function. As an example, it is reported
that RhoB protein expression is higher in T-acute lymphoblastic
leukemia (T-All) cells compared to normal T cells, and it is shown
to be significantly associated with leukemia cells (73). In the present study, the inhibition
of RhoB increased cell apoptosis by ≥300%.
Matrix remodeling-associated protein 5 (MXRA5), also
known as adhesion protein with leucine-rich repeats and
immunoglobulin domains related to perlecan (Adlican), is one of
matrix remodeling related proteins (74). The MXRA5 gene encodes a protein with
a molecular weight of 312 kDa. The MXRA family contains three genes
(MXRA5, MXRA7 and MXRA8), both of which are thought to be involved
in cell adhesion and matrix remodeling (75). The increased expression of MXRA5 has
been reported in many kinds of tumors, such as colorectal cancer,
ovarian cancer and esophageal cancer (76,77).
Furthermore, somatic mutation of MXRA5 has been reported, and this
mutation has been confirmed in various malignant tumors such as
lung, skin, brain, ovary and wall pleura (78).
Among thrombospondin, a family of extracellular
matrix proteins, thrombospondin-1 (THBS 1) is the first member
identified and its major roles are platelet aggregation,
angiogenesis, and tumorigenesis (79). Also in ESCC, THBS 1 can activate the
TGF-β signaling pathway, leading to the transcription of Cyr 61 and
CTGF (78–80). In addition, its overexpression is
thought to be significantly associated with TNM progression
(P=0.029) and lymph node metastasis (P=0.026) in clinicopathologic
studies. In the analysis of prognosis, it was shown that
overexpression of THBS protein is a prognostic predictor in ESCC
patients (P=0.042) (80).
The results in this study suggest that the large
number of genes affected by demethylation of H3 in ESCC Lys4 may be
greatly implicated in the development of ESCC cancer. The
correlation between these gene groups and carcinogenesis and
progression of ESCC needs to be verified in further studies, but
the present results will be helpful for clarifying the
mechanism.
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
Funding
The present study was supported by JSPS KAKENHI
(grant no. JP 24689053).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
IH designed, analyzed and conducted all of the
experiments and wrote the manuscript. MT, AK and II performed and
analyzed the results of the ChIP-seq experiments. YA, KM, YM, HS,
NS, TS and HM contributed to the study conception and design, and
the acquisition, analysis and interpretation of data. FI and YI
contributed to the acquisition, analysis and interpretation of
data, drafted the manuscript and revised it critically for
important intellectual content. KI performed RT-qPCR experiments.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liang H, Fan JH and Qiao YL: Epidemiology,
etiology, and prevention of esophageal squamous cell carcinoma in
China. Cancer Biol Med. 14:33–41. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang Y: Epidemiology of esophageal
cancer. World J Gastroenterol. 19:5598–5606. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Meng J, Zhang J, Xiu Y, Jin Y, Xiang J,
Nie Y, Fu S and Zhao K: Prognostic value of an immunohistochemical
signature in patients with esophageal squamous cell carcinoma
undergoing radical esophagectomy. Mol Oncol. 12:196–207. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Scheepers JJ, van der Peet DL, Veenhof AA,
Heijnen B and Cuesta MA: Systematic approach of postoperative
gastric conduit complications after esophageal resection. Dis
Esophagus. 23:117–121. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Baba Y, Saeki H, Nakashima Y, Oki E,
Shigaki H, Yoshida N, Watanabe M, Maehara Y and Baba H: Review of
chemotherapeutic approaches for operable and inoperable esophageal
squamous cell carcinoma. Dis Esophagus. 30:1–7. 2017.
|
|
9
|
Smyth EC, Lagergren J, Fitzgerald RC,
Lordick F, Shah MA, Lagergren P and Cunningham D: Oesophageal
cancer. Nat Rev Dis Primers. 3:170482017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin DC, Wang MR and Koeffler HP: Genomic
and epigenomic aberrations in esophageal squamous cell carcinoma
and implications for patients. Gastroenterology. 154:374–389. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hamm CA and Costa FF: Epigenomes as
therapeutic targets. Pharmacol Ther. 151:72–86. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hoshino I, Matsubara H, Hanari N, Mori M,
Nishimori T, Yoneyama Y, Akutsu Y, Sakata H, Matsushita K, Seki N
and Ochiai T: Histone deacetylase inhibitor FK228 activates tumor
suppressor Prdx1 with apoptosis induction in esophageal cancer
cells. Clin Cancer Res. 11:7945–7952. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hoshino I, Matsubara H, Akutsu Y,
Nishimori T, Yoneyama Y, Murakami K, Komatsu A, Sakata H,
Matsushita K and Ochiai T: Gene expression profiling induced by
histone deacetylase inhibitor, FK228, in human esophageal squamous
cancer cells. Oncol Rep. 18:585–592. 2007.PubMed/NCBI
|
|
14
|
Kano M, Seki N, Kikkawa N, Fujimura L,
Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M and
Matsubara H: miR-145, miR-133a and miR-133b: Tumor-suppressive
miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J
Cancer. 127:2804–2814. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Isozaki Y, Hoshino I, Nohata N, Kinoshita
T, Akutsu Y, Hanari N, Mori M, Yoneyama Y, Akanuma N, Takeshita N,
et al: Identification of novel molecular targets regulated by tumor
suppressive miR-375 induced by histone acetylation in esophageal
squamous cell carcinoma. Int J Oncol. 41:985–994. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Takeshita N, Mori M, Kano M, Hoshino I,
Akutsu Y, Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, et
al: miR-203 inhibits the migration and invasion of esophageal
squamous cell carcinoma by regulating LASP1. Int J Oncol.
41:1653–1661. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Takeshita N, Hoshino I, Mori M, Akutsu Y,
Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, Akanuma N, et
al: Serum microRNA expression profile: miR-1246 as a novel
diagnostic and prognostic biomarker for oesophageal squamous cell
carcinoma. Br J Cancer. 108:644–652. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Akanuma N, Hoshino I, Akutsu Y, Murakami
K, Isozaki Y, Maruyama T, Yusup G, Qin W, Toyozumi T, Takahashi M,
et al: MicroRNA-133a regulates the mRNAs of two invadopodia-related
proteins, FSCN1 and MMP14, in esophageal cancer. Br J Cancer.
110:189–198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hoshino I, Akutsu Y, Murakami K, Akanuma
N, Isozaki Y, Maruyama T, Toyozumi T, Matsumoto Y, Suito H,
Takahashi M, et al: Histone demethylase LSD1 inhibitors prevent
cell growth by regulating gene expression in esophageal squamous
cell carcinoma cells. Ann Surg Oncol. 23:312–320. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
McGrath J and Trojer P: Targeting histone
lysine methylation in cancer. Pharmacol Ther. 150:1–22. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Allfrey VG and Mirsky AE: Structural
modifications of histones and their possible role in the regulation
of RNA synthesis. Science. 144:5591964. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shi Y, Lan F, Matson C. Mulligan P,
Whetstine JR, Cole PA, Casero RA and Shi Y: Histone demethylation
mediated by the nuclear amine oxidase homolog LSD1. Cell.
119:941–953. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lv T, Yuan D, Miao X, Lv Y, Zhan P, Shen X
and Song Y: Over-expression of LSD1 promotes proliferation,
migration and invasion in non-small cell lung cancer. PLoS One.
7:e350652012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kashyap V, Ahmad S, Nilsson EM, Helczynski
L, Kenna S, Persson JL, Gudas LJ and Mongan NP: The lysine specific
demethylase-1 (LSD1/KDM1A) regulates VEGF-A expression in prostate
cancer. Mol Oncol. 7:555–566. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao ZK, Yu HF, Wang DR, Dong P, Chen L,
Wu WG, Ding WJ and Liu YB: Overexpression of lysine specific
demethylase 1 predicts worse prognosis in primary hepatocellular
carcinoma patients. World J Gastroenterol. 18:6651–6656. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lim S, Janzer A, Becker A, Zimmer A,
Schüle R, Buettner R and Kirfel J: Lysine-specific demethylase 1
(LSD1) is highly expressed in ER-negative breast cancers and a
biomarker predicting aggressive biology. Carcinogenesis.
31:512–520. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kerenyi MA, Shao Z, Hsu YJ, Guo G, Luc S,
O'Brien K, Fujiwara Y, Peng C, Nguyen M and Orkin SH: Histone
demethylase Lsd1 represses hematopoietic stem and progenitor cell
signatures during blood cell maturation. Elife. 2:e006332013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ueda R, Suzuki T, Mino K, Tsumoto H,
Nakagawa H, Hasegawa M, Sasaki R, Mizukami T and Miyata N:
Identification of cell-active lysine specific demethylase
1-selective inhibitors. J Am Chem Soc. 131:17536–17537. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hamada S, Suzuki T, Mino K, Koseki K,
Oehme F, Flamme I, Ozasa H, Itoh Y, Ogasawara D, Komaarashi H, et
al: Design, synthesis, enzyme-inhibitory activity, and effect on
human cancer cells of a novel series of jumonji domain-containing
protein 2 histone demethylase inhibitors. J Med Chem. 53:5629–5638.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Etani T, Suzuki T, Naiki T, Naiki-Ito A,
Ando R, Iida K, Kawai N, Tozawa K, Miyata N, Kohri K and Takahashi
S: NCL1, a highly selective lysine-specific demethylase 1
inhibitor, suppresses prostate cancer without adverse effect.
Oncotarget. 6:2865–2878. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pitroda SP, Wakim BT, Sood RF, Beveridge
MG, Beckett MA, MacDermed DM, Weichselbaum RR and Khodarev NN:
STAT1-dependent expression of energy metabolic pathways links
tumour growth and radioresistance to the Warburg effect. BMC Med.
7:682009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Y, Liu T, Meyer CA, Eeckhoute J,
Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W and
Liu XS: Model-based analysis of ChIP-Seq (MACS). Genome Biol.
9:R1372008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ji X, Li W, Song J, Wei L and Liu XS:
CEAS: Cis-regulatory element annotation system. Nucleic Acids Res
34 (Web Server Issue). W551–W554. 2006. View Article : Google Scholar
|
|
35
|
Shin H, Liu T, Manrai AK and Liu XS: CEAS:
Cis-regulatory element annotation system. Bioinformatics.
25:2605–2606. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Quinlan AR and Hall IM: BEDTools: A
flexible suite of utilities for comparing genomic features.
Bioinformatics. 26:841–842. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Anders S and Huber W: Differential
expression analysis for sequence count data. Genome Biol.
11:R1062010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zheng YC, Yu B, Jiang GZ, Feng XJ, He PX,
Chu XY, Zhao W and Liu HM: Irreversible lsd1 inhibitors:
Application of tranylcypromine and its derivatives in cancer
treatment. Curr Top Med Chem. 16:2179–2188. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Theisen ER, Gajiwala S, Bearss J, Sorna V,
Sharma S and Janat-Amsbury M: Reversible inhibition of lysine
specific demethylase 1 is a novel anti-tumor strategy for poorly
differentiated endometrial carcinoma. BMC Cancer. 14:7522014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Maes T, Mascaró C, Tirapu I, Estiarte A,
Ciceri F, Lunardi S, Guibourt N, Perdones A, Lufino MMP,
Somervaille TCP, et al: ORY-1001, a potent and selective covalent
KDM1A inhibitor, for the treatment of acute leukemia. Cancer Cell.
33:495–511.e12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Maes T, Carceller E, Salas J, Ortega A and
Buesa C: Advances in the development of histone lysine demethylase
inhibitors. Curr Opin Pharmacol. 23:52–60. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Maiques-Diaz A and Somervaille TC: LSD1:
Biologic roles and therapeutic targeting. Epigenomics. 8:1103–1116.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kidger AM, Rushworth LK, Stellzig J,
Davidson J, Bryant CJ, Bayley C, Caddye E, Rogers T, Keyse SM and
Caunt CJ: Dual-specificity phosphatase 5 controls the localized
inhibition, propagation, and transforming potential of ERK
signaling. Proc Natl Acad Sci USA. 114:E317–E326. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yan X, Liu L, Li H, Huang L, Yin M, Pan C,
Qin H and Jin Z: Dual specificity phosphatase 5 is a novel
prognostic indicator for patients with advanced colorectal cancer.
Am J Cancer Res. 6:2323–2333. 2016.PubMed/NCBI
|
|
45
|
Hwang JH, Joo JC, Kim DJ, Jo E, Yoo HS,
Lee KB, Park SJ and Jang IS: Cordycepin promotes apoptosis by
modulating the ERK-JNK signaling pathway via DUSP5 in renal cancer
cells. Am J Cancer Res. 6:1758–1771. 2016.PubMed/NCBI
|
|
46
|
Asanoma K, Liu G, Yamane T, Miyanari Y,
Takao T, Yagi H, Ohgami T, Ichinoe A, Sonoda K, Wake N and Kato K:
Regulation of the mechanism of TWIST1 transcription by BHLHE40 and
BHLHE41 in cancer cells. Mol Cell Biol. 35:4096–4109. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Furukawa H, Makino T, Yamasaki M, Tanaka
K, Miyazaki Y, Takahashi T, Kurokawa Y, Nakajima K, Takiguchi S,
Mori M and Doki Y: PRIMA-1 induces p53-mediated apoptosis by
upregulating Noxa in esophageal squamous cell carcinoma with TP53
missense mutation. Cancer Sci. 109:412–421. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Saha MN, Jiang H, Yang Y, Reece D and
Chang H: PRIMA-1Met/APR-246 displays high antitumor activity in
multiple myeloma by induction of p73 and Noxa. Mol Cancer Ther.
12:2331–2341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zandi R, Selivanova G, Christensen CL,
Gerds TA, Willumsen BM and Poulsen HS: PRIMA-1Met/APR-246 induces
apoptosis and tumor growth delay in small cell lung cancer
expressing mutant p53. Clin Cancer Res. 17:2830–2841. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liang Y, Besch-Williford C and Hyder SM:
PRIMA-1 inhibits growth of breast cancer cells by re-activating
mutant p53 protein. Int J Oncol. 35:1015–1023. 2009.PubMed/NCBI
|
|
51
|
Li XL, Zhou J, Chan ZL, Chooi JY, Chen ZR
and Chng WJ: PRIMA-1met (APR-246) inhibits growth of colorectal
cancer cells with different p53 status through distinct mechanisms.
Oncotarget. 6:36689–36699. 2015.PubMed/NCBI
|
|
52
|
Lu T, Zou Y, Xu G, Potter JA, Taylor GL,
Duan Q, Yang Q, Xiong H, Qiu H, Ye D, et al: PRIMA-1Met suppresses
colorectal cancer independent of p53 by targeting MEK. Oncotarget.
7:83017–83030. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lehmann S, Bykov VJ, Ali D, Andrén O,
Cherif H, Tidefelt U, Uggla B, Yachnin J, Juliusson G, Moshfegh A,
et al: Targeting p53 in vivo: A first-in-human study with
p53-targeting compound APR-246 in refractory hematologic
malignancies and prostate cancer. J Clin Oncol. 30:3633–3639. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Miyazaki T, Kato H, Nakajima M, Faried A,
Takita J, Sohda M, Fukai Y, Yamaguchi S, Masuda N, Manda R, et al:
An immunohistochemical study of TIMP-3 expression in oesophageal
squamous cell carcinoma. Br J Cancer. 91:1556–1560. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Apte SS, Olsen BR and Murphy G: The gene
structure of tissue inhibitor of metalloproteinases (TIMP)-3 and
its inhibitory activities define the distinct TIMP gene family. J
Biol Chem. 270:14313–14318. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Visse R and Nagase H: Matrix
metalloproteinases and tissue inhibitors of metalloproteinases:
Structure, function, and biochemistry. Circ Res. 92:827–839. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ahonen M, Baker AH and Kahari VM:
Adenovirus-mediated gene delivery of tissue inhibitor of
metalloproteinases-3 inhibits invasion and induces apoptosis in
melanoma cells. Cancer Res. 58:2310–2315. 1998.PubMed/NCBI
|
|
58
|
Smith MR, Kung H, Durum SK, Colburn NH and
Sun Y: TIMP-3 induces cell death by stabilizing TNF-alpha receptors
on the surface of human colon carcinoma cells. Cytokine. 9:770–780.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Baker AH, George SJ, Zaltsman AB, Murphy G
and Newby AC: Inhibition of invasion and induction of apoptotic
cell death of cancer cell lines by overexpression of TIMP-3. Br J
Cancer. 79:1347–1355. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Bond M, Murphy G, Bennett MR, Amour A,
Knauper V, Newby AC and Baker AH: Localization of the death domain
of tissue inhibitor of metalloproteinase-3 to the N terminus.
Metalloproteinase inhibition is associated with proapoptotic
activity. J Biol Chem. 275:41358–41363. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ahonen M, Poukkula M, Baker AH, Kashiwagi
M, Nagase H, Eriksson JE and Kähäri VM: Tissue inhibitor of
metalloproteinases-3 induces apoptosis in melanoma cells by
stabilization of death receptors. Oncogene. 22:2121–2134. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Coutinho-Camillo CM, Lourenço SV, Nonogaki
S, Vartanian JG, Nagai MA, Kowalski LP and Soares FA: Expression of
PAR-4 and PHLDA1 is prognostic for overall and disease-free
survival in oral squamous cell carcinomas. Virchows Arch.
463:31–39. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Neef R, Kuske MA, Pröls E and Johnson JP:
Identification of the human PHLDA1/TDAG51 gene: down-regulation in
metastatic melanoma contributes to apoptosis resistance and growth
deregulation. Cancer Res. 62:5920–5929. 2002.PubMed/NCBI
|
|
64
|
Nagai MA, Fregnani JH, Netto MM, Brentani
MM and Soares FA: Down-regulation of PHLDA1 gene expression is
associated with breast cancer progression. Breast Cancer Res Treat.
106:49–56. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Oberst MD, Beberman SJ, Zhao L, Yin JJ,
Ward Y and Kelly K: TDAG51 is an ERK signaling target that opposes
ERK-mediated HME16C mammary epithelial cell transformation. BMC
Cancer. 8:1892008. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Park CG, Lee SY, Kandala G, Lee SY and
Choi Y: A novel gene product that couples TCR signaling to
Fas(CD95) expression in activation-induced cell death. Immunity.
4:583–591. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Gomes I, Xiong W, Miki T and Rosner MR: A
proline- and glutamine-rich protein promotes apoptosis in neuronal
cells. J Neurochem. 73:612–622. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hossain GS, van Thienen JV, Werstuck GH,
Zhou J, Sood SK, Dickhout JG, de Koning AB, Tang D, Wu D, Falk E,
et al: TDAG51 is induced by homocysteine, promotes
detachment-mediated programmed cell death, and contributes to the
cevelopment of atherosclerosis in hyperhomocysteinemia. J Biol
Chem. 278:30317–30327. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ju JA and Gilkes DM: RhoB: Team oncogene
or team tumor suppressor? Genes (Basel). 9(pii): E672018.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Parri M and Chiarugi P: Rac and Rho
GTPases in cancer cell motility control. Cell Commun Signal.
8:232010. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Vega FM and Ridley AJ: Rho GTPases in
cancer cell biology. FEBS Lett. 582:2093–2101. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Porter AP, Papaioannou A and Malliri A:
Deregulation of Rho GTPases in cancer. Small GTPases. 7:123–138.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Bhavsar PJ, Infante E, Khwaja A and Ridley
AJ: Analysis of Rho GTPase expression in T-ALL identifies RhoU as a
target for Notch involved in T-ALL cell migration. Oncogene.
32:198–208. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Poveda J, Sanz AB, Fernandez-Fernandez B,
Carrasco S, Ruiz-Ortega M, Cannata-Ortiz P, Ortiz A and
Sanchez-Niño MD: MXRA5 is a TGF-β1-regulated human protein with
anti-inflammatory and anti-fibrotic properties. J Cell Mol Med.
21:154–164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
He Y, Chen X, Liu H, Xiao H, Kwapong WR
and Mei J: Matrix-remodeling associated 5 as a novel tissue
biomarker predicts poor prognosis in non-small cell lung cancers.
Cancer Biomark. 15:645–651. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang GH, Yao L, Xu HW, Tang WT, Fu JH, Hu
XF, Cui L and Xu XM: Identification of MXRA5 as a novel biomarker
in colorectal cancer. Oncol Lett. 5:544–548. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Buckanovich RJ, Sasaroli D,
O'Brien-Jenkins A, Botbyl J, Hammond R, Katsaros D, Sandaltzopoulos
R, Liotta LA, Gimotty PA and Coukos G: Tumor vascular proteins as
biomarkers in ovarian cancer. J Clin Oncol. 25:852–861. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Xiong D, Li G, Li K, Xu Q, Pan Z, Ding F,
Vedell P, Liu P, Cui P, Hua X, et al: Exome sequencing identifies
MXRA5 as a novel cancer gene frequently mutated in non-small cell
lung carcinoma from Chinese patients. Carcinogenesis. 33:1797–1805.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Huang T, Sun L, Yuan X and Qiu H:
Thrombospondin-1 is a multifaceted player in tumor progression.
Oncotarget. 8:84546–84558. 2017.PubMed/NCBI
|
|
80
|
Zhou ZQ, Cao WH, Xie JJ, Lin J, Shen ZY,
Zhang QY, Shen JH, Xu LY and Li EM: Expression and prognostic
significance of THBS1, Cyr61 and CTGF in esophageal squamous cell
carcinoma. BMC Cancer. 9:2912009. View Article : Google Scholar : PubMed/NCBI
|