Introduction
Glioma is a type of primary tumor that starts in the
brain or spine and accounts for 2% of tumors in adults. The 5-year
case fatality rate is the highest of any cancer except for
pancreatic and lung carcinoma (1–3).
Treatment is often a combined approach, using surgery, radiation
therapy and chemotherapy, however, these treatments cannot
significantly improve the survival rate beyond 5 years and, over
the past 30 years, the median survival duration of glioblastoma has
been <1 year (4,5).
Glioma is a disease involving abnormal genes.
Epidermal growth factor receptor (EGFR) mutation, amplification and
overexpression are early molecular events for glioma progression.
EGFR facilitates the cell cycle of tumor cells, inhibits cell
apoptosis, maintains telomerase activity and promotes tumor
angiogenesis and tumor invasion through downstream pathways
(6–9).
microRNAs are a class of endogenous small non-coding
RNAs that regulate gene expression by targeting mRNAs for
translational repression, degradation, or both, through target
sites in the 3′ untranslated regions (3′ UTR) and modulate cell
differentiation, proliferation and apoptosis (10–13).
Kefas et al first reported complementarity in at least three
regions in the 3′ UTR between miR-7 and EGFR (14). Previous studies by our group
indicated that miR-7 significantly inhibited glioma cell line
proliferation and partially silenced EGFR post-transcriptional
translation and reduced EGFR activity and function (15). However, animal experiments showed
that neither liposome- or adenovirus-transfected miR-7 can
effectively decrease the survival rate of rats with glioma,
although survival duration was slightly prolonged. Subsequent
experiments revealed that due to the blood-brain barrier and
intercellular retention, target genes in the tumor bed were poorly
targeted, significantly limiting the function of miR-7. Thus, a
gene delivery material that can pass through the blood-brain
barrier and is independent of intercellular communication is
needed.
Poly(amido amine) (PAMAM) has a large volume and a
large number of surface functional groups. The high degree of
branching and the specific monodispersity property allow the
terminal functional groups to include anion, cation and hydrophobic
groups, significantly improving biocompatibility, bioavailability
and targeting. In addition, due to its high solubilization, delayed
release and low toxicity, PAMAM has been used as a drug delivery
system (16–18). Folic acid (FA) is rarely expressed
in epithelial cells of newly generated normal tissues, but is
highly expressed in a variety of tumor cells and positively
correlates with tumor malignancy. Therefore, FA has become a target
for tumor cell tracing (19,20).
The present study utilized PAMAM as a vector and FA/FA receptor as
a target axis and found that miR-7 efficiently silenced
glioma-specific genes and achieved favorable effects in treating
glioma in vivo and in vitro.
Materials and methods
Cell culture conditions
U251 human glioma cells were kindly provided by the
Tianjin Neuro-Oncology Institute, China. All cells were maintained
in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad,
CA, USA) supplemented with 10% fetal bovine serum (FBS;
Invitrogen), 2 mM glutamine (Sigma, St. Louis, MO, USA), 100 units
of penicillin/ml (Sigma) and 100 μg of streptomycin/ml (Sigma), at
37°C with 5% CO2.
Animals
A total of 174 male athymic mice, weighing 80 g,
were purchased from the Animal Center of the Academy of Military
Medical Sciences, Beijing (License no. 0195), China, and housed in
the Animal Experimental Center, Tianjin Medical University, with a
humidity of 50±5% at 20–25°C.
FA/PAMAM complex preparation and
transmission electron microscopy observations
FA (Sigma) was linked to PAMAM-D (Sigma) through two
continuous steps. Briefly, 0.032 78 g FA was reacted with 0.199 79
g EDC in 20 ml dimethyl sulphoxide and the FA activated solution
was gradually dropped into a PAMAM-methanol and dimethyl sulphoxide
solution. The complex produced was observed using transmission
electron microscopy.
Cell transfection
The miR-7 sequence was 5′-UUGUUUUA GUGAUCAAGAAGGU-3′
and the reference antisense strand was
5′-GTGAATGCGATGCCCTCCGAGG-3′. The 5′ end was labeled with
fluorescein isothiocyanate. There were three groups in the
experiments: normal control, FA/PAMAM/nonsense-treated and
FA/PAMAM/miR-7 treated. When U251 cells reached 60–70% confluency,
serum concentration was decreased to 5% and PAMAM and miR-7 or
nonsense control were added at a ratio of 16:1 and cultured at 37°C
in 5% CO2 for 4 h. The culture medium was replaced with
complete culture medium supplemented with 10% FBS and cells were
incubated for an additional 48 h. Cell transfection was observed
using a fluorescence microscope.
Reverse transcription (RT)-real-time
PCR
RT reactions and real-time PCR were conducted using
the mirVana™ qRT-PCR miRNA detection kit (Ambion, Austin, TX, USA).
Amplification was performed using an MJ-real-time PCR (Bio-Rad
Laboratories, Hercules, CA, USA) with a protocol of 40 cycles at
95°C for 3 min, 95°C for 15 sec and 60°C for 30 sec. miR-7 upstream
primer: 5′-AAAAAGAACACGTGGAAGGAT AG-3′; downstream primer:
5′-CCGCCTAACGTACCGCG AATTT-3′. A human 18 rRNA TaqMan probe was
used as an internal reference, with upstream primer: 5′-AACTTTCGAT
GGTAGTCGCCG-3′ and downstream primer: 5′-CCTTGGA TGTGGTAGCCGTTT-3′.
Both RT and PCR primers were purchased from Ambion. 5S RNA was used
for normalization. Relative quantification was conducted using
amplification efficiencies derived from cDNA standard curves. Data
were analyzed using Opticon Monitor Analysis software V2.02 (MJ
Research Inc., Waltham, MA, USA) and are shown as fold-change
(2−ΔΔCt).
Western blot analysis
Parental and transfected cells were washed three
times with pre-chilled phosphate-buffered saline (PBS). The cells
were then solubilized in 1% Nonidet P-40 lysis buffer (20 mM Tris,
pH 8.0, 137 mM NaCl, 1% Nonidet P-40, 10% glycerol, 1 mM
CaCl2, 1 mM MgCl2, 1 mM phenylmethylsulfonyl
fluoride, 1 mM sodium fluoride, 1 mM sodium orthovanadate and a
protease inhibitor mixture). Lysates (40 μg) were separated by 8%
SDS-acrylamide electrophoresis. Separated proteins were transferred
to polyvinylidene fluoride membranes (Millipore, Bedford, MA, USA)
and incubated with primary antibodies against EGFR (1:500 dilution;
Santa Cruz Biotechnology, Santa Cruz, CA, USA), Akt-2 (1:1,000
dilution; for Ser473, Santa Cruz Biotechnology), and PI3K
(Zhongshan Bio Corp., Beijing, China), followed by incubation with
horseradish peroxidase-conjugated secondary antibody (Zymed
Laboratories Inc., San Francisco, CA, USA). Specific proteins were
detected using a SuperSignal protein detection kit (Pierce,
Rockford, IL, USA). The membrane was stripped and reprobed with a
primary antibody against β-actin (Santa Cruz Biotechnology).
Optical density was measured and analyzed using Quantity One
software.
Cell growth curves
Cells were seeded into 24-well plates at
1×104 cells/well. The number of cells was quantified
following culture for 1, 2, 3, 4, 5 and 6 days to produce cell
growth curves.
Cell cycle detection
Single cell suspensions were obtained following
trypsinization and cells were then washed with PBS three times,
fixed with absolute alcohol and incubated with RNaseA. Cells were
stained with propidium iodide for 30 min and then analyzed by flow
cytometry. Samples were collected and analyzed using CellQuest
software, and a proliferation index was calculated according to
[PIx =
(S+G2M)/(G0G1+S+G2M)].
Apoptosis detection using the TUNEL
method
Cells were mounted in PBS, immersed in
permeabilization solution for 5 min, incubated with 25 μl
TUNEL-labeling reaction mixture in a humidified box at 37°C for 60
min and then washed with PBS. Nuclei were counterstained with
Hoechst 33258 at a dilution of 1:1,000. Cell apoptosis rate (number
of apoptotic cells/total number of cells × 100%) was quantified
using a fluorescence microscope.
Tumor cell migration detection
Matrigel (60 μl) was placed on the polycarbonate
membrane of the upper chamber of a Transwell at 37°C for 30 min and
the gel was allowed to solidify. Cells were seeded into the upper
Transwell chamber at 1×104 cells/well and 600 μl DMEM
supplemented with 10% FBS was added to the lower chamber. After 48
h, cells that had not permeated the membrane in the upper chamber
were removed using a wet cotton bud and the membrane was stained
with hematoxylin. The number of cells permeating the membrane was
quantified under an inverted microscope.
Soft agar assay for clone formation
Six-well plates were precoated with agar (0.7%)
overnight. Stably transfected stem cell suspension was prepared,
quantified and mixed with 0.35% low melting-point agarose prepared
using DMEM containing 10% FBS, and then rapidly placed in the
6-well plate and allowed to grow in a conventional incubator.
Complete medium (200 μl) was added to every well the following day
and the medium was replaced every 7 days thereafter. After 3 weeks,
the number of colonies with cells >40 was counted using an
inverted microscope.
Establishment of a brain-glioma mouse
model
In the liposome/miR-7 group, the serum concentration
of the culture medium was decreased to 5%, and 1 μg plasmid was
mixed with 100 μl LipoVec™ for 20 min and then placed in the
culture medium. After 4 h, the medium was replaced with medium
containing 20% serum. In the FA/PAMAM/miR-7 group, the serum
concentration of the culture medium was decreased to 5%. PAMAM and
miR-7 (16:1) were added to U251 cells and incubated at 37°C in 5%
CO2. After 4 h of incubation, medium was replaced with
medium supplemented with 10% FBS. In the U251 control group
(n=144), cells in the log phase of growth were harvested and
suspended to prepare a single cell suspension. Mice were
anesthetized by intraperitoneal injection with 10% chloral hydrate
(3.0 ml/kg) and then fixed on a super-clean bench with stereotactic
apparatus. Cell suspension (5 μl) containing 1×106 U251
cells was injected over 5 min into the right caudate nucleus. The
needle was maintained in place for at least 3 min following
injection and the incision was then sutured. All experimental
procedures were carried out according to the regulations and
internal biosafety and bioethics guidelines of Tianjin Medical
University and the Tianjin Municipal Science and Technology
Commission.
Cell transplantation and complex
aggregation in the tumor
Drugs were administered 8 days after cell
transplantation at three sites: caudal vein (n=24); carotid artery
(n=24); and tumor site (n=24). Every mouse was administered 10 μl
FA/PAMAM solution containing 5 μg nonsense sequence or miR-7
oligonucleotide. The mice were sacrificed 10 days after cell
transplantation, i.e. 48 h after drug administration, and the brain
was harvested to prepare frozen sections of tumor tissues.
FA/PAMAM/miR-7 complex aggregation in the tumor was observed by
fluorescence microscopy.
Comparison of glioma inhibition between
two miR-7 delivery systems, both in vivo and in vitro gene
transfection efficiency
Cells were obtained 48 h following gene transfection
of liposome/miR-7, FA/PAMAM/miR-7 and U251 control groups. The
percent of positive cells was determined by fluorescence microscopy
and transfection efficiency was calculated.
miR-7 levels
Total-RNA was extracted from cells, transcribed with
reverse transcriptase, followed by PCR using the same conditions as
described above.
MRI
Seventy-two mice were examined. Each mouse was
administered 10 μl FA/PAMAM solution containing 5 μg miR-7
oligonucleotide or liposome dilution. General survival of animals
was observed and survival duration was recorded. MRI was conducted
at 7, 14 and 21 days following cell transplantation and tumor
volumes were compared. The animal studies were approved by...
In situ cell death detection
The apoptotic cell death in the tumor specimens of
the mice that had died due to the tumor was detected using a cell
death detection kit [ApopTag® In Situ Apoptosis
Detection kit (S7100), Chemicon International, Inc., Temecula, CA,
USA].
Immunohistochemistry
Paraffin-embedded tissue sections were used to
examine proliferating cell nuclear antigen, matrix
metalloproteinase 2 and matrix metalloproteinase 9 levels.
Transfected cells were seeded on coverslips, fixed with 4%
paraformaldehyde (Sigma), treated with 3%
H2O2 for 10 min and incubated overnight at
4°C with the primary antibodies (1:200 dilutions) overnight at 4°C,
followed by biotin-labeled secondary antibody (1:100 dilutions) for
1 h at room temperature. Sections were then incubated with
avidin-biotin-peroxidase complex and diaminobenzidine,
counterstained with hematoxylin and visualized using light
microscopy.
Statistical analysis
Data are expressed as the means ± SD. Data were
analyzed using analysis of variance and the χ2 test
using SPSS13.0 (Windows). P<0.05 was considered to indicate
statistically significant differences.
Results
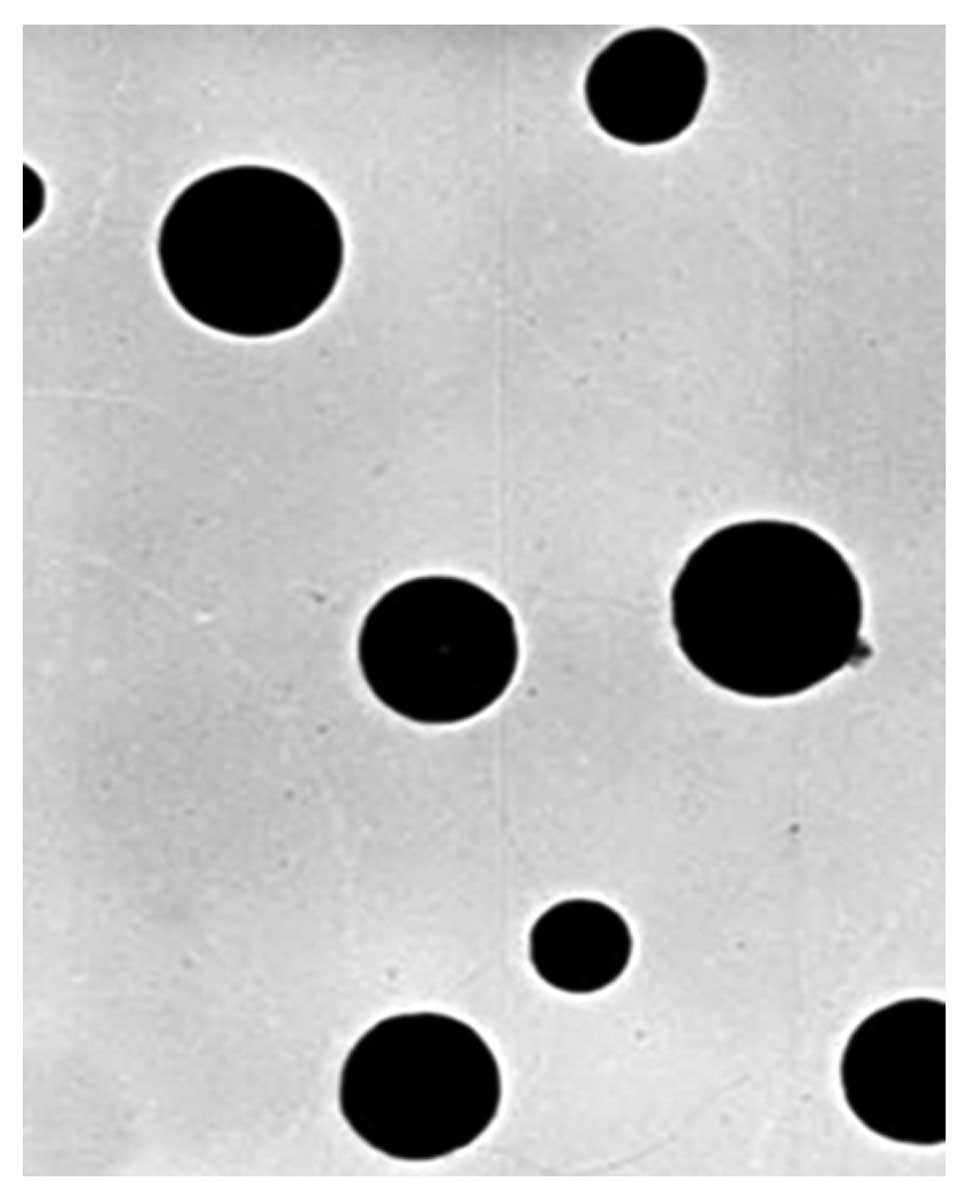
FA/PAMAM complex appearance
FA/PAMAM formed solid spheres that were ~50 nm in
diameter and evenly distributed; the PAMAM/miR-7 or PAMAM/nonsense
sequence particles were 60–80 nm in diameter and were evenly
distributed (Fig. 1).
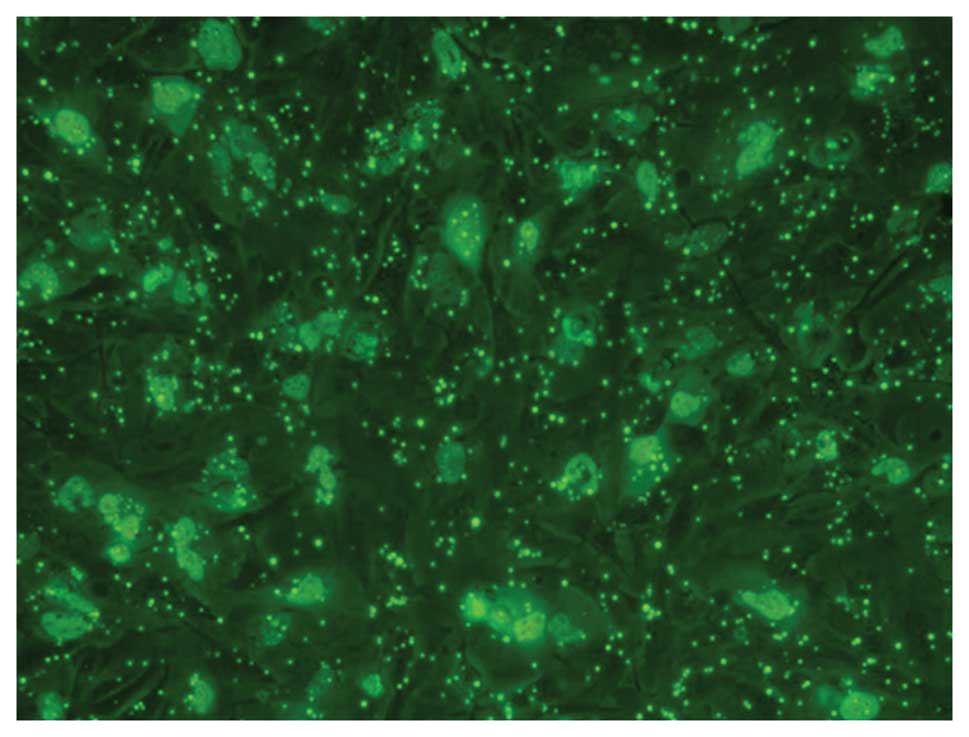
Gene transfection efficiency
Using fluorescence microscopy with excitation light
of 550 nm, FITC-labeled complex particles appeared green and were
distributed in the cell membrane or cytoplasm of U251 cells
(Fig. 2).
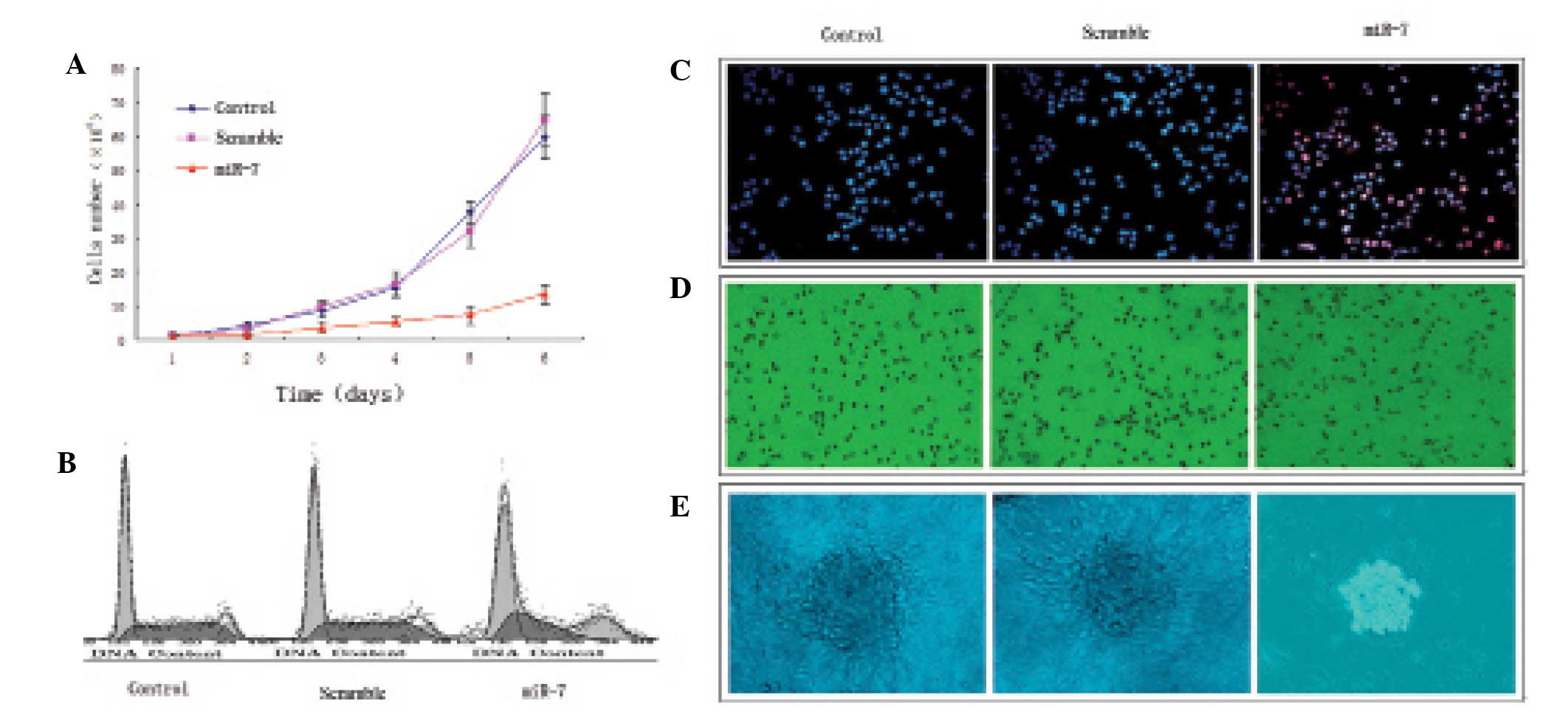
miR-7 transfection suppresses U251 glioma
cell proliferation
U251 glioma cells grew rapidly in the control and
nonsense sequence groups and there was no significant difference
(P>0.05); miR-7 transfection significantly suppressed U251
glioma cell proliferation compared with the control and nonsense
sequence groups (P<0.05; Fig.
3A). The percent of S-phase cells in the control and nonsense
sequence groups was significantly more than that in the miR-7
transfection group, (P<0.05; Table
I, Fig. 3B). In addition, few
apoptotic cells were observed in the control and nonsense sequence
groups (P>0.05), but the apoptosis rate was significantly
increased in the miR-7-transfected cells compared with the control
and nonsense sequence groups (P<0.05; Fig. 3C). A larger number of tumor cells in
the control (79.44±11.34%) and nonsense sequence groups
(83.67±9.14%) migrated to the lower chamber of the migration assay
compared with the miR-7-transfected group (31.79±6.27%); P<0.05;
Fig. 3D). A large number of cell
clones was observed in the control and nonsense sequence groups,
with many pseudopodia, but small-sized cell clones in the absence
of pseudopodia were observed in the miR-7 transfection group.
 | Table IComparison of U251 glioma cell cycle
among groups. |
Table I
Comparison of U251 glioma cell cycle
among groups.
| Group | G1 phase
(%) | S phase (%) | G2-M phase
(%) | PIx |
|---|
| Control | 56.44 | 34.12 | 9.44 | 43.56 |
| Nonsense | 54.27 | 35.49 | 10.24 | 45.73 |
| miR-7
transfection | 72.14 | 14.79 | 13.07 | 27.86a |
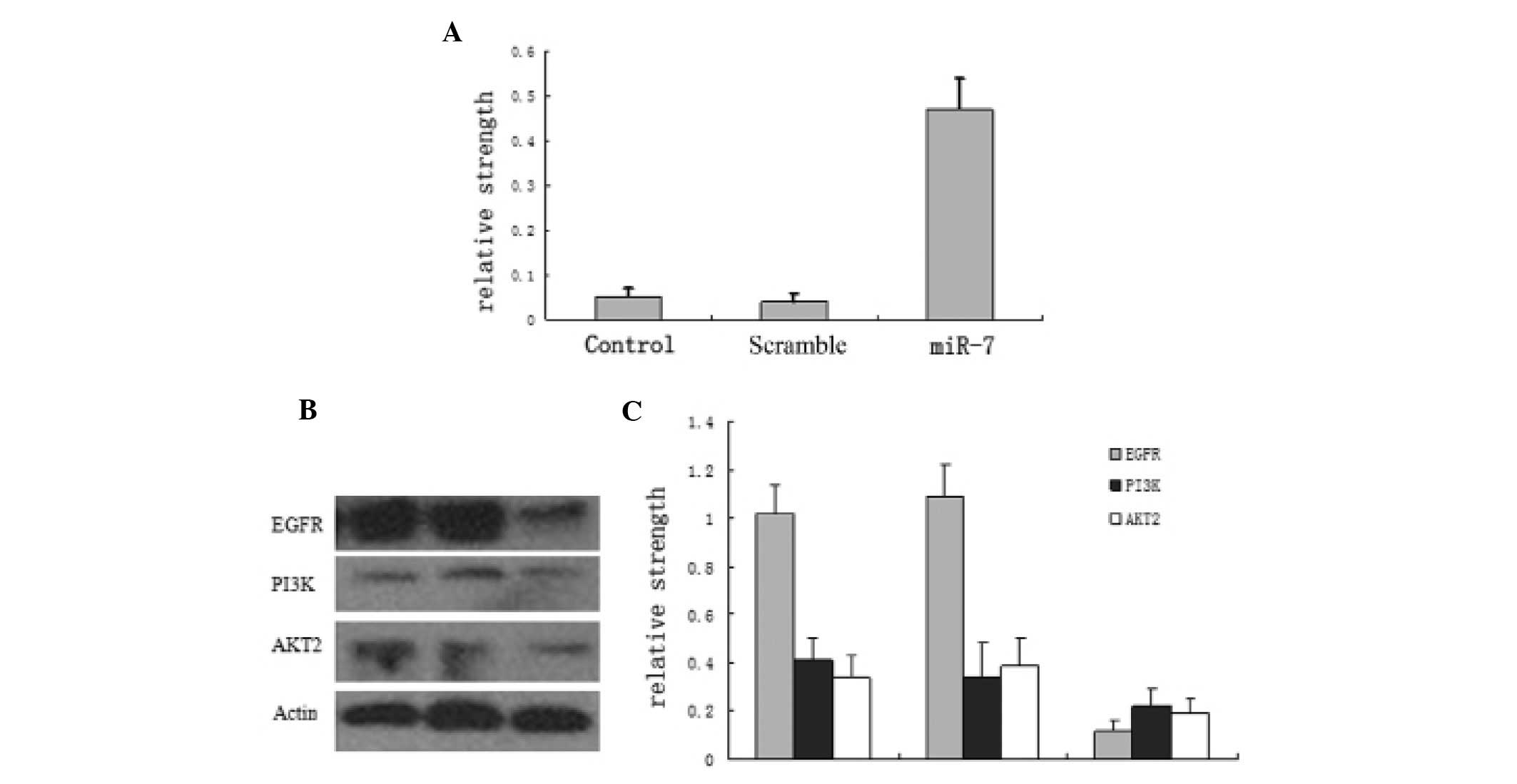
PAMAM increases miR-7 expression and
partially suppresses EGFR pathway protein levels
miR-7 expression was low in normal U251 cells and in
FA/PAMAM/nonsense-treated cells, with no significant difference
(P>0.05), but it was significantly increased in
FA/PAMAM/miR-7-transfected cells (P<0.05; Fig. 4A). In addition, a high level of EGFR
and high levels of PI3K and AKT2 were observed in normal U251 cells
and in FA/PAMAM/nonsense-treated cells, but EGFR, PI3K and AKT2
levels were significantly decreased in FA/PAMAM/miR-7-transfected
cells (P<0.05; Fig. 4B and
C).
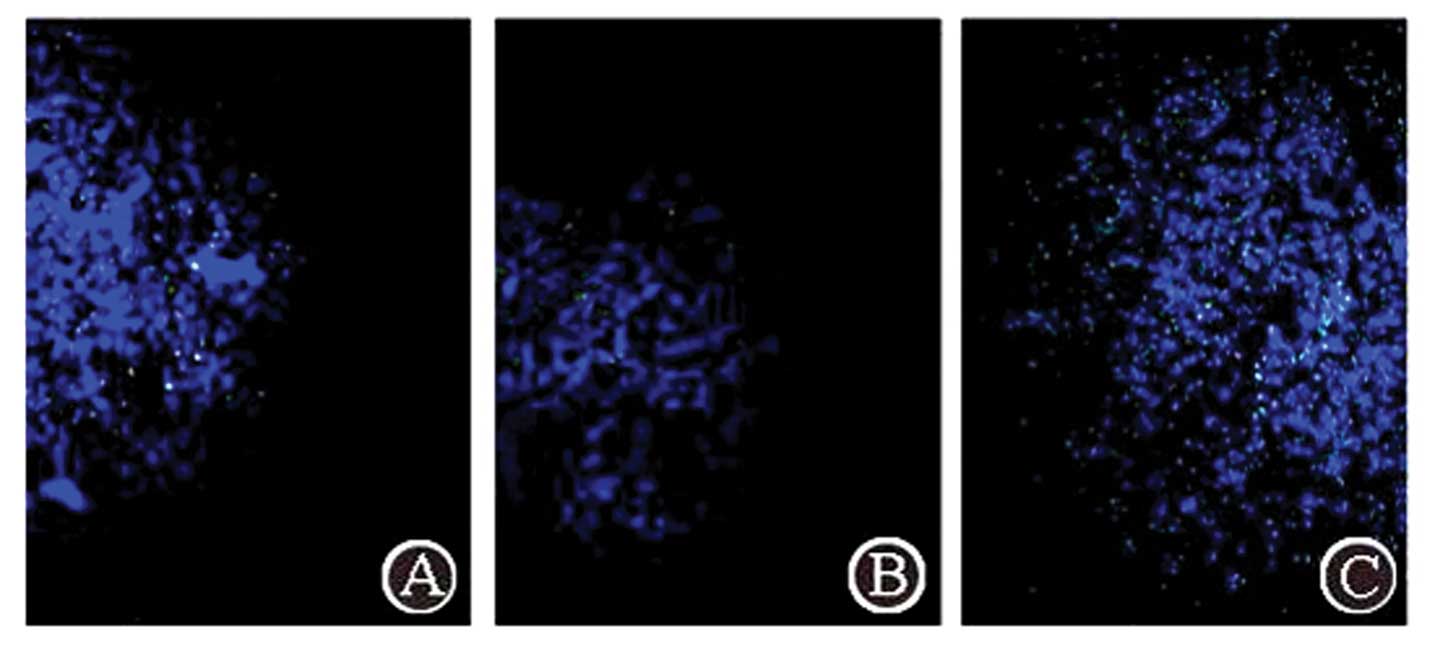
Complex aggregation in the tumor through
three administration routes
Using fluorescence microscopy with excitation light
of 550 nm, FITC-labeled complex particles exhibited green
fluorescence. The FA/PAMAM/miR-7 complex aggregated in the tumor
(Fig. 5). Following caudal vein
administration, a few positive signals were observed in the tumor,
and the positive rate was 15.3±3.7% in 10 random high power fields
of view, but no positive signals were detected in other regions of
the brain. Following carotid artery administration, green
fluorescence signals were significantly enhanced in the tumor
compared with caudal vein administration (P<0.05) and the
positive rate was 27.4±5.4%; no positive signals were detected in
other regions of the brain. Administration at the tumor site
produced the strongest positive signals in the tumor compared with
the other two methods and the positive rate was 91.7±6.1%
(P<0.05).
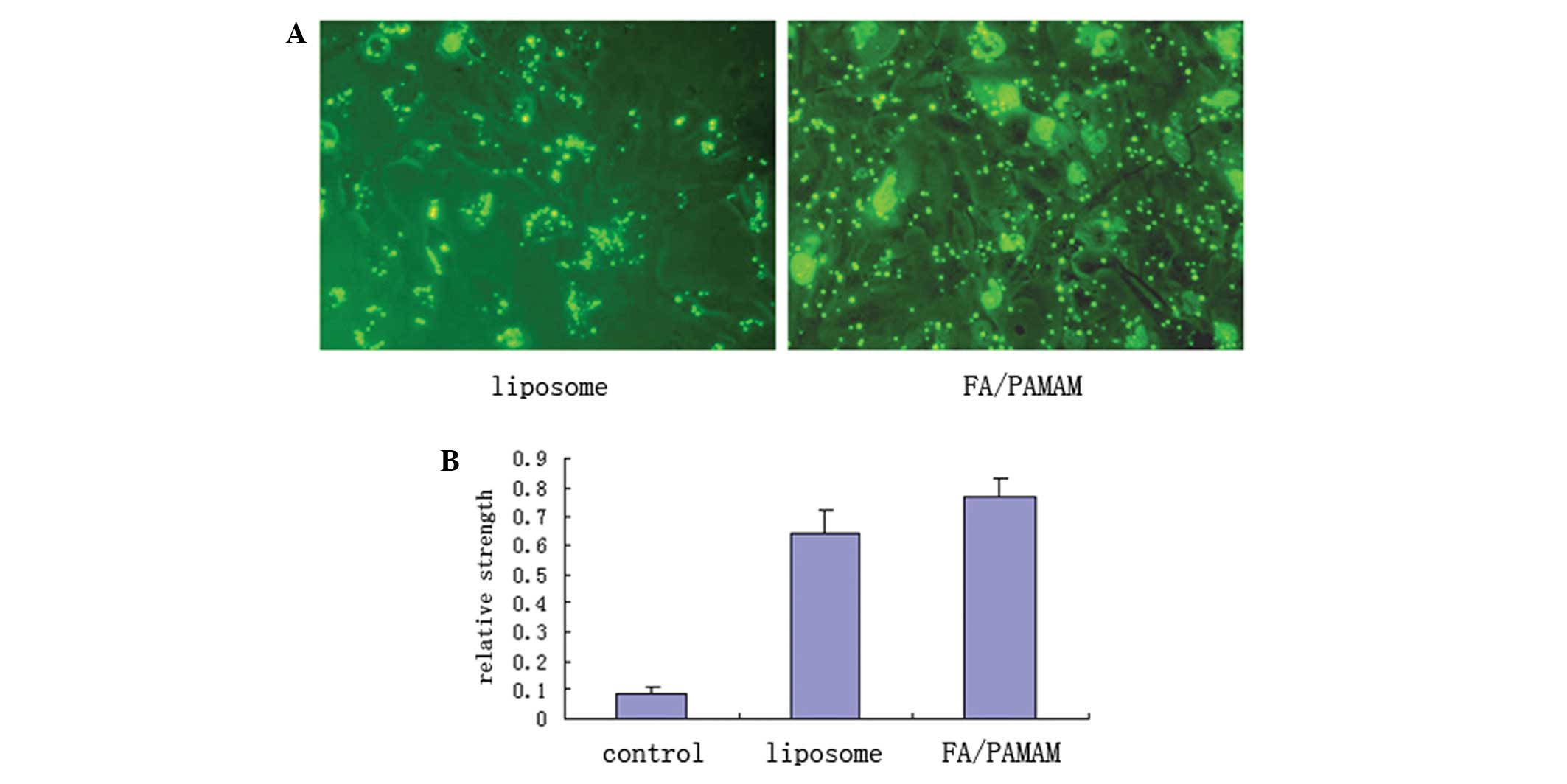
PAMAM produces higher levels of miR-7
transfection compared with liposomes
Using fluorescence microscopy with excitation light
of 550 nm, FITC-labeled green fluorescence was distributed in the
U251 cell membrane or cytoplasm in a bead or spot shape (Fig. 6A). In 10 randomly selected
high-power fields of view, liposome transfection efficiency was
51.4±6.9% and FA/PAMAM transfection efficiency was 87.6±7.8% which
was statistically different (t=11.6, P<0.05). Only a low level
of miR-7 expression was detected in the control group, with a
relative expression intensity of 0.09±0.01. miR-7 expression was
significantly increased in the liposome/miR-7 group with a relative
expression intensity of 0.64±0.08; the relative expression
intensity was 0.77±0.06 in the FA/PAMAM/miR-7 group. There were
significant differences among or between any two groups (P<0.05;
Fig. 6B).
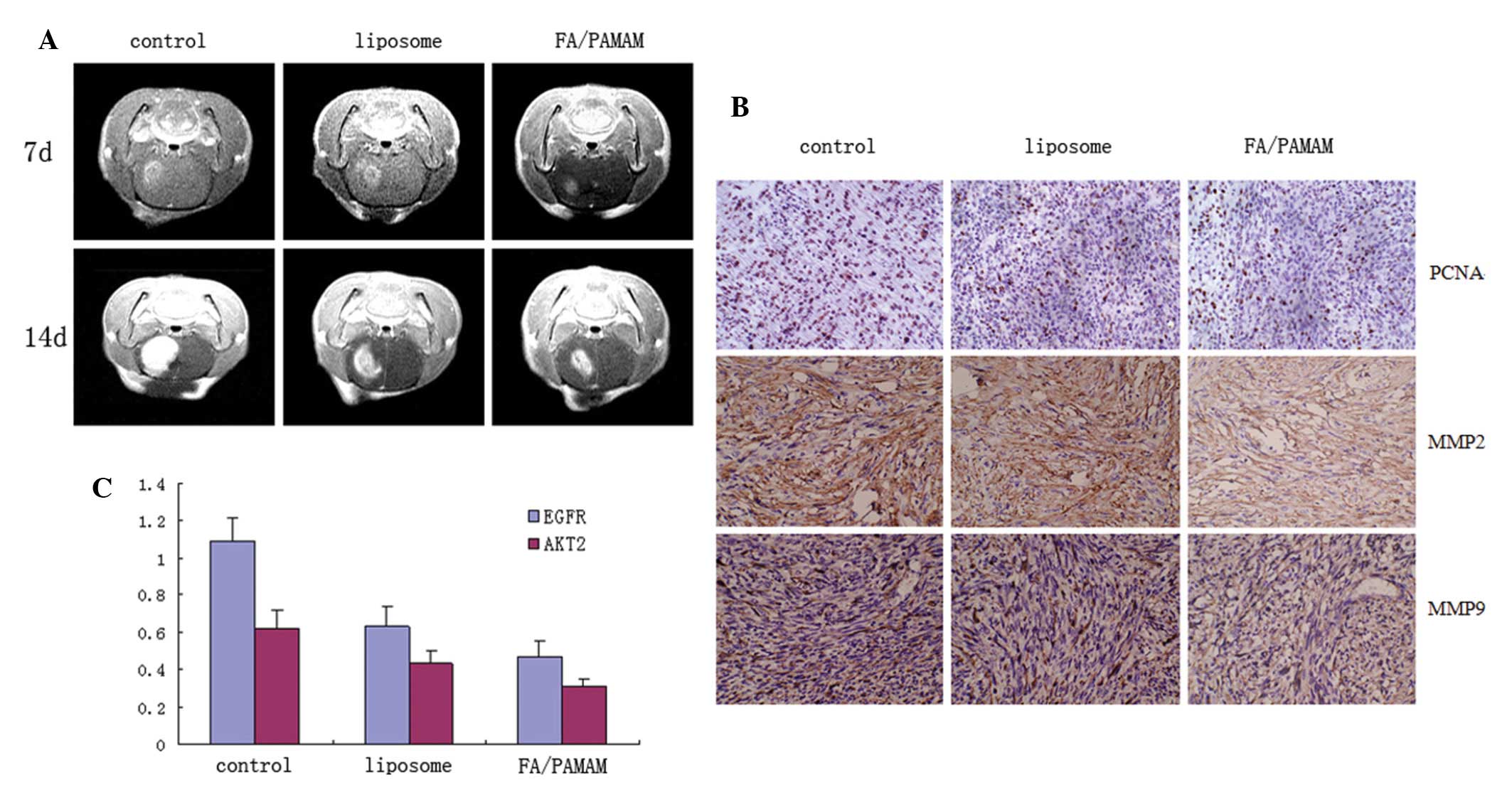
PAMAM/miR-7 suppresses tumor growth more
than liposome/miR-7
Mice in the control group exhibited decreased
activity, depressed emotion, reduced motion to food and water, and
worsened limb hemiplegia. Their mean survival duration was 16.4±2.2
days. The pathological status of mice treated with liposome/miR-7
was delayed and the survival duration was prolonged, with mean
survival duration of 19.4±2.1 days. The pathological status was
significantly delayed in mice treated with FA/PAMAM/miR-7 and they
survived for 23.5±2.4 days on average. There were significant
differences among or between any two groups (P<0.05).
In the control group, intracranial tumor focus was
formed at 7 days after cell transplantation, and the volume of the
tumor grew to 1/2 the size of the hemisphere at 14 days, with
evident edema surrounding the tumor. Intracranial tumor focus was
also observed in the liposome/miR-7 group at 7 days after cell
transplantation, and the volume of the tumor grew to 1/2 the size
of the hemisphere at 14 days, but the volume of the tumor was
smaller than that of the control group. Tumor size was smaller in
the PAMAM/miR-7 group compared with the control and liposome/miR-7
groups at 7 and 14 days and edema surrounding the tumor improved
(Fig. 7A).
The in situ cell apoptosis rate was 5.3±0.9%
in the control group, 11.4±2.4% in the liposome/miR-7 group, and
17.7±3.7% in the FA/PAMAM/miR-7 group, with significant differences
among or between any two groups (P<0.05).
Proliferating cell nuclear antigen, matrix
metalloproteinase 2 and 9 positive rate was 57.3±7.4%, 45.4±6.9%
and 55.1±7.3%, respectively, in the control group, 49.3±5.9%,
31.7±7.1% and 39.4±6.4%, respectively, in the liposome/miR-7 group,
and 34.6±5.4%, 24.5±4.1% and 25.4±5.1%, respectively, in the
FA/PAMAM/miR-7 group, with significant differences among or between
any two groups (P<0.05; Fig.
7B).
The EGFR and AKT2 relative expression intensity was
1.09±0.12 and 0.62±0.1 in the control group, 0.63±0.11 and
0.43±0.07 in the liposome/miR-7 group, and 0.47±0.09 and 0.31±0.04
in the FA/PAMAM/miR-7 group, with significant differences among or
between any two groups (P<0.05; Fig.
7C).
Discussion
The PI3K pathway is a classic pathway in glioma EGFR
signal transduction: ligand→EGFR→PI3K→protein kinase C→IκB kinase,
which phosphorylates inhibitory protein-κB, resulting in nuclear
factor κB migration to the nucleus (21). Nuclear factor κB is an important
transcription factor, and the cascade reaction of signal
transduction can induce intranuclear DNA synthesis and
transformation, impacting glioma cell proliferation,
differentiation and migration (22). In the present study, a miR-7
fragment was designed to silence EGFR and was transfected into U251
glioma cells using FA/PAMAM as a vector. The 5′ end of the miR-7
fragment was labeled with FITC to mark cell transfection
efficiency. Results showed that green fluorescein was evenly
distributed in cell membranes or cytoplasm and the positive rate
was 87.6±7.8%. This was significantly higher than liposome-mediated
miR-7 transfection (58.5±5.4%), indicating the high efficiency of
FA/PAMAM. qRT-PCR results suggested that the level of miR-7 was
significantly increased 48 h following FA/PAMAM/miR-7 transfection.
Moreover, the levels of three major members of the EGRF pathway,
EGFR, PI3K and AKT2, were significantly reduced, indicating that
miR-7 transfection can effectively silence the EGFR pathway in
glioma cells.
Subsequently, the potential for cell proliferation
activity, invasion ability and tumorigenesis was investigated.
Results showed that following miR-7 transfection, U251 cell
proliferation was significantly decreased, and a high percent of
cells was in the G1 phase. Moreover, the invasion
ability of cells was significantly reduced. Soft agar cell clone
formation has frequently been used to simulate semisolid growth
environment of the in vivo extracellular matrix and to
assess colony growth potential of tumor cells (23). In the present study, the clone
formation ability of miR-7 transfected cells was significantly
decreased, indicating reduced oncogenicity of miR-7 transfected
U251 glioma cells.
In addition, the present study investigated the
glioma targeting of FA/PAMAM in vivo. FA/PAMAM/miR-7
administered through the caudal vein, carotid artery or tumor in
situ aggregated in the tumor bed in the brain, but not in
tissues surrounding the tumor, indicating good targeting of the
FA/FA receptor axis. The positive rate of FA/PAMAM/miR-7
aggregation in the tumor bed was 15.3±3.7%, 27.4±5.4% and 91.7±6.1%
with delivery through the caudal vein, carotid artery or tumor
in situ, respectively, while the positive rate of
liposome/miR-7 aggregation in the tumor bed was 5.4±0.7%, 11.4±2.1%
and 64.5±7.4% through the caudal vein, carotid artery or tumor
in situ, respectively, indicating high targeting efficiency
of FA/PAMAM as a small molecule drug carrier. Data comparison also
showed significant differences in the amount of drug aggregation in
the tumor bed among the three administration routes, the amount of
drug was maximal in the tumor in situ group, followed by the
carotid artery and then the caudal vein groups. This may result
from the drug being retained in some organs or tissues. Therefore,
the reduction of drug retainment following vessel perfusion is
important in developing small molecule target drugs.
To assess the treatment effect of FA/PAMAM/miR-7 on
in vivo and in vitro glioma, the present study
utilized liposomes as a control. Results showed that FA/PAMAM
significantly improved miR-7 transfection efficiency compared with
liposomes, manifested by higher miR-7 expression in transfected
U251 cells. In vivo experiments showed that the survival of
animals was prolonged in the FA/PAMAM/miR-7 group compared with the
liposome/miR-7 group. In particular, MRI results showed slow glioma
proliferation in the early stage, but rapid growth in the middle
and later stages in the liposome/miR-7 group, but continuously slow
proliferation in the FA/PAMAM/miR-7 group. The second MRI screen
(14 days following U251 glioma cell transplantation) showed that
the tumor size was significantly smaller in the FA/PAMAM/miR-7
group compared with the control and liposome/miR-7 groups. These
results were consistent with the delayed drug release property of
FA/PAMAM. Furthermore, to investigate the effect of miR-7 on
silencing target genes, the present study detected EGFR and AKT2
protein levels. Results showed better target gene silencing in the
FA/PAMAM/miR-7 group, manifested as significantly lower EGFR and
AKT2 protein levels compared with the control and liposome/miR-7
groups. The proliferating cell nuclear antigen positive rate can
directly reflect tumor cell proliferation activity, matrix
metalloproteinase 2 and 9 levels represent tumor cell invasion
ability (8). Results from the
present study indicate that FA/PAMAM/miR-7 significantly suppressed
tumor cell proliferation and invasion ability. As there were no
complementary sequences in miR-7 with proliferating cell nuclear
antigen, matrix metalloproteinase 2 and 9, we assume that the
protein suppression results from EGFR and AKT2 silencing
effects.
In conclusion, PAMAM exhibited better gene
transfection efficiency and target gene silencing compared with
liposomes. Moreover, PAMAM can maintain a longer action to
significantly suppress tumor cells. However, FA/PAMAM/miR-7
transfection did not completely remove glioma. The survival
duration of animals was only prolonged and the survival rate of 30
days remains very low. Therefore, further investigations are
required for glioma gene therapy.
Acknowledgements
This study was supported by the China National
Natural Scientific Fund (81000901), the Tianjin Science and
Technology Committee (09JCYBJC09500), the Key Laboratory Project of
Tianjin Programs for Science and Technology (10SYSYJC28800), the
Key Project of Chinese National Programs for Fundamental Research
and Development (973 Program, 2010CB529405), and the Tianjin Health
Bureau Science and Technology Projects (2011KZ24).
References
|
1
|
Nakamura M, Shimada K, Nakase H, et al:
Clinicopathological diagnosis of gliomas by genotype analysis.
Brain Nerve. 61:773–780. 2009.(In Japanese).
|
|
2
|
Mladkova N and Chakravarti A: Molecular
profiling in glioblastoma: prelude to personalized treatment. Curr
Oncol Rep. 11:53–61. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huang H, Mahler-Araujo BM, Sankila A, et
al: APC mutations in sporadic medulloblastomas. Am J Pathol.
156:433–437. 2000. View Article : Google Scholar
|
|
4
|
Gilbertson RJ: Medulloblastoma: signalling
a change in treatment. Lancet Oncol. 5:209–218. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Misaki K, Marukawa K, Hayashi Y, et al:
Correlation of gamma-catenin expression with good prognosis in
medulloblastomas. J Neurosurg. 102:197–206. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ellison DW, Onilude OE, Lindsey JC, et al:
beta-Catenin status predicts a favorable outcome in childhood
medulloblastoma: the United Kingdom Children’s Cancer Study Group
Brain Tumour Committee. J Clin Oncol. 23:7951–7957. 2005.PubMed/NCBI
|
|
7
|
Oikonomou E, Barreto DC, Soares B, et al:
Beta-catenin mutations in craniopharyngiomas and pituitary
adenomas. J Neurooncol. 73:205–209. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sekine S, Shibata T and Kokubu A:
Craniopharyngiomas of adamantinomatous type harbor beta-catenin
gene mutations. Am J Pathol. 161:1997–2001. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gonzalez S, Pisano DG and Serrano M:
Mechanistic principles of chromatin remodeling guided by siRNAs and
miRNAs. Cell Cycle. 7:2601–2608. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Novakova J, Slaby O, Vyzula R, et al:
MicroRNA involvement in glioblastoma pathogenesis. Biochem Biophys
Res Commun. 386:1–5. 2009. View Article : Google Scholar
|
|
12
|
Si ML, Zhu S, Wu H, et al: miR-21-mediated
tumor growth. Oncogene. 26:2799–2803. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tong AW and Nemunaitis J: Modulation of
miRNA activity in human cancer: a new paradigm for cancer gene
therapy? Cancer Gene Ther. 15:341–355. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kefas B, Godlewski J, Comeau L, et al:
microRNA-7 inhibits the epidermal growth factor receptor and the
Akt pathway and is down-regulated in glioblastoma. Cancer Res.
68:3566–3572. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang H: Nanoparticle-mediated
brain-specific drug delivery, imaging, and diagnosis. Pharm Res.
27:1759–1771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tomalia DA: A new class of polymers:
starburst-denfritic macromolecules. Polym J. 17:117–132. 1985.
View Article : Google Scholar
|
|
17
|
Malik N, Wiwattanapatapee R, Klopsch R, et
al: Dendrimers: relationship between structure and biocompatibility
in vitro, and preliminary studies on the biodistribution of
125I-labelled polyamidoamine dendrimers in vivo. J Control Release.
65:133–148. 2000.PubMed/NCBI
|
|
18
|
Huang S, Fu L, Zhang X, et al: Syntheses
of polyamidoamine dendrimers starting from a hexadimensional core
and application in gene transfer. Science in China Series B:
Chemistry. 46:271–279. 2003. View
Article : Google Scholar
|
|
19
|
Tang MX, Redemann CT and Szoka FC Jr: In
vitro gene delivery by degraded polyamidoamine dendrimers.
Bioconjug Chem. 7:703–714. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Luo D, Haverstick K, Belcheva N, et al:
Poly(ethylene glycol)-conjugated PAMAM dendrimer for biocompatible,
high-efficiency DNA delivery. Macromolecules. 35:3456–3462. 2002.
View Article : Google Scholar
|
|
21
|
Belda-Iniesta C, de Castro Carpeño J,
Sereno M, et al: Epidermal growth factor receptor and glioblastoma
multiforme: molecular basis for a new approach. Clin Transl Oncol.
10:73–77. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Loew S, Schmidt U, Unterberg A, et al: The
epidermal growth factor receptor as a therapeutic target in
glioblastoma multiforme and other malignant neoplasms. Anticancer
Agents Med Chem. 9:703–715. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zi X, Guo Y, Simoneau AR, et al:
Expression of Frzb/secreted Frizzled-related protein 3, a secreted
Wnt antagonist, in human androgen-independent prostate cancer PC-3
cells suppresses tumor growth and cellular invasiveness. Cancer
Res. 65:9762–9770. 2005. View Article : Google Scholar
|