Introduction
Cervical cancer is a malignant neoplasm of the
cervix. It may present with vaginal bleeding but is often
asymptomatic until the cancer is in its advanced stages (1,2). The
methods of treatment for this cancer consist of surgery (including
local excision) in the early stages, and chemotherapy and
radiotherapy in the advanced stages. Radiotherapy uses certain
types of radiation, including X-rays, γ-rays or particles, to
shrink tumors or eliminate cancer cells by damaging DNA and
subsequently causing cell death or the inability to proliferate. In
recent years, attempts have been made to identify
radiation-sensitive genes for the purpose of elucidating the
complex mechanisms of cellular responses to ionizing radiation, or
for the identification of biomarkers for radiosensitive individuals
(3–5). Previous studies have performed
microarray analysis on various types of normal and cancer cells,
including HeLa cells (6,7). The expression of the immediate early
response 5 (IER5) gene has been revealed to change in a
dose- and time-dependent manner in various types of cells,
following radiation (6).
However, the effect of the atypical expression of
IER5 in HeLa cells on radiosensitivity and tumor growth
remain unclear. In order to understand the role of the IER5
gene in radiotherapy for cervical cancer in the present study, RNA
interference technology was used to silence IER5 in HeLa
cells, and overexpression plasmids were used to upregulate the
gene. The results of these experiments indicated that the
expression of IER5 is involved in the radiation-induced cell
death. In addition, 5-week-old BALB/C nude mice were inoculated
with three types of HeLa cells (normal, IER5-knockdown and
IER5-overexpression) and the sizes of the resulting tumors
were recorded. Based on these investigations and observations, the
response of IER5 expression to γ-ray radiation was examined, as was
its involvement in cell cycle checkpoint control and cell
survival.
Materials and methods
Cell culture
Three versions of HeLa cells (obtained by the
Laboratory of Beijing Institute of Radiation Medicine), including
unmodified cells, cells transfected with small interfering (si)RNA
targeting IER5 mRNA (IER5-siRNA-HeLa) and cells
overexpressing IER5 (IER5-overexpression-HeLa), were
cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen;
Thermo Fisher Scientific, Inc.), supplemented with 10% fetal bovine
serum (PAN-Seratech, Inc.), 100 U/ml penicillin and 100 g/ml
streptomycin. The cells were cultured in a humid atmosphere with 5%
CO2 at 37°C.
Irradiation
The exponentially growing cells were irradiated with
1.7 Gy/min for 2.4 min (total 4Gy) or 1.2 min (total 2 Gy), using a
60Co γ-ray source at room temperature. For the mock
radiation control, the cells were placed in the radiation room for
the same duration as the corresponding treatment groups, but the
cobalt source remained underwater and was barricaded. Following
irradiation, the cells were cultured, harvested and prepared for
the subsequent experiments.
RNA isolation and reverse
transcription (RT)
Total RNA was isolated from the irradiated and
mock-irradiated cells using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) according to the
manufacturer's protocol. The total RNA concentration was determined
using spectrometry, and the quality was determined by 1%
formaldehyde agarose gel electrophoresis. Total RNA was reverse
transcribed into cDNA using a ProtoScript™ First Strand cDNA
Synthesis kit (New England BioLabs, Inc., Ipswich, MA, USA),
according to the manufacturer's protocol. The synthesized cDNA was
stored at −80°C and analyzed directly by quantitative polymerase
chain reaction (qPCR) analysis.
The sequences of siRNAs targeting the IER5
gene (GenBank accession no. NM_016545) were analyzed using the
Basic Local Alignment Search Tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to ensure
they did not target any other gene transcripts. Two siRNAs
targeting IER5 mRNA were screened and the oligonucleotides
were synthesized. The sequences encoding IER5 siRNA were as
follows (targeting sequences are underlined): IER5−27 siRNA,
sense 5′-GATCCGCCTCATCAGCATCTTCGGTTTCAAGAGAACCGAAGATGCTGATGAGGTTTTTTGGAAA-3′
and antisense 5′-AGCTTTTCCAAAAAACCTCATCAGCATCTTCGGTTCTCTTGAAACCGAAGATGCTGATGAGGCG-3′;
IER5−16 siRNA, sense 5′-GATCCGCTGCATAAGAACCTCCTGTTCAAGAGACAGGAGGTTCTTATGCAGCTTTTTTGGAAA-3′
and antisense 5′-AGCTTTTCCAAAAAAGCTGCATAAGAACCTCCTGTCTCTTGAACAGGAGGTTCTTATGCAGCG-3′.
HindIII and BamHI restriction sites were added
upstream and downstream of each oligonucleotide. Following
annealing, these duplex oligonucleotides were inserted between the
HindIII and BamHI sites of the pSilencer™ 3.1 vector
(Ambion; Thermo Fisher Scientific, Inc.) to generate pSilencerIER5
siRNA vectors. All constructs were sequence-verified prior to use.
A vector containing the following non-specific siRNA (NSsiRNA) was
used as the experimental control: NSsiRNA, sense
5′-GATCCCACTACCGTTGTTATAGGTGTTCAAGAGACACCTATAACAACGGTAGTTTTTTTTGGAAA-3′
and antisense 5′-AGCTTTTCCAAAAAAAACTACCGTTGTTATAGGTGTCTCTTGAACACCTATAACAACGGTAGTGG-3′.
Cell transfection
For vector DNA transfection, the HeLa cells were
plated in 60-mm Petri dishes at a density of 5×105 cells
per dish. The cells were transfected with the pSilencerIER5 siRNA
vector using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA)
reagent 24 h after plating, according to the manufacturer's
protocol. The transfected cultures were sub-cultured into new 60-mm
dishes after 24 h and maintained in conditioned DMEM supplemented
with 250 µg/ml hygromycin B (Roche Diagnostics, Basel, Switzerland)
to select for transfected clones.
Construction of IER5-overexpression
vectors
Based on the GenBank IER5 gene sequence, the
following primers were designed for PCR amplification of a 986-bp
segment: Sense 5′-GACGAATTCAATGGAGTTCAAGCTG-3′ and antisense
5′-GTAGCACCGGAAGACTAGATCTCAG-3′. The plasmid for the overexpression
of IER5 (defined as IER5-overexpression) was
constructed by PCR, and the 986-bp amplicon was inserted into the
EcoRI-XbaI site of the pCMV-3×FLAG vector
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) using Lipofectamine
2000 (Sigma-Aldrich; Merck KGaA), according to the manufacturer's
protocol. Briefly, HeLa cells (~1.5×104) were plated in
6-well plates and growth medium was removed from the plates when
the cells reached ~80% confluence. A total of 500 µl transfection
mixture containing 1.25 µg plasmid DNA and 3.75 µl Lipofectamine
2000 reagent was added to the plates. The cells were incubated at
37°C for 5 h, gently overlaid with 2 ml pre-warmed complete growth
medium and incubated for an additional 5 days. The transfected
cells were then cultured in medium containing 400 µg/ml neomycin
(G418) for 14 days for stress selection. The selective media were
replaced every 3–4 days. The surviving transfected cells localized
in distinct ‘islands’ were maintained with growth medium.
Individual clones were transferred to 96-well plates for
proliferation using standard techniques (cloning cylinders). The
vector without the IER5 gene fragment was used as a control
(Non-IER5-overexpressing). Stable transfectants were used in
the subsequent experiments.
Determination of mRNA and protein
expression levels of IER5
For confirmation, the expression levels of
IER5 in the transfectants were measured using RT-qPCR and
western blot analyses. The mRNA levels were quantified by RT-qPCR
analysis using an ABI 7300 Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The reaction mixture
contained 10 µl DyNAmo™ SYBR® Green qPCR Master mix
containing the modified Thermus brockianus hot-start DNA
polymerase (Finnzymes Oy, Espoo, Finland), 0.2 µM each primer and 2
µl cDNA template (obtained by reverse transcription, as described
above), to a final volume of 20 µl. The RT-qPCR was performed in
accordance with the conditions: 95°C 5 min; 35 PCR cycles (95°C, 5
sec; 57°C, 15 sec; 72°C, 15 sec); extension at 72°C for 5 min and
the cycle at which the fluorescent signal crossed the detection
threshold was denoted as the cycle threshold. Relative mRNA levels
normalized to endogenous β-actin mRNA levels are presented relative
to IER5 mRNA levels using the 2-ΔΔCq method (8). Each PCR was run in triplicate in three
independent experiments. The primers for amplification of human
IER5 were as follows: Forward 5′-CCGGGAACGTGGCTAACC-3′ and
reverse 5′-TTCCGTAGGAGTCCCGAGAA-3′; and those for human β-actin
were as follows: Forward 5′GCGCGGCTACAGCTTCA-3′ and reverse
5′-CTTAATGTCACGCACGATTTCC-3′.
For the western blot analysis, the cells were
harvested, washed three times with PBS and then lysed using RIPA
buffer. The protein concentrations were determined using a BCA
protein assay kit (Thermo Scientific Pierce, Micro BCA™ Protein
Assay kit). Equal quantities of total protein (60 µg per lysate)
were separated by 10% sodium dodecylsulfate-polyacrylamide gel
electrophoresis and electroblotted onto a nitrocellulose membrane.
The nitrocellulose membrane was then placed in 5% blocking buffer
made up with skim milk powder at room temperature for 1–2 h on the
shaker. The membrane was first hybridized with antibody against
IER5 (cat. no. ARP56939-P050; Santa Cruz Biotechnology, Inc., Santa
Cruz, CA, USA) and β-actin (cat. no. 4970; Cell Signaling
Technology, Inc., Beverly, MA, USA) overnight at 4°C. The dilution
ratio of antibody to IER5 and β-actin was 1:1,000 and 1:5,000,
respectively. The hybridized nitrocellulose membrane was washed
with 1×TBST (0.1% Tween) for 5 times, each time for 8 min, followed
by incubation with the sheep anti-mouse (cat. no. A0408; Beyotime
Institute of Biotechnology, Beijing, China) diluted to 1:3,000 for
1 h at room temperature. The specific band that reflected the IER5
protein was visualized with enhanced chemiluminescence (ECL)
reagents (GE Healthcare, Chicago, IL, USA), followed by subsequent
exposure of the blot onto a film (Eastman Kodak Company, Rochester,
NY). The protein loading was standardized using β-actin. The films
were developed using ECL methods.
Cell proliferation and clonogenic
survival analysis
For the cell proliferation assays,
1.5×104 cells per well were seeded in 6-well culture
plates, cultured for 24 h and subjected to γ-ray irradiation. The
cell numbers in three wells were counted every day following
radiation. The culture medium was replaced on day 3. Three
independent experiments were performed, and the mean cell numbers
were used to generate a growth curve.
For the clonogenic survival assays, exponentially
growing cells were collected and diluted into appropriate
concentrations and exposed to a 60Co γ-ray source at a
dose rate of 1.74 Gy min−1. The corresponding controls
were mock-irradiated. Immediately following radiation, an
appropriate number of cells (100–2,000, depending on the radiation
dose) were plated into 60-mm diameter Petri dishes. Each experiment
was performed in triplicate. The culture medium was replaced 1 week
post-radiation. After 12 days of culture, the cells were mixed with
methanol for 5 min at room temperature, stained with Giemsa
solution for 3 min at 37°C, and colonies that consisted of >50
cells were counted by laser confocal microscope (magnification,
×400; Olympus Coorperation).
Cell cycle analysis by flow
cytometry
Following irradiation with a dose of 2 Gy, the cells
were harvested at the designed time point and mixed with 75%
ethanol. The cells were then resuspended in PBS with 0.1% saponin
and 1 µg/ml RNase A (Sigma-Aldrich; Merck KGaA), incubated for 20
min at 37°C and stained with 25 µg/ml propidium iodide (PI;
Sigma-Aldrich; Merck KGaA). Cell cycle distribution was evaluated
by flow cytometry (>10,000 cells per sample). For the detection
of cell mitosis, the fixed cells were treated with 0.5% Triton ×100
for 15 min and incubated in PBS containing 0.5 µg/ml mouse
monoclonal antibody against H3 pSer10 (cat. no. 53348; Cell
Signaling Technology, Inc., Danvers, MA, USA) for 30 min at room
temperature. Following two washing steps with PBS, the cells were
incubated with fluorescein isothiocyanate-conjugated goat
anti-mouse secondary antibody (cat. no. TA130013; OriGene
Technologies, Inc., Beijing, China) for 30 min at room temperature,
and resuspended in PBS containing 10 µg/ml PI and 10 µg/ml RNase A
at 37°C for 1 h in the dark. The volume ratio of antibody against
H3 pSer10 and the secondary antibody to 1% bovine serum albumin
solution was 1:400 and 1:200, respectively. The stained cells were
analyzed by flow cytometry. Two independent experiments were
performed.
Tumorigenic ability of cells
Adult male BALB/c-nu host mice (5 weeks old; ~15 g
weight; Beijing Vital River Laboratory Animal Technology Co., Ltd.)
were housed and cared for in compliance with the regulations of the
Ministry of Health and the Experimental Animal Center of Capital
Medical University (Beijing, China). The Committee for Animal Use
at the Capital Medical University approved all experimental
procedures performed in the present study. Water and food were
available ad libitum in the cages. The ambient temperature
was 22°C, relative humidity was 60% and the air was changed every
12h. The mice were allowed to acclimate for 1 week prior to
treatment.
The three cell lines (IER5-siRNA-HeLa, HeLa and
IER5-overexpression-HeLa) were cultured in DMEM containing 10%
fetal bovine serum in an incubator with 5% CO2 at 37°C.
The exponentially growing cells were trypsinized with 0.25% trypsin
digestion and washed twice with 1X PBS. Following centrifugation at
1,000 × g for 3 min at room temperature, the three cell lines were
suspended and 1×107 cells per cell line were injected
into the front armpits of BALB/c-nu nude mice (the inoculation
sites on the nude mouse skin were disinfected with tincture of
iodine and alcohol prior to inoculation) using the sterile
syringes, each with 0.5 ml cell suspension (containing
~1×107 cells).
The above experiment was repeated three times with
three different groups. Each animal was inoculated with
1×107 cells, which grew into tumors under their skin. On
day 28 of inoculation of the nude mice with the above-mentioned
cells, the mice were removed for weighing and injected with sodium
pentobarbital into the abdominal cavity at a dose of 50 mg/kg. In
their anesthetized state, the mice were sacrificed by cervical
dislocation, following which the tumors were weighed on being
removed from under the skin.
Statistical analysis
One-way analysis of variance (ANOVA) and two-tailed
t-tests were used to compare differences among groups. The
Student-Newman-Keuls test was used as the post hoc test following
ANOVA. P<0.05 was considered to indicate a statistically
significant difference. Statistical analyses were performed using
SPSS 19.0 software (IBM Corp., Armonk, NY, USA). Data are presented
as the mean ± standard error of the mean in all figures.
Results
Cell lines with atypical expression of
IER5
Two siRNA molecules targeting IER5 were
designed and separately transfected into HeLa cells, and a number
of stably transfected clones (27siRNA-C1,
27siRNA-C2, 27siRNA-C3,
27siRNA-C4, 16siRNA-C1 and
NSsiRNA-C1) were selected. The total RNA was isolated
and used to quantify the expression level of IER5 by RT-qPCR
analysis. As demonstrated in Table
I, the expression of IER5 in clone 27siRNA-C2
was suppressed the most. This clone was derived from HeLa cells
transfected with the IER5−27 siRNA vector. In addition,
western blot analysis revealed that the protein expression of IER5
in clone 27SiRNA-C2 was markedly lower than that in the
NSsiRNA-C1 and untransfected HeLa cells. Therefore, the
27siRNA-C2 clone was selected for subsequent
investigation of the influence of the inhibited expression of
IER5 on the cellular response to radiation and was labeled
IER5-siRNA-HeLa. Subsequently, the 27siRNA-C2,
NSsiRNA-C1 and untransfected HeLa cells were separately
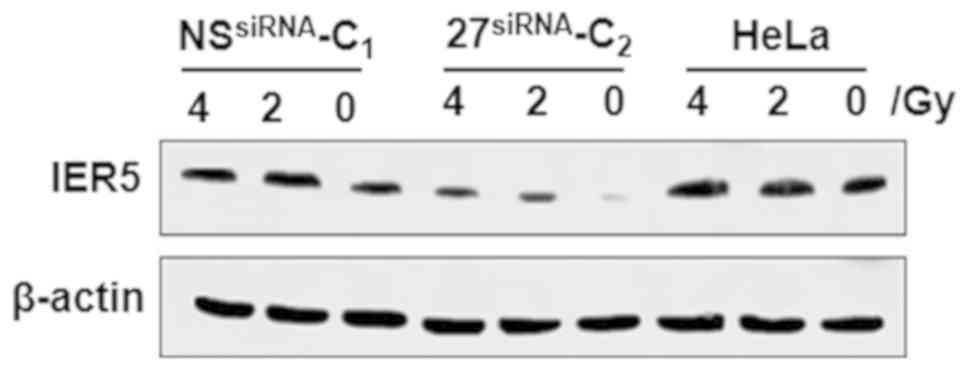
exposed to radiation. As shown in Fig.
1, western blot analysis revealed that the NSsiRNA-C1 and
untransfected HeLa cells had higher protein expression of IER5,
whereas the 27siRNA-C2 HeLa cells exhibited no notable
increase in IER5 protein.
 | Table I.mRNA expression levels of IER5
in IER5-silenced HeLa cells determined by reverse
transcription-quantitative polymerase chain reaction analysis. |
Table I.
mRNA expression levels of IER5
in IER5-silenced HeLa cells determined by reverse
transcription-quantitative polymerase chain reaction analysis.
| Sample | IER5 Cq | β-actin Cq | ΔCq | ΔΔCq | siRNA/control
(2−ΔΔCq) |
|---|
|
27siRNA-C1 | 23.139 | 19.476 | 3.914 | 4.372 |
2−4.372 |
|
27siRNA-C2 | 24.276 | 15.985 | 8.291 | 8.794 |
2−8.794 |
|
27siRNA-C3 | 23.076 | 15.594 | 7.482 | 7.940 |
2−7.94 |
|
27siRNA-C4 | 23.595 | 21.390 | 2.205 | 2.663 |
2−2.663 |
|
16siRNA-C1 | 24.675 | 21.877 | 2.798 | 3.256 |
2−3.256 |
|
NSsiRNA-C1 | 23.614 | 31.524 | −7.910 | −6.452 |
2+6.452 |
| Control | 31.342 | 31.800 | −0.458 | <0.001 | 2°=1 |
Three IER5-overexpression clones,
3overexpression-C1, 3overexpression-C2 and
3overexpression-C3, were selected. The total RNA from
each clone was isolated and used to quantify the expression of
IER5 by RT-qPCR analysis. Table
II demonstrates that the expression of IER5 was highest
in clone 3overexpression-C2, which was generated from
cells transfected with IER5-overexpression-3. Therefore, clone
3overexpression-C2 was selected for the subsequent
experiments, and labeled as IER5-overexpression-HeLa cells.
 | Table II.mRNA expression levels of IER5
in IER5-overexpressed HeLa cells determined by reverse
transcription-quantitative polymerase chain reaction analysis. |
Table II.
mRNA expression levels of IER5
in IER5-overexpressed HeLa cells determined by reverse
transcription-quantitative polymerase chain reaction analysis.
| Sample | IER5 Cq | β-actin Cq | ΔCq | ΔΔCq |
IER5-overexpression-HeLa/control
(2−ΔΔCq) |
|---|
|
3overexpression-C1 | 17.245 | 22.142 | −4.897 | −4.001 |
24.001 |
|
3overexpression-C2 | 17.823 | 23.639 | −5.816 | −4.920 |
24.920 |
|
3overexpression-C3 | 18.502 | 21.674 | −3.172 | −2.276 |
22.276 |
| Control | 32.3672 | 33.258 | −0.896 | <0.001 | 2°=1 |
Overexpression of IER5 decreases the
proliferation and survival of HeLa cells following radiation
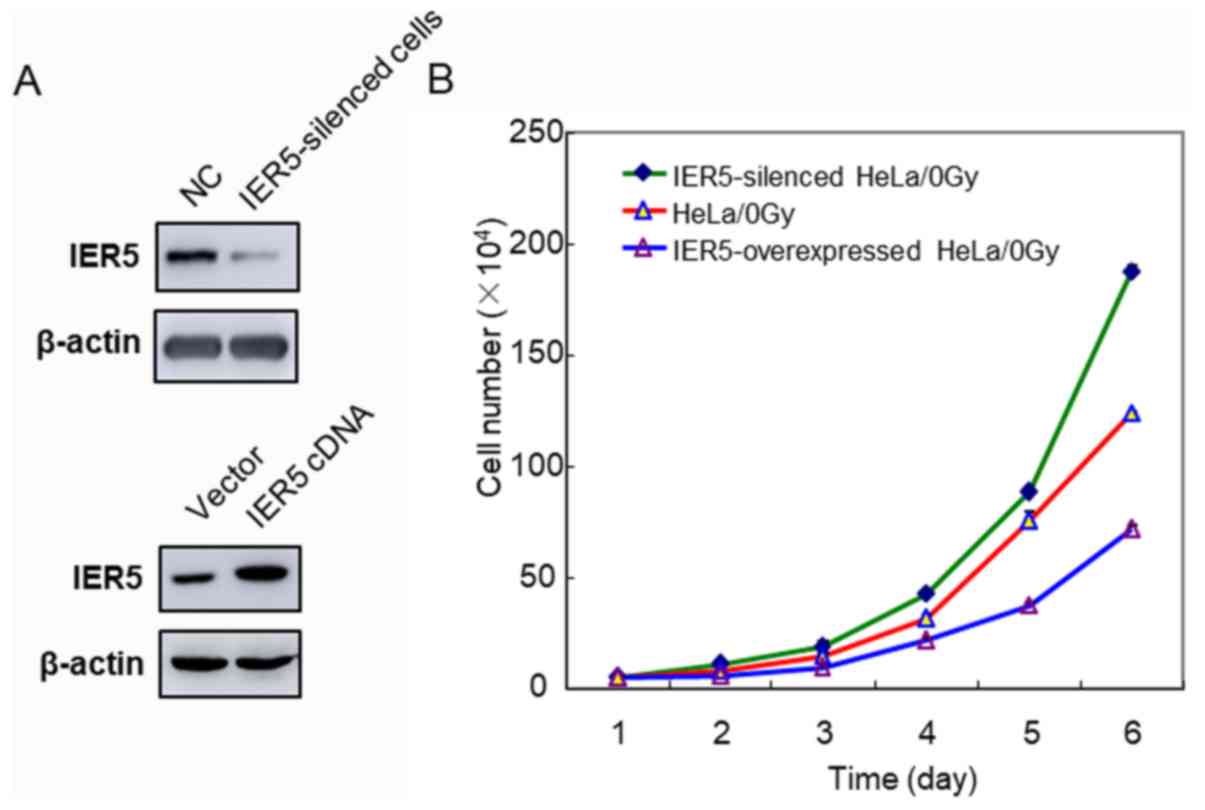
First, the proliferation abilities of the
IER5-siRNA-HeLa and IER5-overexpression-HeLa cells were
investigated. On day 6 post-seeding, the IER5-siRNA-HeLa cells
exhibited significantly higher proliferation than the normal HeLa
cells (Fig. 2A and B). By contrast,
the proliferation rate of the IER5-overexpression-HeLa cells was
lower than that of the normal HeLa cells.
Furthermore, the influence of atypical expression of
IER5 on the proliferation of HeLa cells following
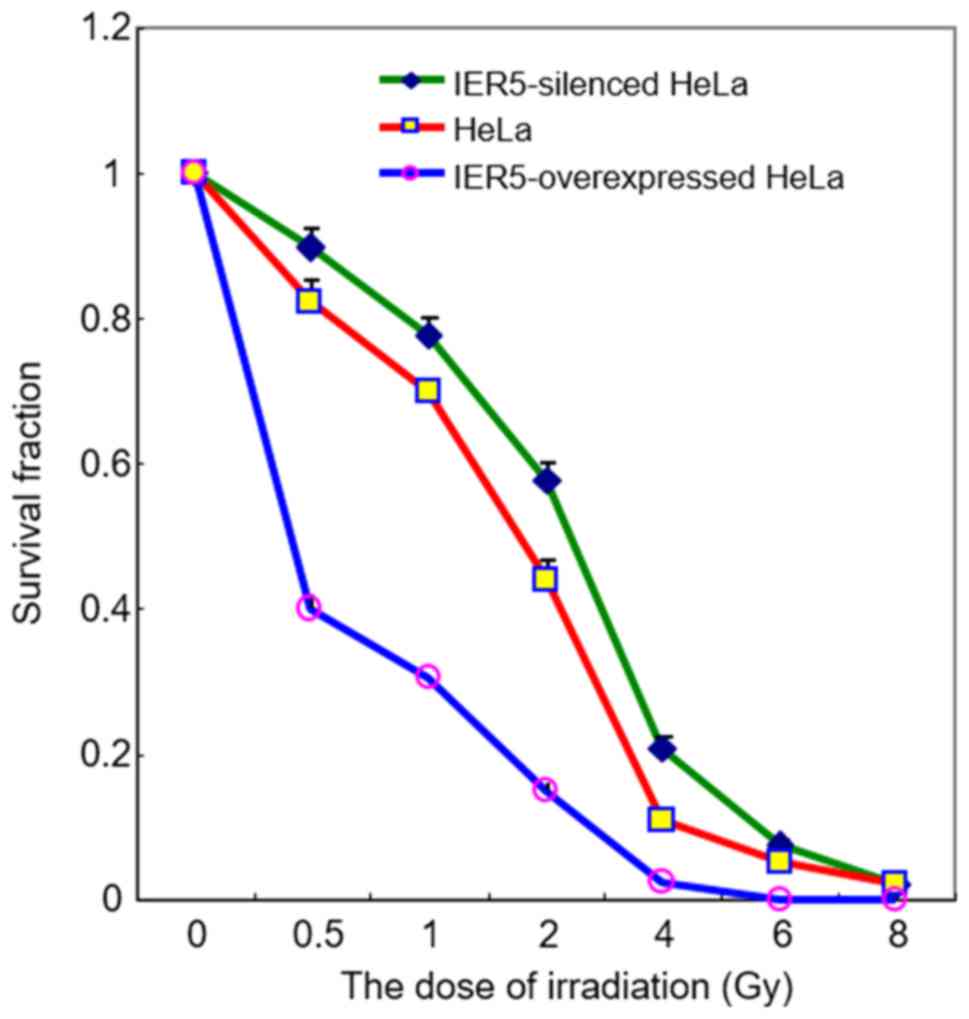
irradiation was examined. The clonogenic survival assay
demonstrated that the overexpression of IER5 significantly
increased the sensitivity of HeLa cells to radiation (with 0.5, 1,
2, 4 and 6 Gy) compared with normal expression of IER5
(Fig. 3). By contrast, the survival
rate of the IER5-siRNA-HeLa cells following radiation was higher
than that of the normal HeLa cells. These results indicate that the
overexpression of IER5 suppressed the growth of HeLa cells
and increased the radiosensitivity of HeLa cells.
Overexpression of IER5 induces
G0/G1 arrest in HeLa cells
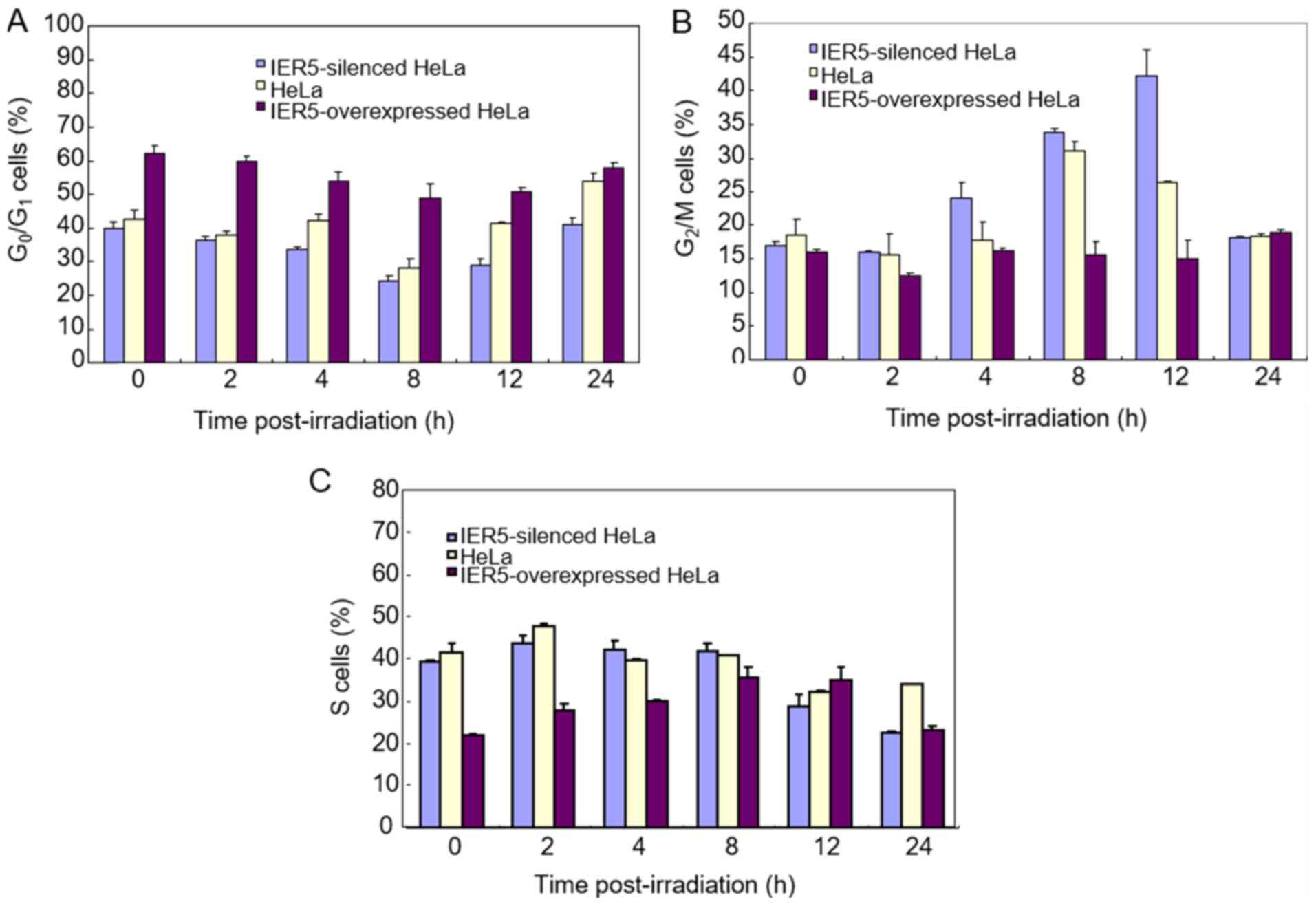
The results of the flow cytometry demonstrated that
a higher percentage of the IER5-overexpression-HeLa cells were in
the G0/G1 phase compared with that for the
normal or IER5-siRNA-HeLa cells, either prior to or following
radiation (Fig. 4A). The fraction of
cells in the G0/G1 phase was the smallest for
the cells with downregulated IER5. As shown in Fig. 4B, the proportion of IER5-siRNA-HeLa
cells in the G2/M phase following radiation was higher
than that for the other two cell types, and the proportion in the
IER5-overexpression-HeLa cells was lowest at 12 h post-radiation.
The number of cells in the S phase was higher in the normal HeLa
cells following irradiation (Fig.
4C).
Inhibition of IER5 promotes
tumorigenesis in nude mice
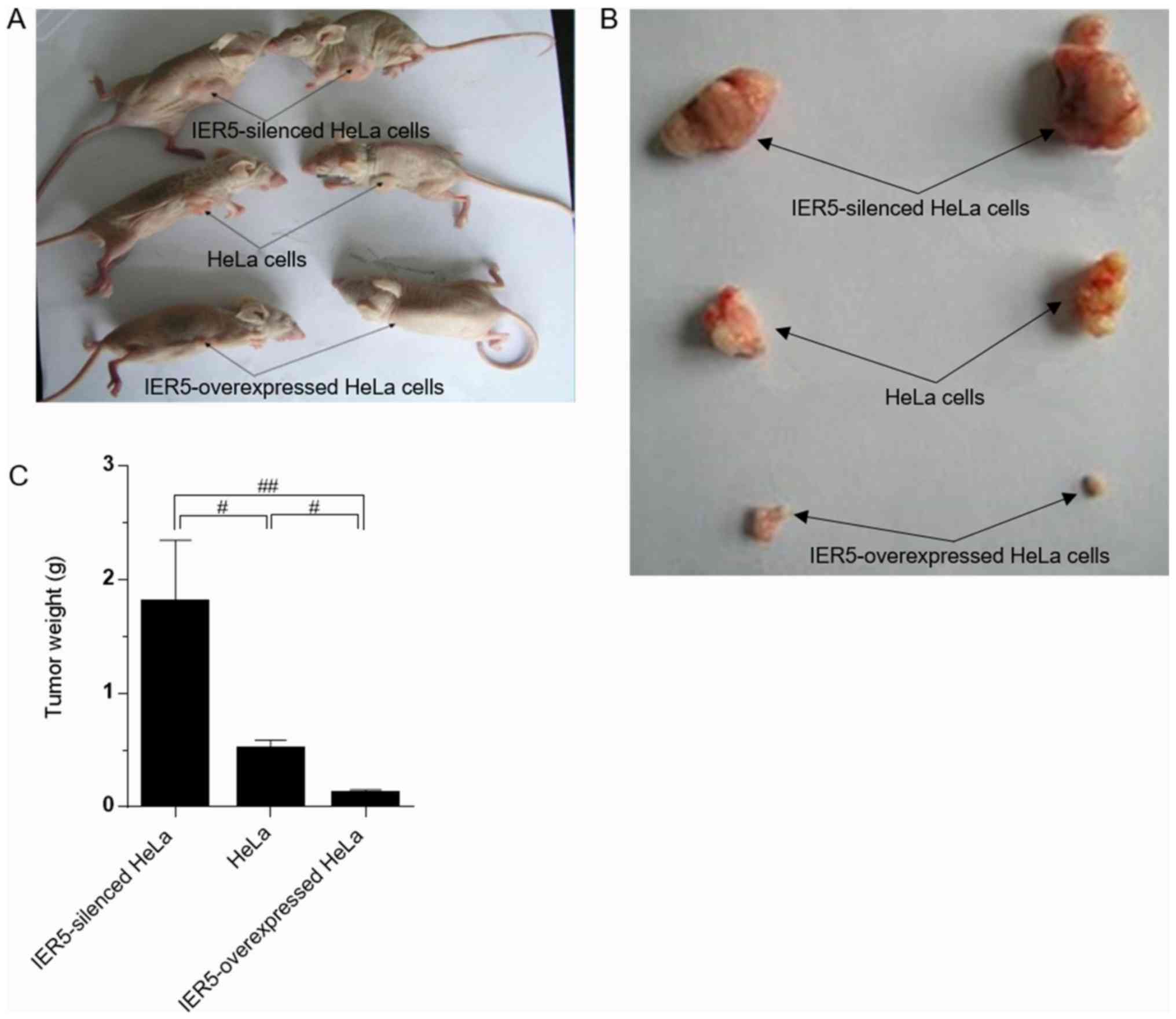
The mice were sacrificed 4 weeks after inoculation
of the three cell types, and any resulting tumors were weighed. The
average weight of the xenograft tumors resulting from the
IER5-siRNA-HeLa cells was markedly higher than that of tumors
derived from the normal or IER5-overexpression-HeLa cells (Fig. 5A-C). The maximum diameter of a single
subcutaneous tumor was 8.3×14.2×11.2 mm (1.32 mm3), and
the average size of the fastest growing type of tumor was 1.0
mm3. The smallest tumors were observed in the mice
inoculated with IER5-overexpression-HeLa cells.
Discussion
Radiotherapy is particularly effective against
tumors, and is one of the three conventional methods of tumor
treatment. This method can effectively prevent the growth of
certain tumors and prolong the life of a patient. However,
radioresistance is a major obstacle in radiation treatment
(9–11). In order to improve the quality and
sensitivity of radiation treatment, substantial research has
focused on identifying radiosensitivity-associated genes (12–18). If
the expression of a gene can respond to radiation at certain doses
and simultaneously cause the apoptosis and/or suppression of tumor
cell proliferation, this can be useful for the treatment of cancer.
Furthermore, the pursuit of radiosensitive genes may help define a
dosage standard for radiotherapy based on radiation-induced
damage.
Based on these reasons, microarray technology was
used to select candidate radiosensitive genes. Using this
state-of-the-art assay, it was found that the mRNA expression of
IER5 was upregulated following radiation (6). IER5, which belongs to the
immediate early response gene family, is located in chromosome 1
(NM-016545), is 2,369 bp long and encodes 308 amino acids (19). The activation of early response genes
is considered to be an important primary response to external
stimulation in cells (20,21). It has been found that IER5 is
overexpressed in the condition of wakefulness and sleep deprivation
(21). During the protein- and
peptide-bound polysaccharide-induced apoptosis of early human
promyelocytic leukemia cells (HL-60), IER5 likely serves a
critical role in the formation of brain vessels, and changes occur
in its expression in the process of valproic acid-induced neural
tube defects, which suggests that this gene likely regulates cell
cycle (22). In addition, chromatin
immunoprecipitation assays have confirmed that radiation induces
the overexpression of IER5 (6,7).
In the present study, experiments were performed in
order understand the effects of radiation on the IER5 gene
and its mechanism of action. First, two stable transfection cell
lines were established by methods of gene silencing and
overexpression. Compared with the vector control cells, higher
percentages of the IER5-siRNA-HeLa cells were in the S and
G2/M phases. These results indicate that IER5 may
be involved in cell proliferation and tumorigenesis. Of note,
IER5 knockdown increased the radioresistance of cells. The
IER5-overexpression-HeLa cells exhibited the opposite
characteristics of the IER5-siRNA-HeLa cells. For example, the
proliferation rate of the IER5-overexpression-HeLa cells was lower.
In addition, the ratio of cells in the G0/G1
phase was higher than that of the normal and IER5-siRNA-HeLa cells.
The results of this experiment suggest that the IER5 gene is
involved in cell cycle progression and influences the course of
cell division. The present findings demonstrated that radiation
inhibited cell proliferation and, among the three cell types
assessed, had the greatest impact on the IER5-overexpression-HeLa
cells, providing further evidence for the role of IER5 in
the DNA damage checkpoint. In addition, the silencing of
IER5 accelerated cell division, leading to a larger
proportion of cells in the G2 phase. This increase may
be caused by more efficient arrest of early S phase cells, although
a failure of the G1/S checkpoint cannot be excluded. It has been
suggested that cell cycle arrest provides the time necessary for
irradiated cells to repair DNA lesions and ensure precise
chromosome segregation prior to continuation of the cell cycle
(6). Therefore, the increased
radioresistance that resulted from suppressing the expression of
IER5 may be attributed, at least in part, to the activation of cell
cycle checkpoints.
The three types of cell lines (normal,
IER5-overexpression-HeLa and IER5-siRNA-HeLa) were injected
subcutaneously into the limbs of nude mice. Tumors of various sizes
resulted from all inoculations. The weights of the xenograft tumors
resulting from the IER5-siRNA-HeLa cells were the highest on
average. This finding was in agreement with the in vitro
experiments, suggesting that the IER5 gene has a biological
function in cell division and proliferation. Although the signal
transduction pathways of IER5 were not examined in the
present study, a previous study revealed the regulating mechanism
of the IER5 gene (23). The
results of this study also support the findings of the present
study to a certain extent.
Radiotherapy is normally used in the treatment of
tumors. The radiation administered not only inhibits tumor cell
proliferation but also causes more tumor cells to undergo
apoptosis. Any genes affected by radiation can cause cancer cells
to undergo apoptosis by physical stimulation; such resulting
sensitivity to radiation is an important finding in cancer
research. The investigations in the present study were designed to
examine whether a gene results in characteristics of tumor growth
inhibition under conditions of radiation. The xenograft mouse model
experiment further demonstrated the consequences of IER5
dysregulation. The tumorigenic capacity of the
IER5-overexpression-HeLa cells in nude mice was the poorest, and
that of the IER5-siRNA-HeLa cells was the highest. The consistency
between the in vivo and in vitro experimental results
has important implications for future investigations of
radiotherapy methods for cervical cancer. Certainly, further
investigations are required, including those into the regulation
mechanism of IER5 and optimization of the effect of
radiotherapy in cervical cancer.
In conclusion, the IER5 gene affects the
radiosensitivity of HeLa cells by decreasing DNA repair and
dysregulating cell cycle checkpoints, and it serves an important
role in radiation-induced cell death. The present study
demonstrated that radiation induced the upregulation of IER5
mRNA, that IER5 gene modulation affected radiosensitivity
and that the overexpression of IER5 decelerated the
proliferation of HeLa cells. These results suggest that IER5
may be a potential radiotherapeutic target for cervical cancer.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the National
Natural Science Foundation of China (grant nos. 31770907 and
31640022), the Beijing Natural Science Foundation (grant no.
7172146) and the Natural Science Foundation of Liaoning Province
(grant no. 2015020287).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KKD, FY, HQJ, ZQY, XDL, YMW, PKZ and CJY performed
the genetic experiments, participated in the sequence alignment and
drafted the manuscript. KKD and XZ conceived the study,
participated in its design and coordination, and helped to draft
the manuscript. LYD, WC, QG and XYJ participated in the design of
the study and performed the statistical analyses. LLY participated
in the sequence alignment analysis. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The Committee for Animal Use at the Capital Medical
University approved all experimental procedures performed in the
present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yu C, Zhu W, Ji Y, Guo J, Pan P, Han J and
Zhou X: A comparative study of intensity-modulated radiotherapy and
standard radiation field with concurrent chemotherapy for local
advanced cervical cancer. Eur J Gynaecol Oncol. 36:278–282.
2015.PubMed/NCBI
|
|
2
|
Fu ZC, Wang FM and Cai JM: Gene expression
changes in residual advanced cervical cancer after radiotherapy:
Indicators of poor prognosis and radioresistance? Med Sci Monit.
21:1276–1287. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chu TY, Yang JT, Huang TH and Liu HW:
Crosstalk with cancer-associated fibroblasts increases the growth
and radiation survival of cervical cancer cells. Radiat Res.
181:540–547. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xiang L, Xie G, Liu C, Zhou J, Chen J, Yu
S, Li J, Pang X, Shi H and Liang H: Knock-down of glutaminase 2
expression decreases glutathione, NADH, and sensitizes cervical
cancer to ionizing radiation. Biochim Biophys Acta. 1833:2996–3005.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yuan W, Xiaoyun H, Haifeng Q, Jing L,
Weixu H, Ruofan D, Jinjin Y and Zongji S: MicroRNA-218 enhances the
radiosensitivity of human cervical cancer via promoting radiation
induced apoptosis. Int J Med Sci. 11:691–696. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ding KK, Shang ZF, Hao C, Xu QZ, Shen JJ,
Yang CJ, Xie YH, Qiao C, Wang Y, Xu LL and Zhou PK: Induced
expression of the IER5 gene by gamma-ray irradiation and its
involvement in cell cycle checkpoint control and survival. Radiat
Environ Biophys. 48:205–213. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kis E, Szatmári T, Keszei M, Farkas R,
Esik O, Lumniczky K, Falus A and Sáfrány G: Microarray analysis of
radiation response genes in primary human fibroblasts. Int J Radiat
Oncol Biol Phys. 66:1506–1514. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang XQ, Chen X, Xie XX, Zhou Q, Li K, Li
S, Shen LF and Su J: Co-expression of CD147 and GLUT-1 indicates
radiation resistance and poor prognosis in cervical squamous cell
carcinoma. Int J Clin Exp Pathol. 7:1651–1666. 2014.PubMed/NCBI
|
|
10
|
Gaffney DK, Jhingran A, Portelance L,
Viswanathan A, Schefter T, Weidhaas J and Small W Jr: Radiation
therapy oncology group gynecologic oncology working group:
Comprehensive results. Int J Gynecol Cancer. 24:956–962. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zheng L, Tang W, Wei F, Wang H, Liu J, Lu
Y, Cheng Y, Bai X, Yu X and Zhao W: Radiation-inducible protein
RbAp48 contributes to radiosensitivity of cervical cancer cells.
Gynecol Oncol. 130:601–608. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Moreno-Acosta P, Gamboa O, Sanchez de
Gomez M, Cendales R, Diaz GD, Romero A, Balart Serra J, Conrado Z,
Levy A, Chargari C and Magné N: IGF1R gene expression as a
predictive marker of response to ionizing radiation for patients
with locally advanced HPV16-positive cervical cancer. Anticancer
Res. 32:4319–4325. 2012.PubMed/NCBI
|
|
13
|
Halle C, Andersen E, Lando M, Aarnes EK,
Hasvold G, Holden M, Syljuåsen RG, Sundfør K, Kristensen GB, Holm
R, et al: Hypoxia-induced gene expression in chemoradioresistant
cervical cancer revealed by dynamic contrast-enhanced MRI. Cancer
Res. 72:5285–5295. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Baiocchi G, Begnami MD, Fukazawa EM,
Oliveira RA, Faloppa CC, Kumagai LY, Badiglian-Filho L, Pellizzon
AC, Maia MA, Jacinto AA, et al: Prognostic value of nuclear factor
kappa B expression in patients with advanced cervical cancer
undergoing radiation therapy followed by hysterectomy. J Clin
Pathol. 65:614–618. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Paul S, Barker CA, Turner HC, McLane A,
Wolden SL and Amundson SA: Prediction of in vivo radiation dose
status in radiotherapy patients using ex vivo and in vivo gene
expression signatures. Radiat Res. 175:257–265. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim WY, Lee JW, Park YA, Choi JJ, Sung CO,
Song SY, Choi CH, Kim TJ, Huh SJ, Kim BG and Bae DS: RAR-beta
expression is associated with early volumetric changes to radiation
therapy in cervical cancer. Gynecol Obstet Invest. 71:11–18. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kabacik S, Mackay A, Tamber N, Manning G,
Finnon P, Paillier F, Ashworth A, Bouffler S and Badie C: Gene
expression following ionising radiation: Identification of
biomarkers for dose estimation and prediction of individual
response. Int J Radiat Biol. 87:115–129. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Filiano AN, Fathallah-Shaykh HM, Fiveash
J, Gage J, Cantor A, Kharbanda S and Johnson MR: Gene expression
analysis in radiotherapy patients and C57BL/6 mice as a measure of
exposure to ionizing radiation. Radiat Res. 176:49–61. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Williams M, Lyu MS, Yang YL, Lin EP,
Dunbrack R, Birren B, Cunningham J and Hunter K: Ier5, a novel
member of the slow-kinetics immediate-early genes. Genomics.
55:327–334. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Göttgens B, Barton LM, Chapman MA,
Sinclair AM, Knudsen B, Grafham D, Gilbert JG, Rogers J, Bentley DR
and Green AR: Transcriptional regulation of the stem cell leukemia
gene (SCL)-comparative analysis of five vertebrate SCL loci. Genome
Res. 12:749–759. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cirelli C and Tononi G: Gene expression in
the brain across the sleep-waking cycle. Brain Res. 885:303–321.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zeng F, Hon CC, Sit WH, Chow KY, Hui RK,
Law IK, Ng VW, Yang XT, Leung FC and Wan JM: Molecular
characterization of Coriolus versicolor PSP-induced apoptosis in
human promyelotic leukemic HL-60 cells using cDNA microarray. Int J
Oncol. 27:513–523. 2005.PubMed/NCBI
|
|
23
|
Nakamura S, Nagata Y, Tan L, Takemura T,
Shibata K, Fujie M, Fujisawa S, Tanaka Y, Toda M, Makita R, et al:
Transcriptional repression of Cdc25B by IER5 inhibits the
proliferation of leukemic progenitor cells through NF-YB and p300
in acute myeloid leukemia. PLoS One. 6:e280112011. View Article : Google Scholar : PubMed/NCBI
|