Introduction
Pancreatic ductal adenocarcinoma (PDAC) accounts for
80–90% of all cases of pancreatic cancer and is an epithelial
malignancy that originates from the cells of the ducts or ductules
(1,2). It is well known that PDAC can
disseminate rapidly to the lymphatic system and distant organs
(3). In previous reports, 43,090
cases of pancreatic cancer-associated mortality were predicted to
occur in the United States in 2017, and it is gradually becoming
the second leading cause of cancer-associated mortality (4,5). At
present, surgical resection, radiochemotherapy and
molecular-targeted therapy are considered to be effective
treatments for patients with PDAC. However, almost all patients
have a typical clinical presentation of incurable disease at the
time of diagnosis (3). In addition,
the 5-year survival rates remain at a steady rate of <8%
(4,6). A previous etiological study (7) suggests that the development of PDAC is
spurred on by genetic and epigenetic events.
Numerous short, non-coding RNAs (ranging between 19
and 24 nt in length), collectively named microRNAs (miRNAs), are
now regarded as key components of the epigenetic machinery
(8). The regulation of endogenous
gene expression by miRNAs is an important mechanism in the
pathogenesis and progression of human malignancies (9). In mammals, miRNAs are considered to
inhibit gene expression through post-transcriptional regulation via
mRNA decay and/or translational repression in a sequence-specific
manner (10). In recent years, the
aberrant expression of miRNAs has been found to serve a critical
role in various fundamental biological and pathological processes,
including proliferation, differentiation, inflammation, apoptosis
and stress response (11).
miR-139-5p, a member of the miRNA family, is located on chromosome
11q13.4 (12). Previous studies have
shown that miR-139-5p is an epigenetically silenced tumor
suppressor miRNA in bladder cancer (13), lung cancer (14), hepatocellular carcinoma (15) and colorectal cancer (16). By contrast, miR-139-5p exerts
pro-oncogenic and pro-metastatic activities in adrenocortical
cancer cells though negatively regulating the expression of NDRG4
(17). Li et al (18) recently reported that reduced
miR-139-5p levels are associated with the pro-survival effect of
liraglutide on pancreatic tissues in diabetic rats and INS-1 cells.
However, the role and underlying mechanism of miR-139-5p in human
PDAC remain to be fully elucidated. Thymopoietin (TMPO), also known
as lamina-associated protein (LAP2), is an inner nuclear membrane
protein that has six alternatively spliced isoforms (19). It has been suggested that a complex
of TMPO and lamin A/C modulates multiple signaling pathways that
balance proliferation and differentiation (20). The interplay between TMPO and
barrier-to-autointegration factor in a sequence-independent manner
may affect the stabilization of chromatin structure (21). TMPO has been identified as an
oncogene in several types of cancer, including breast cancer, lung
cancer and glioblastoma (22).
Specifically, the regulation of cell motility, proliferation, cell
cycle distribution and apoptosis have been described for TMPO in a
various types of digestive tract cancer, glioblastoma and/or other
types of cancer (22,23). However, whether TMPO serves an
important role in PDAC and is regulated by miR-139-5p remains
unclear.
The present study evaluated patterns of miR-139-5p
expression in tumor tissues and tumor cell lines from patients with
PDAC, and attempted to identify PDAC-associated miRNAs. The
oncogene TMPO, a potential target of miR-139-5p, was revealed using
bioinformatics resources. Furthermore, the biological function of
the identified miR-139-5p/TMPO axis was determined to clarify the
molecular mechanisms implicated in the progression and tumor
biology of PDAC.
Materials and methods
Human tissue samples
A total of 40 pairs of fresh human PDAC tissues and
adjacent matched non-tumor tissues were obtained from patients
between August 2016 and October 2017 in the Department of
Hepatobiliary Pancreatic Surgery, Renmin Hospital, Hubei University
of Medicine (Hubei, China). Prior to tissue collection, it was
confirmed that no patients had received chemotherapy or radiation
therapy, and informed consent was obtained from all patients.
Patient information, including age, sex, tumor size, pTNM category
and differentiation, are summarized in Table I. The tumors were staged according to
the American Cancer Association TNM staging system (2010) (24). The collected tissue samples were
immediately snap-frozen in liquid nitrogen and stored at −80°C
until processed. The present study was approved by the Human Ethics
Review Committee of Renmin Hospital, Hubei University of
Medicine.
 | Table I.Clinicopathological features of the
patients with pancreatic ductal adenocarcinoma. |
Table I.
Clinicopathological features of the
patients with pancreatic ductal adenocarcinoma.
| Characteristics | Number of cases
(n=40) |
|---|
| Age, years |
|
| ≤60 | 29 |
|
>60 | 11 |
| Sex |
|
| Male | 25 |
|
Female | 15 |
| Tumor size, cm |
|
| ≤4 | 32 |
|
>4 | 8 |
| pTNM category |
|
|
I/II | 34 |
|
III | 6 |
|
Differentiation |
|
|
Well/moderate | 29 |
|
Poor | 11 |
Cell lines and culture
The human PDAC cell lines (SW1990, ASPC-1, BXPC-3
and PANC-1) and the HPNE human pancreatic duct epithelial cell line
were purchased from the American Type Culture Collection (Manassas,
VA, USA). The SW1990, PANC-1 and HPNE cell lines were cultured in
Dulbecco's modified Eagle's medium (DMEM, Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The ASPC-1 and BXPC-3 were
grown in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.).
All media were supplemented with 10% fetal bovine serum (FBS), 100
IU/ml penicillin and 100 µg/ml streptomycin (Gibco; Thermo Fisher
Scientific, Inc.). All cell lines were maintained in a humidified
incubator containing 5% CO2 at 37°C.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
Total RNA was extracted from the patient tissues or
cells using RNAiso (Takara Biotechnology Co., Ltd., Dalian, China).
The complementary DNA was synthesized using PrimeScript™ RT Reagent
kit (Takara Biotechnology Co., Ltd.) according to the
manufacturer's instructions. For the miR-139-5p quantitative assay,
the Hairpin-it™ miRNA qPCR Quantitation kit (GenePharma Co., Ltd.,
Shanghai, China) was used to evaluate the expression of miR-139-5p,
with U6 as an internal control. For the TMPO mRNA quantitative
assay, the mRNA expression level of TMPO was determined using SYBR
Green Premix Ex Taq (Takara Biotechnology, Co., Ltd.) with GAPDH as
the internal control. The qPCR amplification was performed using an
ABI 7900 Real-Time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.) using the following the thermocycling conditions:
initial denaturation at 95°C for 10 min and 40 cycles of 95°C for
15 sec, and 60°C for 1 min. The relative expression of miR-139-5p
or TMPO was calculated using the 2−ΔΔCq method (25). The primer sequences for miR-139-5p,
U6, TMPO and GAPDH are listed in Table
II.
 | Table II.Primers used for reverse
transcription-quantitative polymerase chain reaction analysis. |
Table II.
Primers used for reverse
transcription-quantitative polymerase chain reaction analysis.
| Gene | Primer sequence
(5′-3′) |
|---|
| hsa-miR-139-5p | F:
TCTACAGTGCACGTGTCTCCAGT |
|
| R:
TGGAGACACGTGCACTGTAGATT |
| U6 | F:
CTCGCTTCGGCAGCACA |
|
| R:
AACGCTTCACGAATTTGCGT |
| TMPO | F:
TGCTCGCCTCCTGCCTGTAG |
|
| R:
GACACAAAGCCAAGCCAGACC |
| GAPDH | F:
TGTTCGTCATGGGTGTGAAC |
|
| R:
ATGGCATGGACTGTGGTCAT |
Cell transfection
The miR-139-5p mimics
(5′-UAAGAUACUUAUGGCUUUGUGAA-3′) and the negative control (miR-NC;
5′-UAAGAUGACUAUGGCUUUGCUGA-3′) were obtained from Guangzhou RiboBio
Co., Ltd. (Guangzhou, China). The pcDNA3.1-TMPO and pcDNA3.1 empty
vector were synthesized by Shanghai GeneChem Co., Ltd. (Shanghai,
China). Prior to transfection, the SW1990 and PANC-1 cells were
seeded in six-well plates at a density of 4×105 cells
per well. For the overexpression of miR-139-5p, the SW1990 and
PANC-1 cells were transfected with miR-139-5p mimics or miR-NC to
generate the miR-139-5p group and miR-NC group, respectively. In
the rescue experiment, the SW1990 cells were transfected with sole
pcDNA3.1 empty vector, pcDNA3.1-TMPO, or together with miR-139-5p
and vector or pcDNA3.1-TMPO, respectively for 48 h, followed by
subsequently experiments. All transfections were performed using
Lipofectamine™ 2000 reagent (Invitrogen; Thermo Fisher Scientific,
Inc.).
Bioinformatics analysis
The online software programs of miRanda (http://www.microrna.org/microrna/home.do), miRDB
(http://www.mirdb.org/), PicTar (http://pictar.mdc-berlin.de/) and TargetScan
(http://www.targetscan.org/vert_71/)
were searched to predict the putative target genes of
miR-139-5p.
Luciferase reporter assay
The psiCHECK-2 luciferase reporter plasmid
containing the wild-type (WT) or mutant type (MUT) TMPO was
generated by Guangzhou RiboBio Co., Ltd.. For the luciferase
reporter assay, the SW1990 and PANC-1 cells were co-transfected
with miR-139-5p mimics or miR-NC together with WT TMPO or MUT TMPO
for 48 h using Lipofectamine 2000, according to the manufacturer's
protocol. Subsequently, the luciferase activity was determined
using a dual-luciferase reporter assay kit (Promega Corporation,
Madison, WI, USA).
Western blot analysis
The transfected cells were washed with PBS and
incubated in lysis buffer with protease inhibitor (Pierce; Thermo
Fisher Scientific, Inc.), and protein concentrations were
determined using a BCA Protein Assay kit (Beyotime Institute of
Biotechnology, Beijing, China). Equal quantities of protein (30 µg)
were separated on 10% SDS-PAGE gels and transferred onto
polyvinylidene difluoride membranes. The membranes were blocked in
TBS/0.1% Tween containing 5% BSA (Beyotime Institute of
Biotechnology) at room temperature, followed by incubation with
primary antibodies against TMPO (1:1,000; cat. no. ab226348; Abcam,
Cambridge, UK) and GAPDH (1:5,000, cat. no. 10494-1-AP, ProteinTech
Group, Inc., Chicago, IL USA) overnight at 4°C. Following
incubation with horseradish peroxidase-conjugated secondary
antibody (1:5,000; cat. no. SC-2054; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) for 2 h at room temperature, the protein
bands were visualized using an Enhanced Chemiluminescence Western
Blotting kit (EMD Millipore, Billerica, MA, USA) and quantified
using Image-pro plus 6.0 software (National Institutes of
Health).
Cell Counting Kit-8 (CCK-8) assay
A CCK-8 assay (Beyotime Institute of Biotechnology)
was used to determine cell proliferation. In brief, cells from the
different groups were seeded at a density of 3×103 cells
in 96-well plates and cultured overnight at 37°C. Subsequently, 10
µl CCK-8 reagents were added to each well on 5 consecutive days and
each well was incubated for a further 2 h at 37°C. Finally, the
absorbance at 450 nm was measured using a microplate reader (Epoch,
BioTek Instruments, Inc., Winooski, VT, USA).
Colony formation assay
The transfected cells were seeded in 6-well plates
at a density of 500 cells per well and cultured for 10 days to form
colonies naturally. The colonies were then fixed in 4%
paraformaldehyde, subsequently stained with 1% crystal violet at
room temperature for 20 min, and then counted under a light
microscope (magnification, ×200).
Cell cycle analysis
The transfected cells were collected, washed with
pre-cooled PBS and fixed in 70% alcohol. Following washing twice
with PBS, the cells were centrifuged at 500 × g at 4°C for 5 min to
collect the precipitate, followed by incubation with 500 µl PBS
containing 50 µg/ml propidium iodide (PI, Nanjing KeyGen Biotech
Co., Ltd., Nanjing, China) solution and 100 µg/ml RNase for 30 min
at room temperature. Finally, the cells were analyzed for their
percentages at the G0/G1, S and G2/M phases by flow cytometry (BD
Biosciences, San Jose, CA, USA).
Analysis of cell apoptosis
The transfected cells were collected and seeded into
6-well plates (3×105/well). The samples were
subsequently digested in EDTA-free trypsin (Beyotime Institute of
Biotechnology) and stained with 5 µl Annexin V-FITC and 5 µl PI
(20%; Invitrogen; Thermo Fisher Scientific, Inc.) in the dark for
15 min at room temperature. The percentage of apoptotic cells,
including early apoptosis and late apoptosis was measured by flow
cytometry (BD Biosciences).
Statistical analysis
Data were analyzed using SPSS statistical software
18.0 (IBM Corp., Armonk, NY, USA) and expressed as the mean ±
standard deviation from at least three independent experiments.
Statistical difference between two groups or multiple groups were
evaluated using Student's t-test or one-way analysis of variance
followed by Dunnett's test, respectively. The association between
the expression of TMPO and miR-139-5p in PDAC tissues was
determined by Spearman's correlation analyses. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-139-5p is downregulated in PDAC
tissues and cell lines
To examine the biological function of miR-139-5p in
PDAC, RT-qPCR analysis was first performed to examine the
expression of miR-139-5p in 40 pairs of human PDAC tissues and
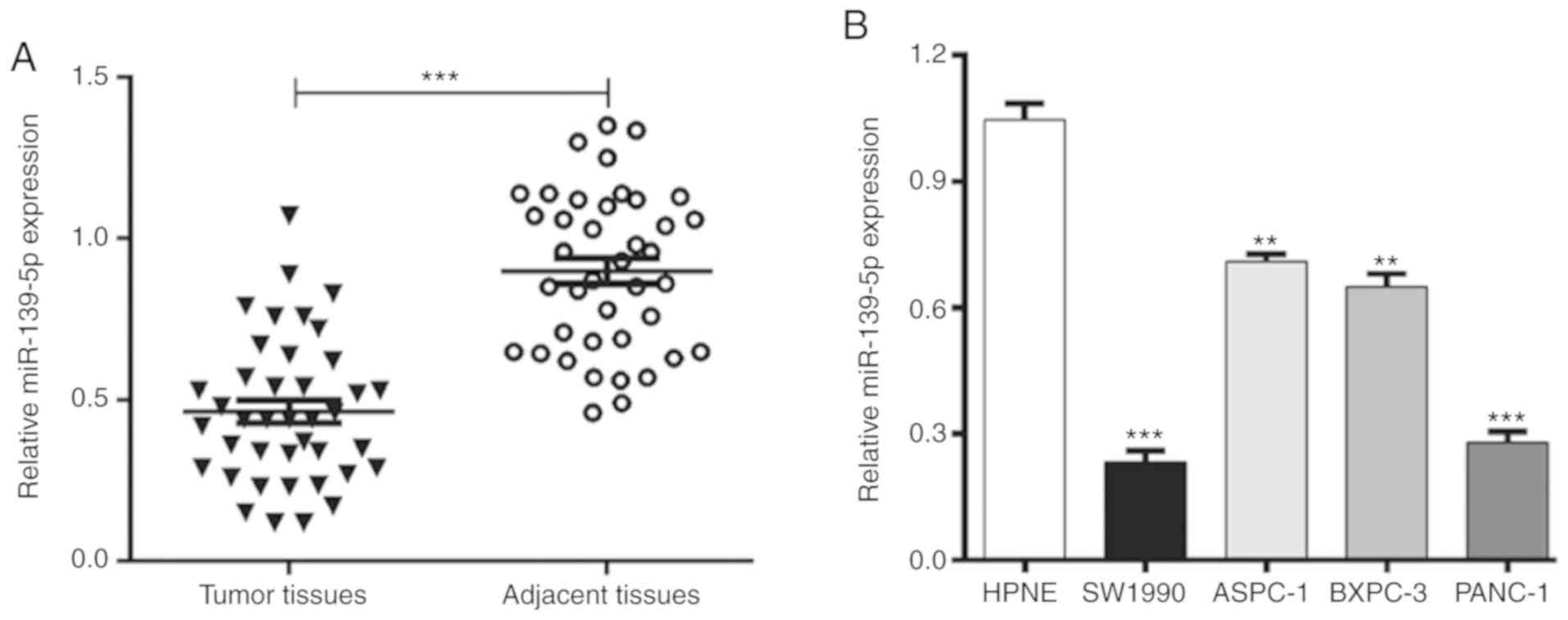
matched adjacent normal tissues. As shown in Fig. 1A, the expression levels of miR-139-5p
were significantly lower in the PDAC tissues than in the matched
adjacent normal tissues (P<0.001). Subsequently, the expression
levels of miR-139-5p were analyzed and compared between several
PDAC cell lines and the normal HPNE pancreatic duct epithelial cell
line. The results showed that the expression of miR-139-5p was
markedly downregulated in all PDAC cell lines (SW1990, ASPC-1,
BXPC-3 and PANC-1) compared with that in the HPNE cells (Fig. 1B). These data indicated that
downregulated miR-139-5p may be a critical regulator in the
progression of PDAC.
TMPO is a direct target of
miR-139-5p
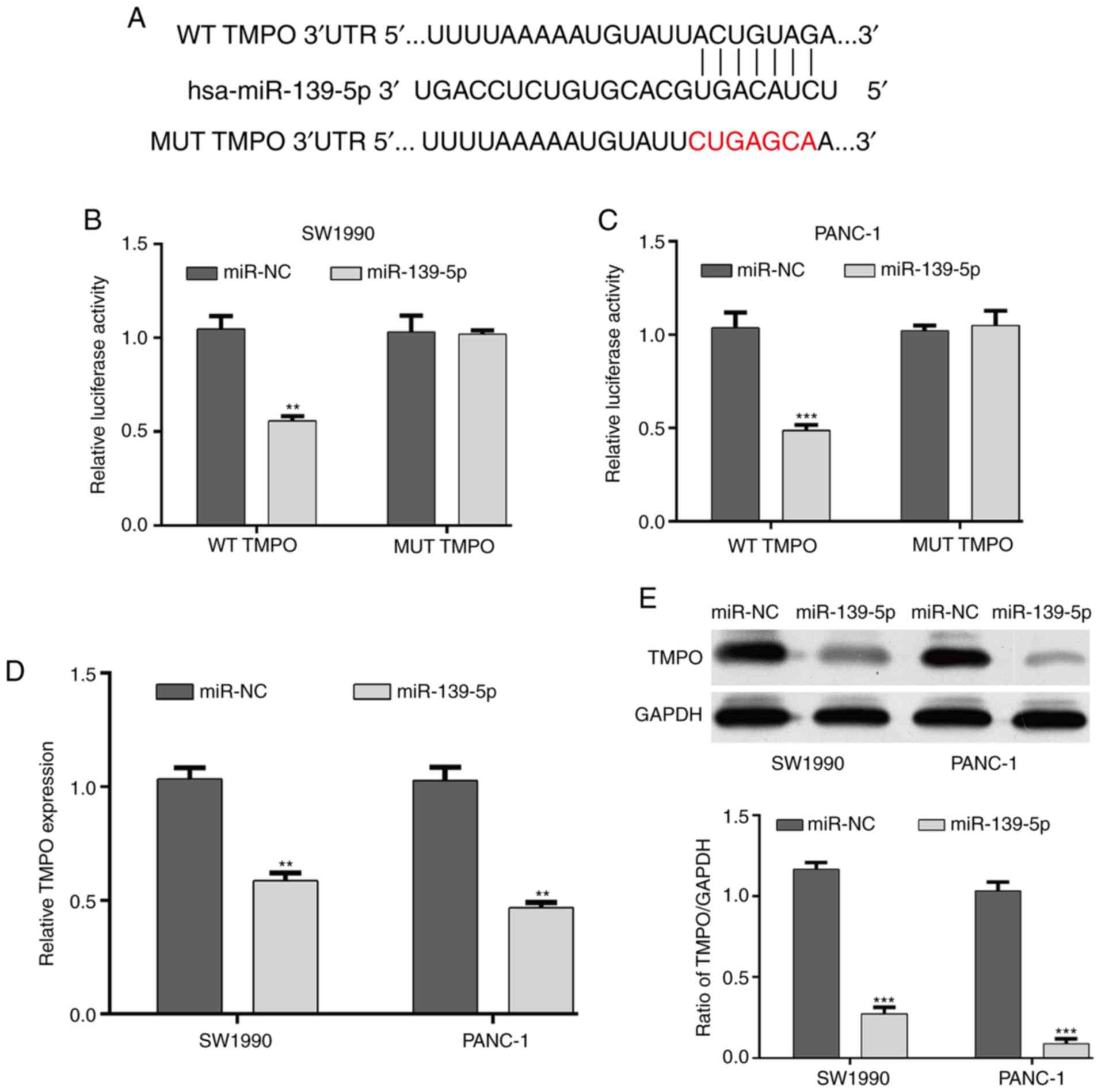
Using online bioinformatics analysis, TMPO was
predicted to be a target gene of miR-139-5p and the binding sites
are shown in Fig. 2A. Luciferase
reporter assays were then performed to confirm whether miR-139-5p
directly targeted TMPO. The results revealed that miR-139-5p
significantly reduced the luciferase activity of a
TMPO-3′-untranslated region (3′-UTR) WT reporter plasmid, but did
not affect the luciferase activity of a TMPO-3′-UTR MUT reporter in
SW1990 (Fig. 2B) and PANC-1
(Fig. 2C) cells. In addition, the
mRNA and protein expression levels of TMPO were determined in
SW1990 and PANC-1 cells under the regulation of miR-139-5p. As
shown in Fig. 2D and E, the
overexpression of miR-139-5p downregulated the expression of TMPO
in SW1990 and PANC-1 cells at the mRNA (P<0.01) and protein
(P<0.001) levels. These results suggested that TMPO was as a
direct target gene of miR-139-5p in PDAC cells.
TMPO is overexpressed in PDAC and
inversely correlated with miR-139-5p
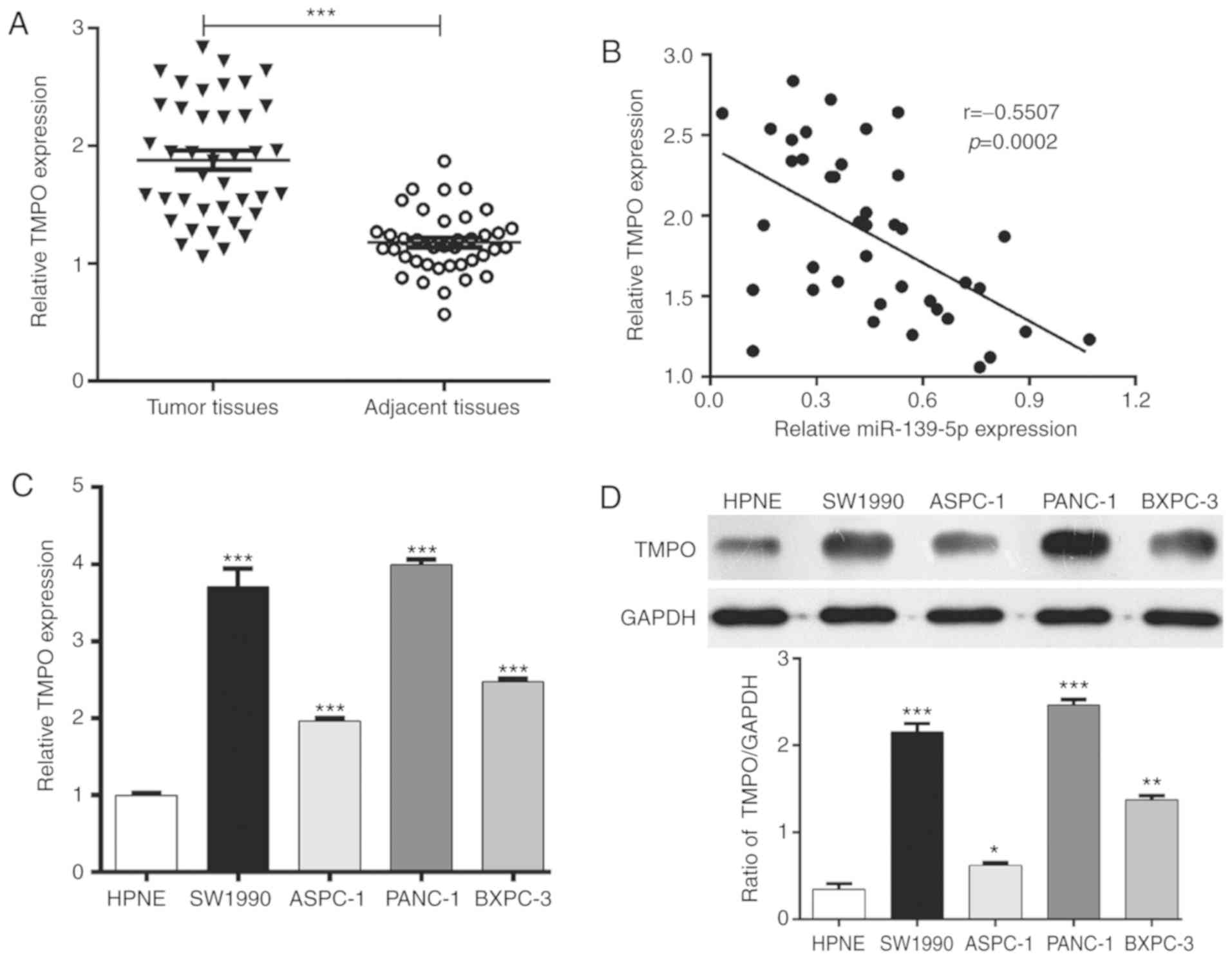
As TMPO is a target of miR-139-5p in PDAC cells, the
present study subsequently investigate the expression of TMPO in 40
pairs of human PDAC tissues and matched adjacent normal tissues
using RT-qPCR analysis. As presented in Fig. 3A, the expression of TMPO was
significantly upregulated in the PDAC tissues compared with that in
the adjacent normal tissues (P<0.001). Notably, the expression
level of miR-139-5p was inversely associated with the expression of
TMPO in the 40 paired PDAC specimens (Fig. 3B, r=−0.5507, P=0.0002). In addition,
RT-qPCR analysis revealed that the expression of TMPO was higher in
the PDAC cell lines compared with that in the HPNE cell line
(Fig. 3C, P<0.001), which was
consistent with the results of the western blot analysis (Fig. 3D). These observations indicated that
TMPO was frequently overexpressed in PDAC.
miR-139-5p affects cell proliferation,
cell cycle and apoptosis in PDAC cells
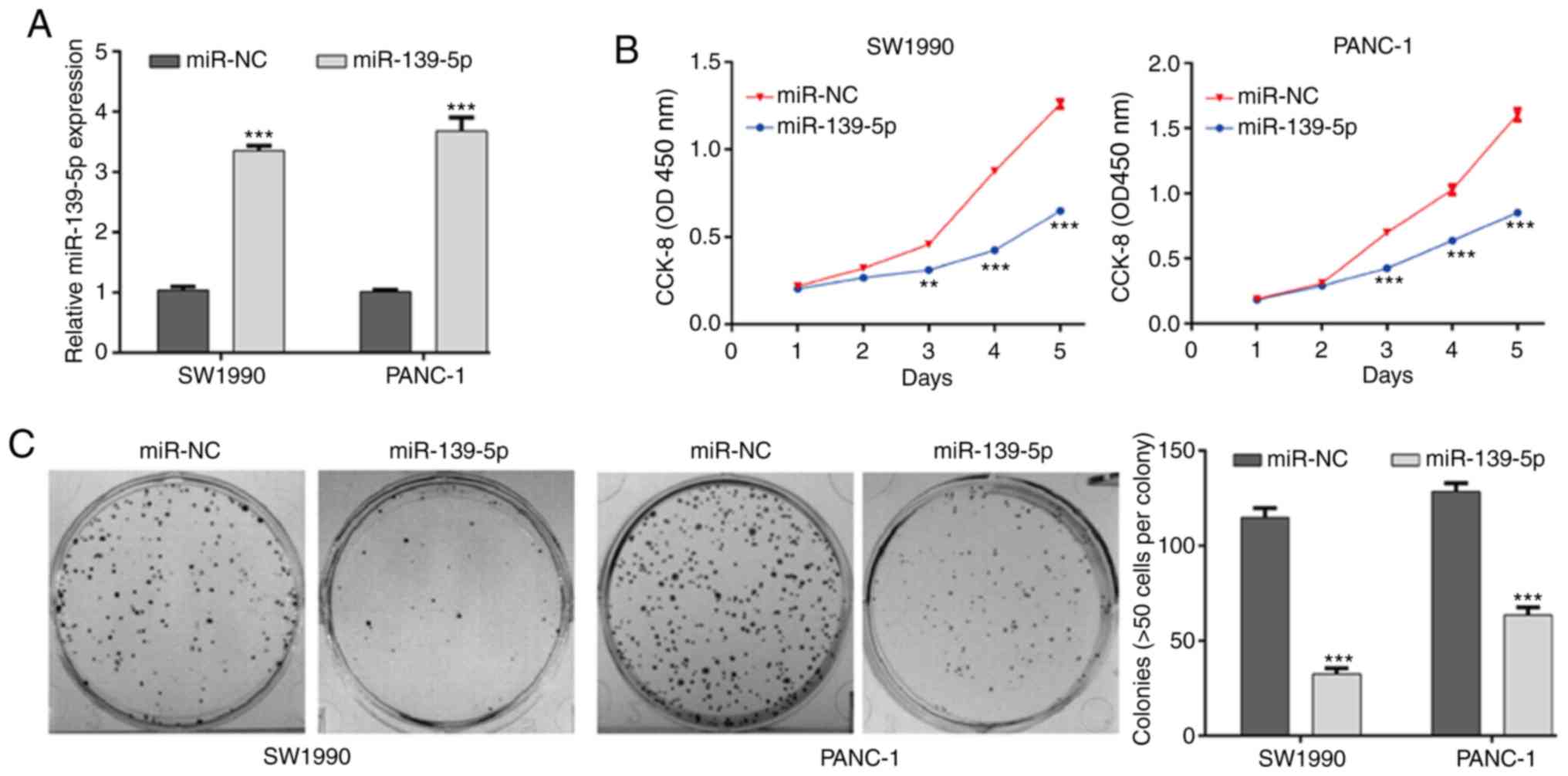
To investigate the functional role of miR-139-5p in
PDAC cells, SW1990 and PANC-1 cells were selected for
gain-of-function assays as they had the lowest expression levels of
miR-139-5p of the four PDAC cell lines. The efficiency of
transfection was validated by RT-qPCR analysis, and the results
showed that miR-139-5p was significantly upregulated in SW1990 and
PANC-1 cells from the miR-139-5p group compared with those in the
miR-NC group (Fig. 4A, P<0.001).
The results of the CCK-8 assay showed that the proliferative
ability was significantly suppressed in the miR-139-5p group
compared with that in the miR-NC group in both SW1990 and PANC-1
cells (Fig. 4B, P<0.001). The
colony formation ability of the PDAC cells was further examined. As
shown in Fig. 4C, the number of
colonies was significantly lower in the miR-139-5p group than in
the miR-NC group in the SW1990 and PANC-1 cells (P<0.001).
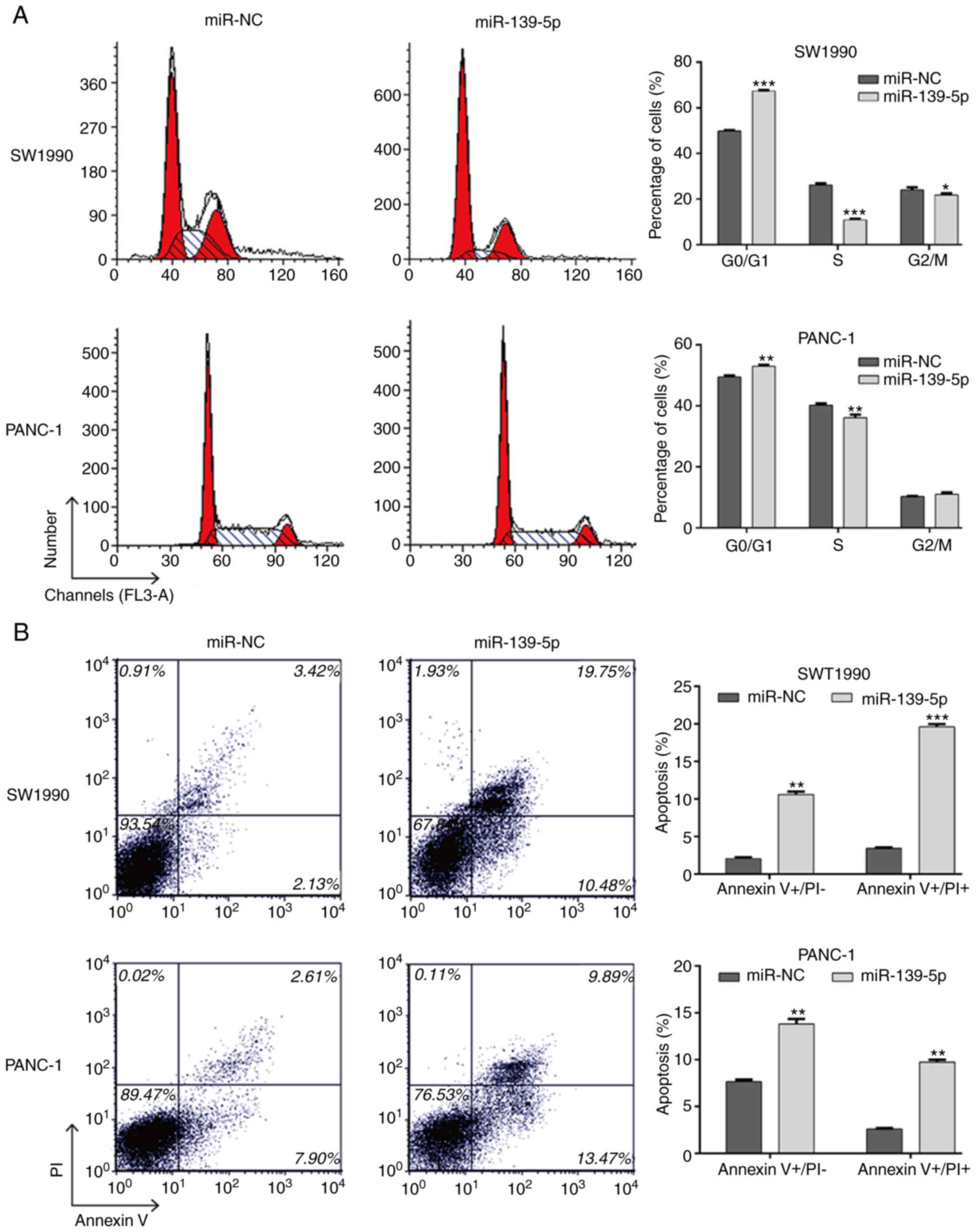
Furthermore, the regulation of miR-139-5p on cell
cycle distribution and apoptosis was analyzed using flow cytometry.
As depicted in Fig. 5A, miR-139-5p
transfection significantly increased the population of cells in the
G0/G1 phase (SW1990: 67.38±0.31 vs. 49.82±0.38%, P<0.001;
PANC-1: 52.92±0.49 vs. 49.48±0.47%, P<0.001) and decreased the
population in the S phase (SW1990: 10.90±0.42 vs. 26.12±0.74%,
P<0.001; PANC-1: 36.08±1.00 vs. 40.25±0.58%, P<0.01) and G2/M
phase (SW1990: 21.72±0.70 vs. 24.06±1.02%, P<0.05) compared with
populations in the miR-NC transfection group. This suggested that
cell cycle was arrested at the G0/G1 phase by the overexpression if
miR-139-5p. As shown in Fig. 5B, the
overexpression of miR-139-5p markedly elevated the percentages of
early apoptotic cells (SW1990: 10.59±0.40 vs. 2.08±0.15%,
P<0.01; PANC-1: 13.82±0.51 vs. 7.66±0.20%, P<0.01) and late
apoptotic cells (SW1990: 19.59±0.38 vs. 3.43±0.08%, P<0.001;
PANC-1: 9.73±0.28 vs. 2.62±0.07%, P<0.01) compared with those in
the miR-NC group. These results revealed that the overexpression of
miR-139-5p exerted a potent apoptotic effect in PDAC cells.
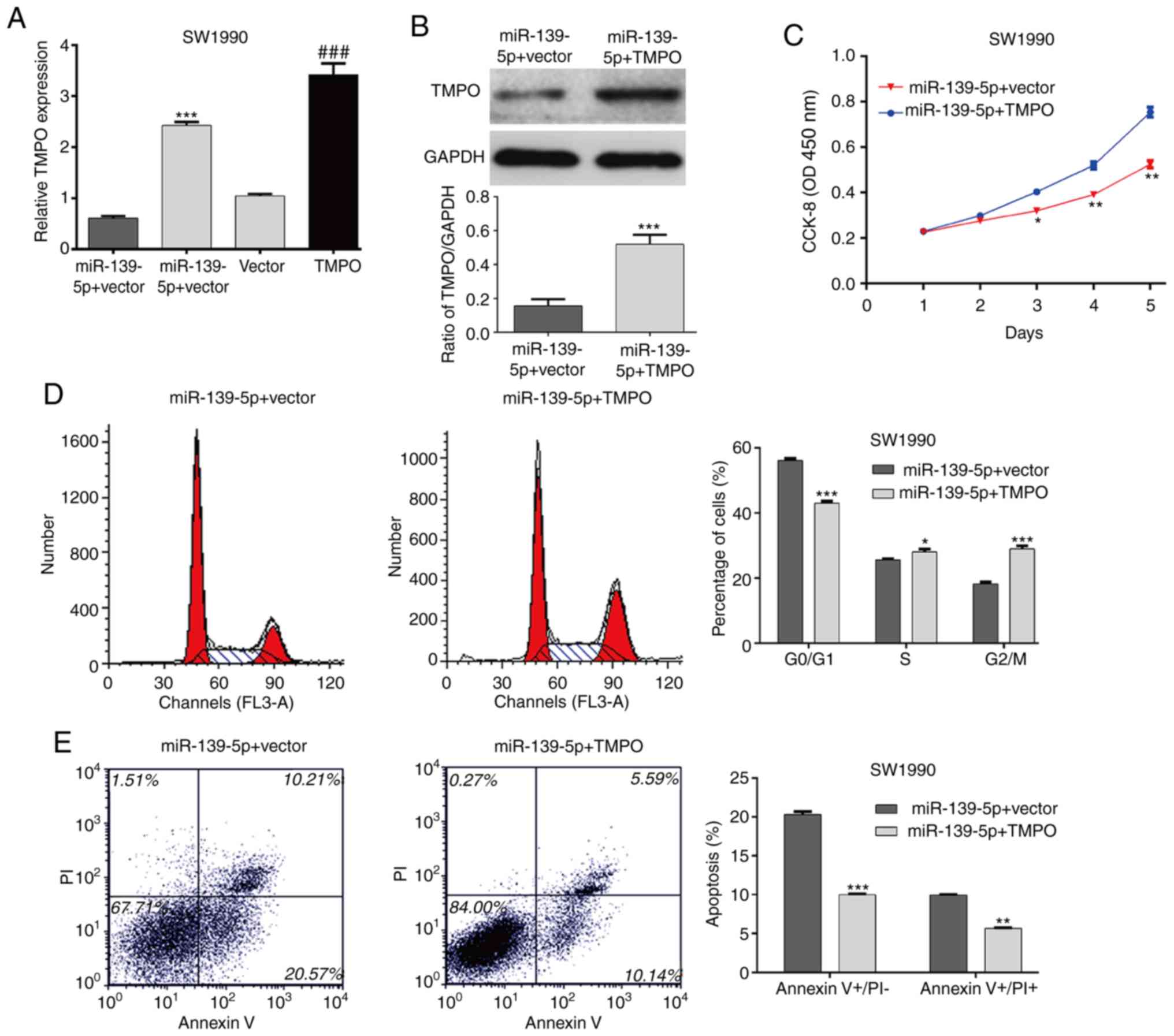
Overexpression of TMPO partially
eliminates the effects of miR-139-5p on cell proliferation, cell
cycle and apoptosis in PDAC cells
To further investigate the role of TMPO in
miR-139-5p-mediated cell proliferation, cell cycle arrest and
apoptosis in PDAC cells, it was first confirmed that pcDNA3.1-TMPO
was successfully transfected into SW1990 cells by RT-qPCR analysis
(Fig. 6A, P<0.001). In addition,
TMPO was overexpressed in SW1990/miR-139-5p mimics cells, as
confirmed by RT-qPCR analysis (Fig.
6A, P<0.001) and western blot analysis (Fig. 6B, P<0.001). The CCK-8 assay
revealed that the ectopic expression of TMPO effectively reversed
the inhibition of proliferation induced by the overexpression of
miR-139-5p in SW1990 cells (Fig.
6C). Consistently, the overexpression of TMPO alleviated the
effects of miR-139-5p-induced cell cycle G0/G1 phase arrest
(Fig. 6D) and apoptosis (Fig. 6E) in SW1990 cells. These results
demonstrated that the anti-proliferative effects of miR-139-5p in
PDAC cells may be mediated through TMPO.
Discussion
miRNAs are assigned as part of the epigenetic
machinery and have been highlighted as critical regulators of gene
expression (26). Aside from DNA
methylation and histone modifications, the biological function and
molecular mechanisms of miRNA-target interactions are of particular
interest to the scientific community (27). Epigenetic modifications have been
shown to be involved in the onset and development of numerous
diseases and can contribute to explaining the clinical fluctuation
of symptoms (28).
The aberrant expression of miR-139-5p in cancer is
frequently reported and linked to the regulation of oncogenic
and/or tumor suppressor genes. The present study revealed critical
findings concerning the biological role of miR-139-5p in the
progression of human PDAC. Firstly, the expression of miR-139-5p
was found to be markedly downregulated in PDAC tissues and cell
lines compared with that in adjacent tissues and pancreatic duct
epithelial cells. To further analyze the functional role of
miR-139-5p in PDAC, its potential gene targets were assessed. A
bioinformatic screen identified TMPO is a potential target of
miR-139-5p, the TMPO 3′-UTR and miR-139-5p 5′-UTR crosstalk was
then confirmed using a luciferase reporter assay. Mechanically, the
enforced expression of miR-139-5p decreased PDAC cell proliferation
and induced G0/G1 arrest and apoptosis through negatively
regulating TMPO, which indicated an antitumor effect of miR-139-5p
in PDAC. MiR-139-5p is also a positive regulator of human
colorectal cancer recurrence and metastasis; the hyperexpression of
miR-139-5p was found to trigger peritoneal dissemination in a mouse
tumor model (29). By contrast,
emerging evidence indicates that miR-139-5p often acts as a tumor
suppressor to regulate cellular events, including proliferation,
aggressiveness and apoptosis, in tumor development and progression
(12). These studies have revealed
the tissue-specific epigenetic regulation of miR-139-5p in
cancer.
TMPO is a chromatin-associated protein that
interacts with A/B/C-type lamins, and localizes along the inner
nuclear membrane of the nuclear envelope (30). The common N-terminal
LAP2-Emerin-MAN1-domain (LEM-D) and a motif like LEM-D enable the
binding between TMPO and chromosome to promote DNA replication
through altering chromatin structure (31). LAP2α, a specific isoform of TMPO, can
form a complex with lamin A/C at their C-terminal tails to reduce
fibroblast proliferation via the retinoblastoma protein-linked
pathways (32). The oncogenic role
of TMPO was also identified in glioblastoma cells; TMPO deficiency
depressed tumor cell proliferation, and induced cell cycle arrest
and apoptosis (22). Consistently,
the present study demonstrated that transfection of pcDNA3.1-TMPO
alleviated the effects of miR-139-5p-induced growth inhibition,
cell cycle G0/G1 phase arrest and apoptosis in PDAC cells.
In conclusion, the present study demonstrated for
the first time, to the best of our knowledge, that the
miR-139-5p/TMPO complex is an important regulator of the
pathogenesis of PDAC; however, clinicopathological analysis of the
expression of miR-139-5p and TMPO in PDAC is lacking. At present,
additional clinical samples are being collected to further
investigate the association between the expression of miR-139-5p or
TMPO and the clinicopathological status of PDAC, and further data
on the prognosis of miR-139-5p and TMPO in patients with PDAC will
be presented. This preliminary study indicates a potential
epigenetic therapy for PDAC.
Acknowledgements
Not applicable.
Funding
The study was supported by Renmin Hospital, Hubei
University of Medicine (Hubei, China; grant no. N0161003).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HT was mainly involved in experimental design and
integral control. HZ and LZ performed the experiments, analyzed
data and wrote the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Human Ethics
Review Committee of Renmin Hospital, Hubei University of Medicine.
Informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sahakyan MA, Kim SC, Kleive D, Kazaryan
AM, Song KB, Ignjatovic D, Buanes T, Røsok BI, Labori KJ and Edwin
B: Laparoscopic distal pancreatectomy for pancreatic ductal
adenocarcinoma: Long-term oncologic outcomes after standard
resection. Surgery. 162:802–811. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hwang CI, Boj SF, Clevers H and Tuveson
DA: Preclinical models of pancreatic ductal adenocarcinoma. J
Pathol. 238:197–204. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dunne RF and Hezel AF: Genetics and
biology of pancreatic ductal adenocarcinoma. Hematol Oncol Clin
North Am. 29:595–608. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rahib L, Smith BD, Aizenberg R, Rosenzweig
AB, Fleshman JM and Matrisian LM: Projecting cancer incidence and
deaths to 2030: The unexpected burden of thyroid, liver, and
pancreas cancers in the United States. Cancer Res. 74:2913–2921.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Moffitt RA, Marayati R, Flate EL, Volmar
KE, Loeza SG, Hoadley KA, Rashid NU, Williams LA, Eaton SC, Chung
AH, et al: Virtual microdissection identifies distinct tumor- and
stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat
Genet. 47:1168–1178. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Delpu Y, Hanoun N, Lulka H, Sicard F,
Selves J, Buscail L, Torrisani J and Cordelier P: Genetic and
epigenetic alterations in pancreatic carcinogenesis. Curr Genomics.
12:15–24. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Imaoka H, Toiyama Y, Okigami M, Yasuda H,
Saigusa S, Ohi M, Tanaka K, Inoue Y, Mohri Y and Kusunoki M:
Circulating microRNA-203 predicts metastases, early recurrence, and
poor prognosis in human gastric cancer. Gastric Cancer. 19:744–753.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Acunzo M, Romano G, Wernicke D and Croce
CM: MicroRNA and cancer-a brief overview. Adv Biol Regul. 57:1–9.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Catalanotto C, Cogoni C and Zardo G:
MicroRNA in control of gene expression: An overview of nuclear
functions. Int J Mol Sci. 17:E17122016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tüfekci KU, Meuwissen RL and Genç S: The
role of microRNAs in biological processes. Methods Mol Biol.
1107:15–31. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang HD, Jiang LH, Sun DW, Li J and Tang
JH: MiR-139-5p: Promising biomarker for cancer. Tumour Biol.
36:1355–1365. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yonemori M, Seki N, Yoshino H, Matsushita
R, Miyamoto K, Nakagawa M and Enokida H: Dual tumor-suppressors
miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in
bladder cancer. Cancer Sci. 107:1233–1242. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun C, Sang M, Li S, Sun X, Yang C, Xi Y,
Wang L, Zhang F, Bi Y, Fu Y and Li D: Hsa-miR-139-5p inhibits
proliferation and causes apoptosis associated with down-regulation
of c-Met. Oncotarget. 6:39756–39792. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hua S, Lei L, Deng L, Weng X, Liu C, Qi X,
Wang S, Zhang D, Zou X, Cao C, et al: miR-139-5p inhibits aerobic
glycolysis, cell proliferation, migration, and invasion in
hepatocellular carcinoma via a reciprocal regulatory interaction
with ETS1. Oncogene. 37:1624–1636. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Q, Liang X, Wang Y, Meng X, Xu Y, Cai
S, Wang Z, Liu J and Cai G: miR-139-5p inhibits the
epithelial-mesenchymal transition and enhances the chemotherapeutic
sensitivity of colorectal cancer cells by downregulating BCL2. Sci
Rep. 6:271572016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Agosta C, Laugier J, Guyon L, Denis J,
Bertherat J, Libé R, Boisson B, Sturm N, Feige JJ, Chabre O and
Cherradi N: MiR-483-5p and miR-139-5p promote aggressiveness by
targeting N-myc downstream-regulated gene family members in
adrenocortical cancer. Int J Cancer. 143:944–957. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li J, Su L, Gong YY, Ding ML, Hong SB, Yu
S and Xiao HP: Downregulation of miR-139-5p contributes to the
antiapoptotic effect of liraglutide on the diabetic rat pancreas
and INS-1 cells by targeting IRS1. PLoS One. 12:e01735762017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Marrero-Rodríguez D, Taniguchi-Ponciano K,
Lopez-Sleman J, Romero-Morelos P, Mendoza-Rodríguez M, Garcia I,
Huerta-Padilla V, Mantilla A, Duarte A, Piña P, et al: Thymopoietin
beta and gamma isoforms as a potential diagnostic molecular marker
for breast cancer: Preliminary data. Pathol Oncol Res.
21:1045–1050. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gesson K, Vidak S and Foisner R:
Lamina-associated polypeptide (LAP)2α and nucleoplasmic lamins in
adult stem cell regulation and disease. Semin Cell Dev Biol.
29:116–124. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dechat T, Vlcek S and Foisner R: Review:
Lamina-associated polypeptide 2 isoforms and related proteins in
cell cycle-dependent nuclear structure dynamics. J Struct Biol.
129:335–345. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang L, Wang G, Chen S, Ding J, Ju S, Cao
H and Tian H: Depletion of thymopoietin inhibits proliferation and
induces cell cycle arrest/apoptosis in glioblastoma cells. World J
Surg Oncol. 14:2672016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim HJ, Hwang SH, Han ME, Baek S, Sim HE,
Yoon S, Baek SY, Kim BS, Kim JH, Kim SY and Oh SO: LAP2 is widely
overexpressed in diverse digestive tract cancers and regulates
motility of cancer cells. PLoS One. 7:e394822012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jung H, Lee HH, Song KY, Jeon HM and Park
CH: Validation of the seventh edition of the American Joint
Committee on Cancer TNM staging system for gastric cancer. Cancer.
117:2371–2378. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ferreira HJ and Esteller M: Non-coding
RNAs, epigenetics, and cancer: Tying it all together. Cancer
Metastasis Rev. 37:55–73. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yan H, Bonasio R, Simola DF, Liebig J,
Berger SL and Reinberg D: DNA methylation in social insects: How
epigenetics can control behavior and longevity. Annu Rev Entomol.
60:435–452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Piletič K and Kunej T: MicroRNA epigenetic
signatures in human disease. Arch Toxicol. 90:2405–2419. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Miyoshi J, Toden S, Yoshida K, Toiyama Y,
Alberts SR, Kusunoki M, Sinicrope FA and Goel A: MiR-139-5p as a
novel serum biomarker for recurrence and metastasis in colorectal
cancer. Sci Rep. 7:433932017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Furukawa K: LAP2 binding protein 1
(L2BP1/BAF) is a candidate mediator of LAP2-chromatin interaction.
J Cell Sci. 112:2485–2492. 1999.PubMed/NCBI
|
|
31
|
Gant TM, Harris CA and Wilson KL: Roles of
LAP2 proteins in nuclear assembly and DNA replication: Truncated
LAP2beta proteins alter lamina assembly, envelope formation,
nuclear size, and DNA replication efficiency in Xenopus
laevis extracts. J Cell Biol. 144:1083–1096. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dorner D, Vlcek S, Foeger N, Gajewski A,
Makolm C, Gotzmann J, Hutchison CJ and Foisner R: Lamina-associated
polypeptide 2alpha regulates cell cycle progression and
differentiation via the retinoblastoma-E2F pathway. J Cell Biol.
173:83–93. 2006. View Article : Google Scholar : PubMed/NCBI
|