Introduction
microRNAs (miRNAs) are small non-coding RNAs (~22
nt) that regulate the expression of target genes by promoting RNA
cleavage or inhibiting RNA translation. Dicer, a key member of the
RNase III family, is an essential component of the miRNA-processing
machinery in the cytoplasm in which the precursors of miRNAs are
processed by Dicer into mature miRNAs (1). The abnormal expression of miRNAs is
associated with the development and progression of cancer (2,3). Dicer
is aberrantly expressed in different types of cancer, and, being an
upstream regulator of miRNAs, Dicer may affect the behavior of
cancer by altering the expression spectrum of miRNAs.
The miRNA expression spectrum varies with cancer
types, although in most types of cancer miRNAs are globally
downregulated (4). However, whether
the reduction in miRNA expression promotes cancer development or
merely reflects the state of undifferentiated tumors is unclear.
Markedly, tumor behaviors have been reported to be altered after
silencing Dicer expression in vitro (5,6), but
little evidence is available regarding the expression patterns of
Dicer in tongue squamous cell carcinoma or the influence of
knocking down Dicer expression on this type of cancer. In the
present study, we first compared Dicer protein and mRNA expression
between 2 carcinoma cell lines, Tca-8113 (7,8) and
UM-1 (8,9) and normal gingival epithelial cells. We
found that Dicer protein levels were lower in the carcinoma cells
than in normal cells, and that Dicer expression was the lowest in
the highly aggressive UM-1 cancer cells. Thus, we knocked down the
expression of Dicer in Tca-8113 cells using siRNA transfection
in vitro to investigate how disrupting Dicer-dependent
maturation of miRNAs affects cell proliferation, cell cycle
patterns and cell migration and invasion to affect cancer
development.
Materials and methods
Cell culture and siRNA transfection
Normal gingival epithelial cells were purchased from
the American Type Culture Collection (ATCC; Manassas, VA, USA).
Tca-8113 and UM-1 were kindly provided by Dr AnXun Wang (The First
Hospital Affiliated with Sun Yat-Sen University, Guangzhou, China).
Cells were grown in minimal essential medium containing dimethyl
sulfoxide (DMSO); media used were MEM for normal gingival
epithelial cells, RPMI-1640 for Tca-8113 cells, and DMEM for UM-1
cells, all supplemented with 10% fetal bovine serum (FBS; Gibco).
Dicer siRNA and negative-control siRNA were synthesized by Shanghai
Jima Co. (Shanghai, China). The target sequences for Dicer siRNA
were: 5′-GCCAAGGAAAUCAGCUAAATT-3′ and 5′-UUUAGCU GAUUUCCUUGGCTT-3′.
The negative-control siRNA sequences were:
5′-UUCUCCGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′.
One day before transfection, Tca-8113 cells were
re-plated in 6-well plates to obtain 50–80% confluence on the day
of transfection. A complex of siRNAs (100 nM) and 5 μl
Lipofectamine RNAiMAX reagent (Invitrogen) was added into each well
according to the manufacturer’s instructions. Transfection
efficiency was estimated 6 h later using fluorescence microscopy.
Transfections were performed in triplicate for each treatment.
Real-time quantitative PCR analysis
Total RNA was extracted using TRIzol reagent
(following the manufacturer’s instructions; Invitrogen) from normal
gingival epithelial cells, Tca-8113 cells, UM-1 cells and 3 groups
of Tca-8113 cells: cells transfected with Dicer-siRNA and
negative-control siRNA and non-transfected cells. From the total
RNA, cDNAs were synthesized using the Takara PrimeScript RT reagent
(Perfect Real-Time kit) under reaction systems for 500 ng and 10
μl. The qPCR primer sequences used in the study were: Dicer forward
primer, 5′-TGTGGGGAGAGGGCTGCTCA-3′ and reverse primer,
5′-GGCACAGGGCCTTTTCCCGA-3′; GAPDH forward primer,
5′-AAGGTGAAGGTCGGAGTCA AC-3′ and reverse primer,
5′-GGGGTCATTGATGGCAACA ATA-3′.
GAPDH was used as an internal control. PCR was
performed in triplicate in a total reaction volume of 20 μl, which
included 10 μl of the 2X reaction mix, 0.4 μl of 50X Rox, 3 μl of
primer (2 μM), 5.6 μl of water and 1 μl of cDNA, with SYBR Premix
Ex Taq kit. The PCR protocol used involved denaturation for 30 sec
at 95°C, followed by amplification for 40 cycles, each cycle
consisting of 5 sec at 95°C, 30 sec at 56°C and 30 sec at 72°C. The
median in each triplicate was used to calculate the relative Dicer
expression level using the comparative ΔCt method [value of
2−ΔCt (Dicer or GAPDH)]; fold-changes in expression were
calculated using 2−ΔΔCt.
Western blotting
Total proteins were extracted from normal gingival
epithelial cells, Tca-8113 cells, UM-1 cells and the 3 groups of
Tca-8113 cells (transfected with Dicer-siRNA and negative-control
siRNA and non-transfected) after transfection for 24 and 48 h.
Cells were washed thrice with phosphate-buffered saline (PBS)
before being lysed on ice in 50 μl 1X extraction buffer prepared
fresh. Following centrifugation for 10 min, supernatants were
collected and aliquots were withdrawn for detecting proteins using
Micro BCA™ protein assay reagent kit according to the
manufacturer’s instructions. SDS-PAGE (10% gels) was used to
resolve proteins (40 μg/lane), which were transferred
electrophoretically for 2 h to polyvinylidene difluoride membranes.
Membranes were blocked for 1 h in PBS-Tween (0.1%) containing 5%
non-fat milk, probed overnight at 4°C with primary antibodies
(rabbit anti-human Dicer, 1:500; Sigma, St. Louis, MO, USA), and
then incubated for 1 h with secondary antibodies (goat anti-rabbit
IgG, 1:1,000; Wuhan Boster Biological Technology, Ltd., Wuhan,
China) for visualizing protein bands using enhanced
chemiluminescence. Results shown are representative from
experiments repeated at least twice.
Immunohistochemical staining
Dicer was labeled in normal gingival epithelial
cells, Tca-8113 and UM-1 cells that had reached 80% confluence.
Labeling with anti-human Dicer (Sigma) and secondary anti-rabbit
IgG (Wuhan Boster Biological Technology) was according to the
manufacturer’s protocols using the SV-002 2-step-method
Immunohistochemistry kit.
MTT assay
Cells were plated in 96-well plates at a density of
2,000 cells/well in 100 μl of culture medium and maintained until
they adhered before carrying out transfections. After transfection
with siRNAs at 37°C for 4 h, cells were incubated with 20 μl of MTT
solution (5 mg/ml) for 24 and 48 h. At the end of the incubation,
media were aspirated, 200 μl of DMSO was added to each well and the
absorbance at 570 nm was measured. Mean absorbance from 3 replicate
wells was determined, and cell viability was defined as (absorbance
of treated cells)/(absorbance of control cells).
Cell cycle analysis
The 3 groups of cells were harvested 24 h after
transfection with siRNA; cells were fixed overnight with 70% cold
ethanol, washed twice with cold PBS, and then incubated in RNase A
(10 mg/ml) for 1 h at 37°C. Propidium iodide (PI) 100 μg/ml was
added and mixed for 30 min and then the cells were used for flow
cytometric analysis.
Cell adhesion test, Erasion Trace test
and Transwell cell-invasive assay
Matrigel (50 μl) diluted with RPMI-1640 (1:5) was
added to 96-well plates and maintained at 4°C overnight. The next
day, plates were sealed at 37°C for 1 h after replacing the fluid
in the wells with 100 μl of RPMI-1640 containing 1% BSA.
Experimental and control cells were plated without serum in the
wells and incubated for 1 h, and the cells that did not adhere to
the Matrigel were washed off using PBS. Next, 20 μl of MTT with 100
μl of RPMI-1640 was added to each well and incubated for 4 h. At
the end of the incubation, the solution was aspirated, 200 μl of
DMSO was added to each well, and the absorbance at 570 nm was
measured; mean absorbance was determined from 3 replicate
wells.
When attached, transfected cells were 80% confluent
in 24-well plates, the culture medium was replaced with basic
medium, and a 100-μl pipette tip was used to scrape cells in the
middle of the wells. The scraped cells were washed off using PBS
and the plates were placed in a 37°C incubator for 48 h. Distances
migrated by cells across the middle of the wells were calculated by
examining cells from 5 fields selected randomly under the
microscope.
To the top chamber of a Transwell chamber, 30 μl of
Matrigel diluted with RPMI-1640 (1:3) was added and maintained at
37°C and allowed to polymerize. Transfected cells were trypsinized
and adjusted to a concentration of 5×105 cells/ml in
RPMI-1640, and 200 μl of the resuspended cell solution was added to
the top chamber above the Matrigel. The bottom chamber was filled
with 600 μl of RPMI-1640 containing 10% serum. The Transwell plates
were incubated at 37°C for 24 h and then the top chamber was
removed and the Matrigel with unmigrated cells was gently scraped
with a wet cotton swab. Cells were stained with 0.1% crystal violet
for 5 min, washed with PBS to remove excess stain, and the average
number of cells per field that had migrated was quantified under
the microscope. The assay was repeated using the Transwell chamber
without Matrigel.
Statistical analysis
SPSS-13.0 software, variance analysis, and 2
independent t-tests were used to analyze the differences in Dicer
expression between the 3 types of cells and to evaluate the
biological responses in treated and control cells.
Results
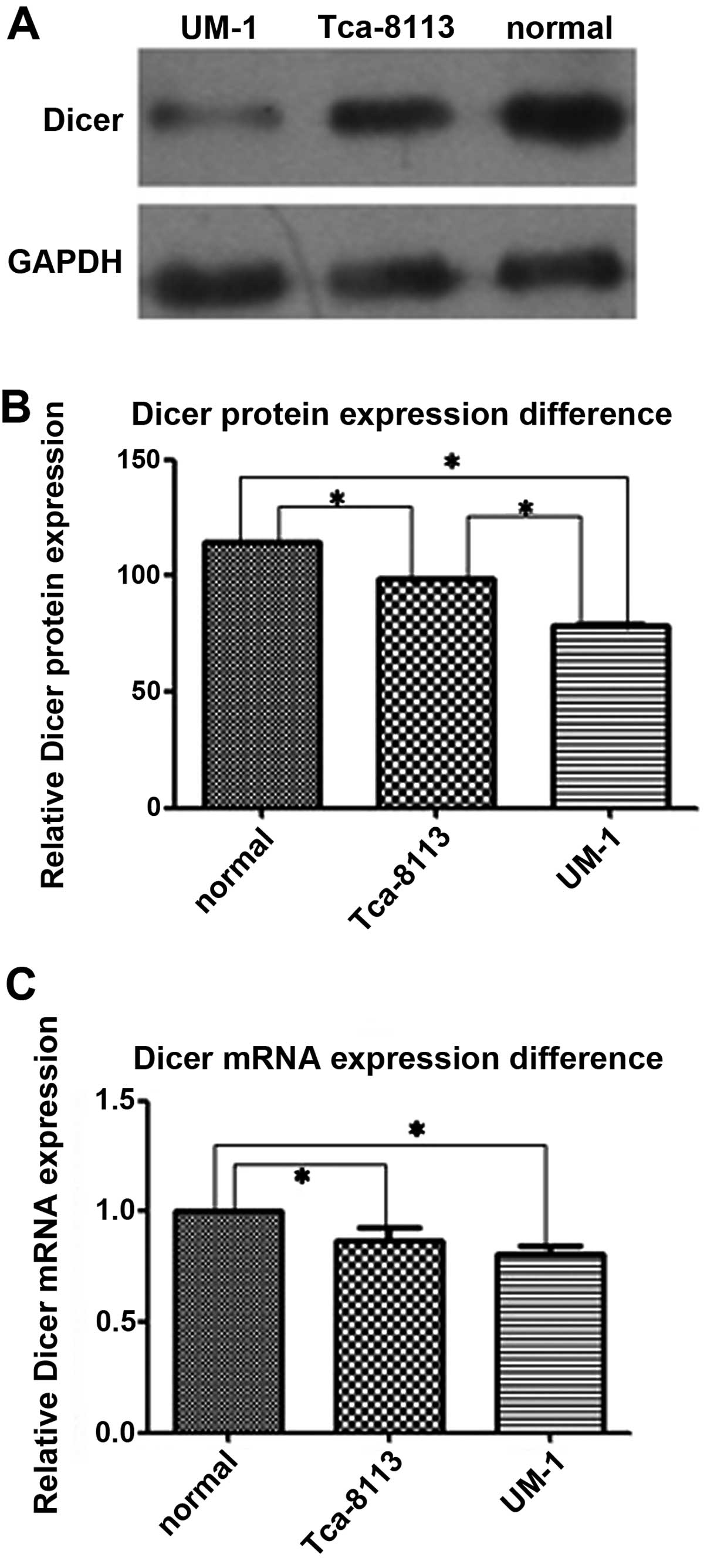
Dicer is downregulated in tongue squamous
cell carcinoma cell lines
The relative expression levels of Dicer mRNA and
protein were evaluated in normal oral gingival epithelial cells and
2 tongue squamous cell carcinoma cell lines, Tca-8113 and UM-1
cells (Figs. 1 and 2). Real-time qPCR showed that more Dicer
mRNA was present in normal oral gingival cells than in Tca-8113
cells (P=0.020) and UM-1 cells (P=0.002). Western blotting showed
that the amount of Dicer protein was significantly less in the 2
tongue squamous cell lines than in normal gingival epithelial cells
(P=0.000 for both Tca-8113 and UM-1 cells). Dicer mRNA levels in
Tca-8113 and UM-1 were not significantly different (P=0.166), but
the amount of Dicer protein in the highly aggressive carcinoma cell
line UM-1 was less than in Tca-8113 cells (P=0.000). Lastly, these
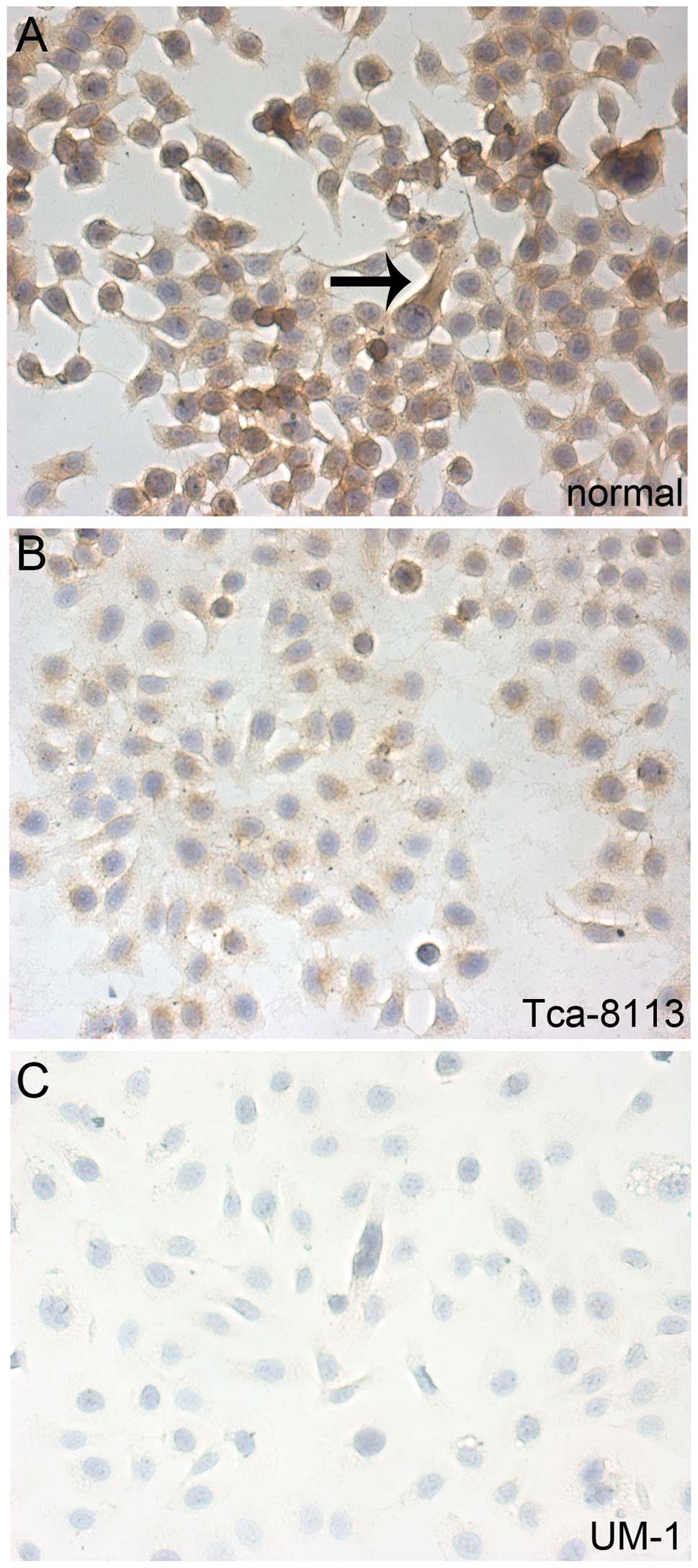
results were further confirmed by the immunohistochemical staining
for Dicer in the cells.
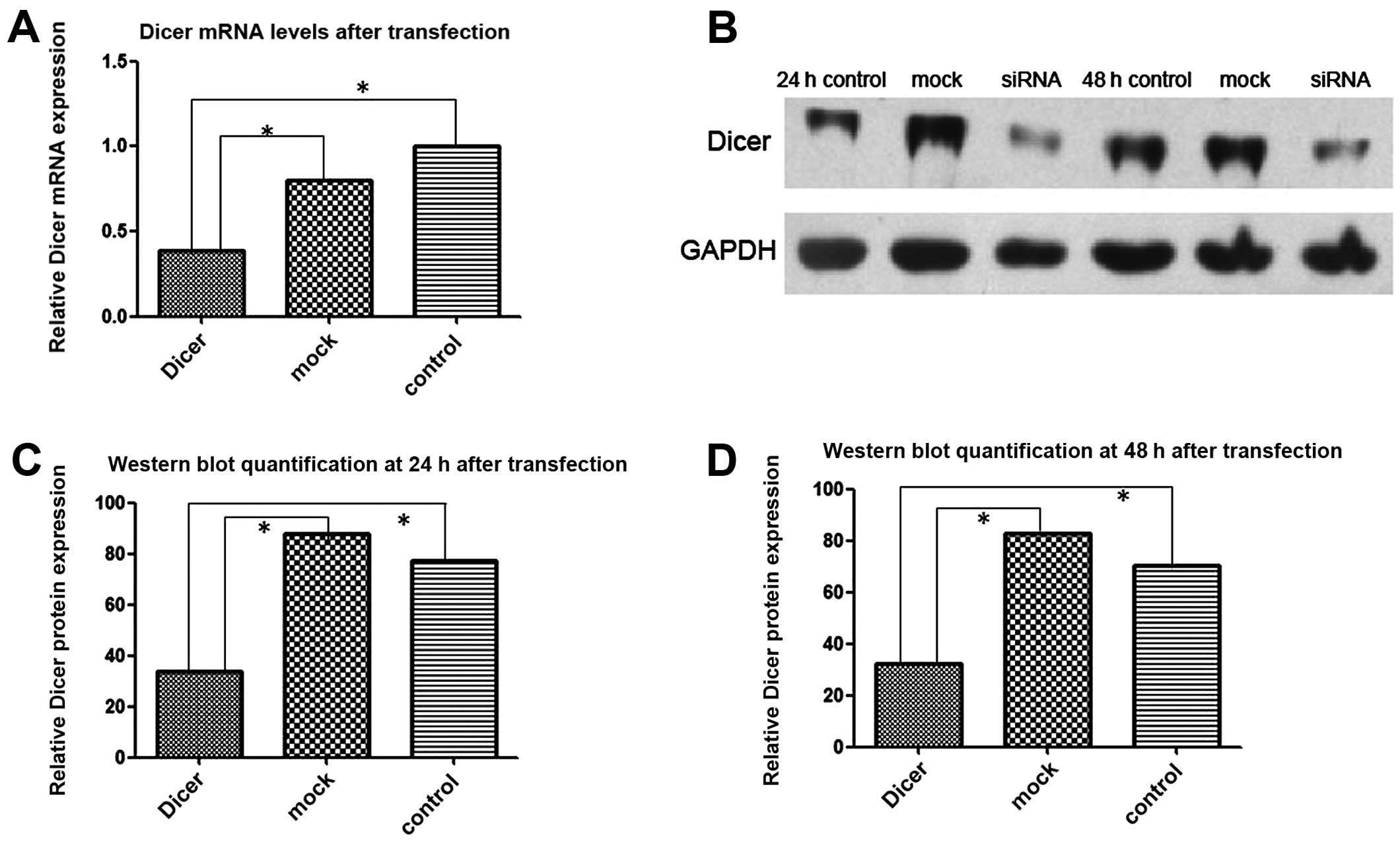
RNAi knocks down Dicer expression in
Tca-8113 cells
FAM-labeled siRNAs were transfected into Tca-8113
cells and 6 h later the transfection efficiency was determined
using fluorescence microscopy. Approximately 80% of the transfected
cells emitted green fluorescence upon excitation. Total cellular
RNAs from Dicer siRNA-transfected cells and control cells
(negative-control-siRNA transfected and non-transfected) were
extracted 24 h after transfection. RT-PCR showed that the level of
Dicer mRNA in the Dicer-siRNA group was significantly lower than in
the 2 control groups (P=0.000). Similarly, western blotting
demonstrated that the Dicer protein amount was substantially lower
in the Dicer-siRNA group than in the 2 control groups at 24 and 48
h after transfection (Fig. 3).
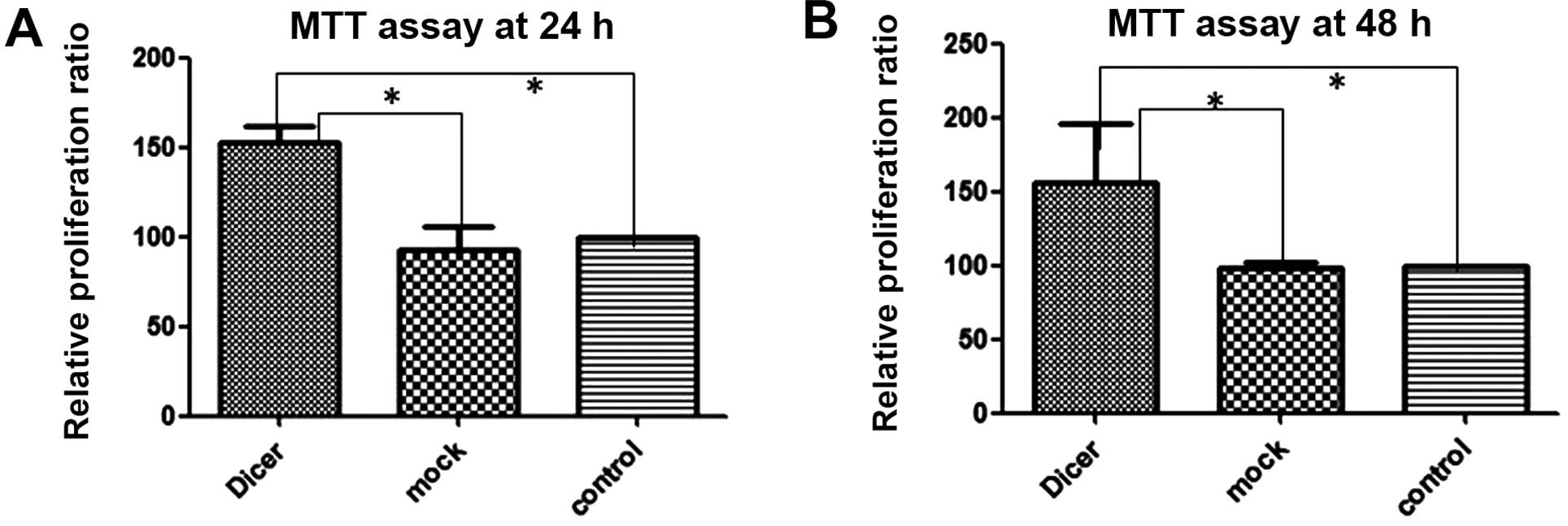
Silencing Dicer promotes cell
proliferation and cell cycle
To investigate the impact of Dicer on the
proliferative capacity of cells, we measured the effects of
Dicer-siRNA transfection on cell proliferation using the MTT assay
and on cell cycle progression by using flow cytometry following PI
staining. The MTT assay (Fig. 4)
demonstrated that transfection with Dicer-siRNA increased Tca-8113
cell proliferation at 24 and 48 h after transfection relative to
the 2 controls (negative-control siRNA transfected and
non-transfected cells). Moreover, cell cycle analysis revealed that
in Tca-8113 cells transfected with Dicer-siRNA, the rate of S+G2
phases increased sharply accompanied by a reduction in G1 phase
(Fig. 5; Table I), indicating that the depletion of
Dicer accelerated the cell cycle (to S and G2 phases).
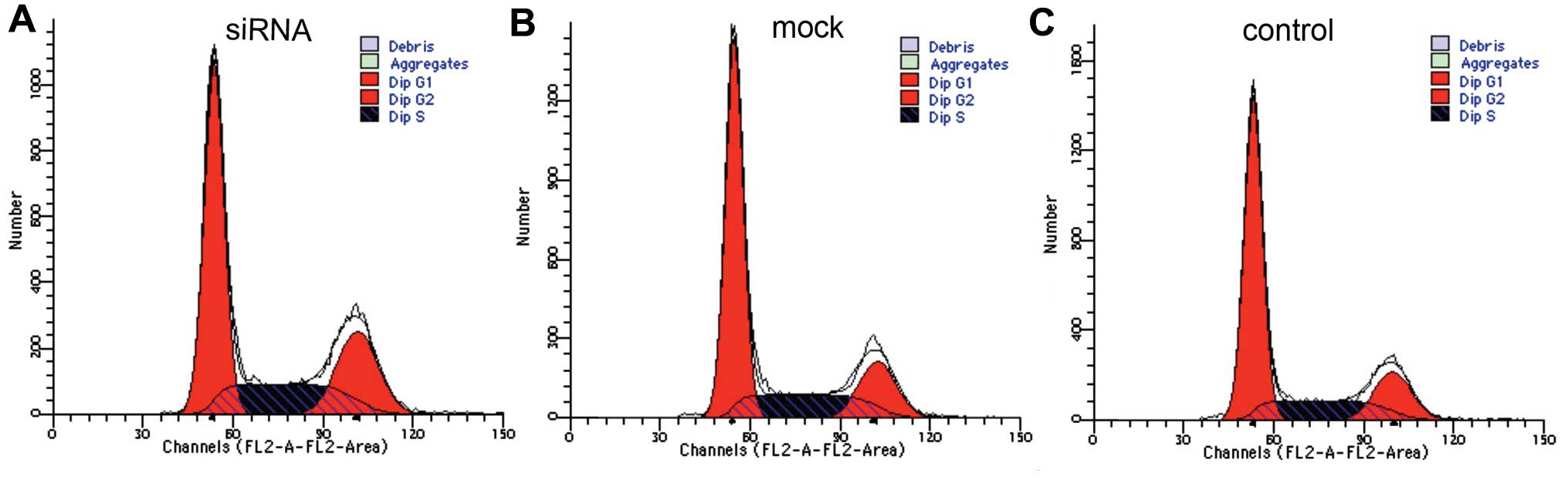 | Figure 5Silencing Dicer expression alter cell
cycle distribution. Cell cycle analysis using flow cytometry
demonstrated that Dicer downregulation increased the proliferative
capacity of Tca-8113 cells. Cells in S+G2 phases made up 48.47,
39.58 and 38.73%, and cells in the G1 phase made up 51.42, 60.20
and 61.23%, in Tca-8113 cells transfected with Dicer-siRNA,
negative-control siRNA, and non-transfected cells, respectively.
Thus, the cell cycle in the experimental group (Dicer-siRNA) had
advanced to S phase. (A) siRNA, Tca-8113 cells transfected with
Dicer-siRNA; (B) mock, Tca-8113 cells transfected with negative
siRNA; (C) control, non-transfected Tca-8113 cells. |
 | Table ICell cycle distribution. |
Table I
Cell cycle distribution.
| Cell cycle | Control | Mock | siRNA | P-value |
|---|
| G1 | 61.23±0.15 | 60.20±0.04 | 51.42±0.23 | 0.000/0.001 |
| S+G2 | 38.73±0.21 | 39.58±0.35 | 48.47±0.07 | 0.004/0.014 |
Silencing Dicer expression increases cell
migration and invasion
To examine how silencing Dicer expression affects
the migratory and invasive abilities of Tca-8113 cells, we
quantified cell migration using cell adhesion and Erasion Trace
tests and the Transwell cell-invasive assay. Compared to the
control cells, more cells that had been transfected with
Dicer-siRNA adhered to the Matrigel, migrated through the Transwell
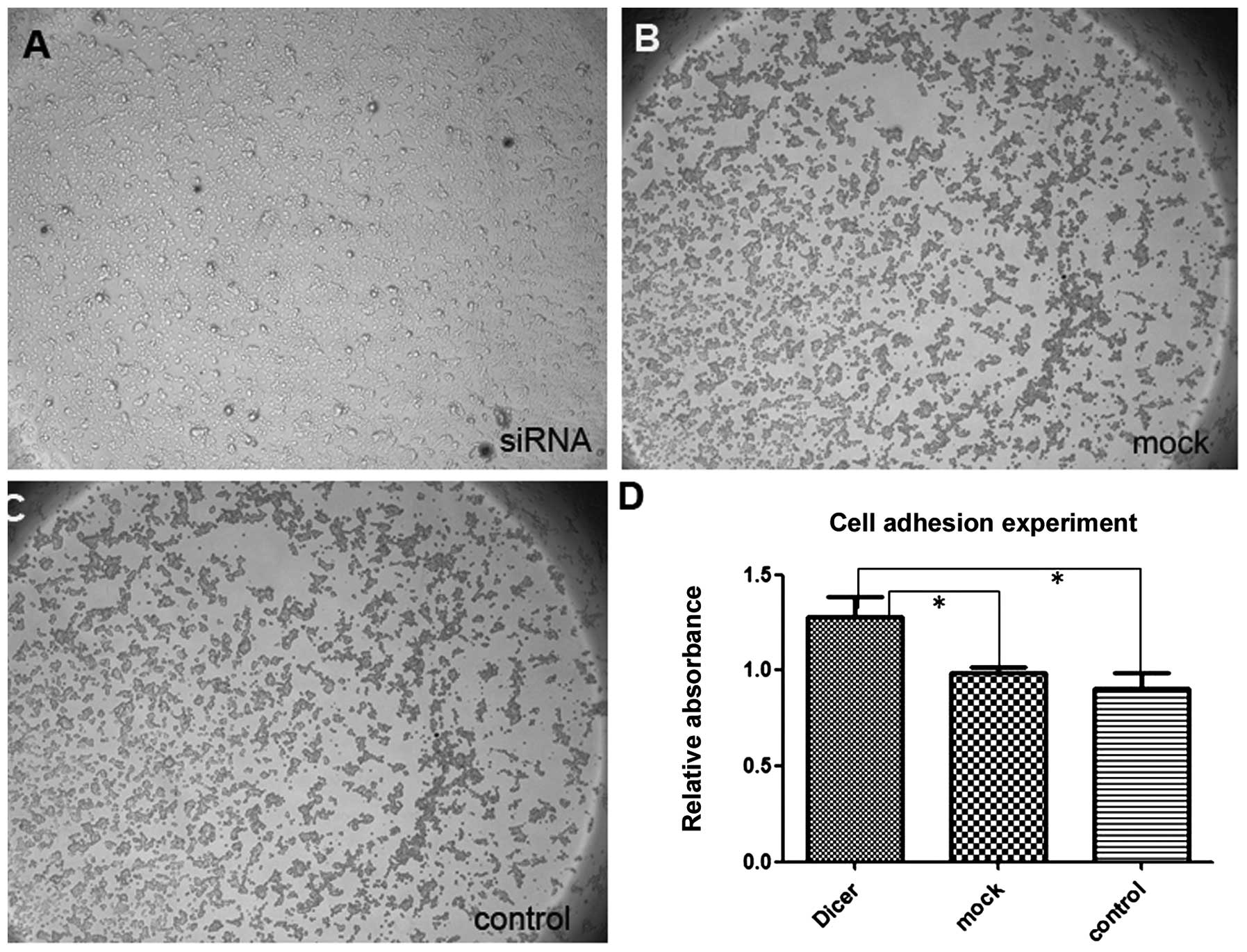
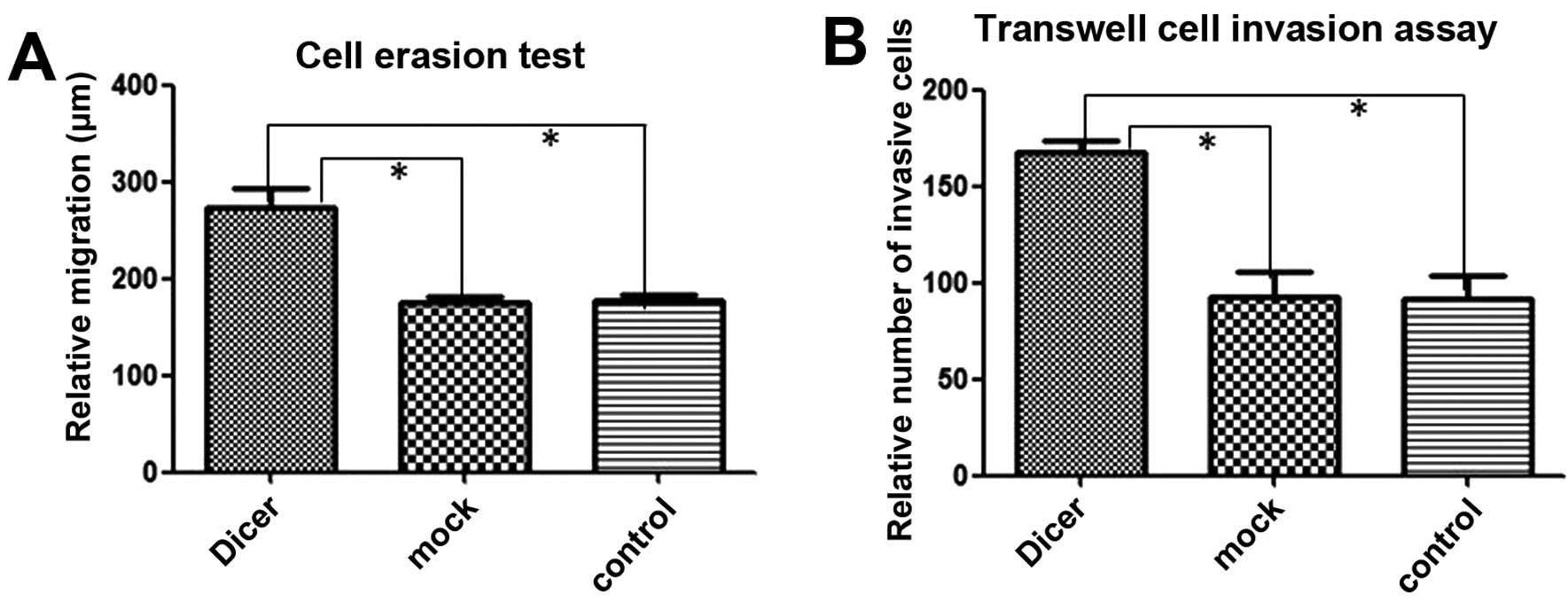
membrane and covered the erased trace better. Absorbance at 570 nm
in the Matrigel adhesion test for Dicer-siRNA cells was 1.277±0.109
compared to 0.990±0.026 and 0.905±0.079 for negative-control siRNA
cells and non-transfected cells, respectively (P=0.011, P=0.009;
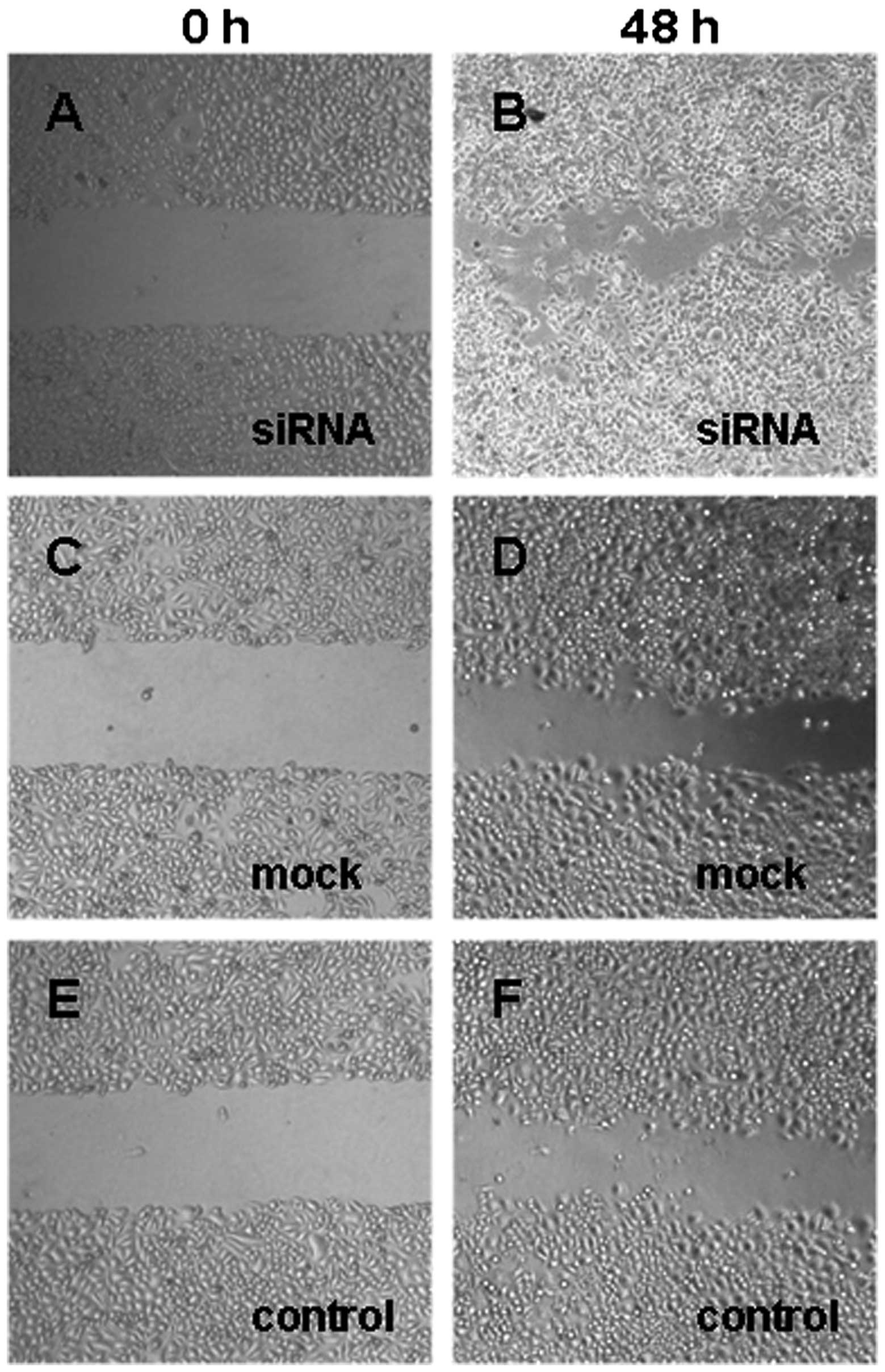
Fig. 6). Similarly, Dicer-siRNA
cells migrated more and covered the erased trace to 273.42±18.95 μm
compared to 175.76±5.81 μm and 178.14±5.01 μm for cells transfected
with the negative-control siRNA and non-transfected cells,
respectively (P=0.000, P=0.000; Figs.
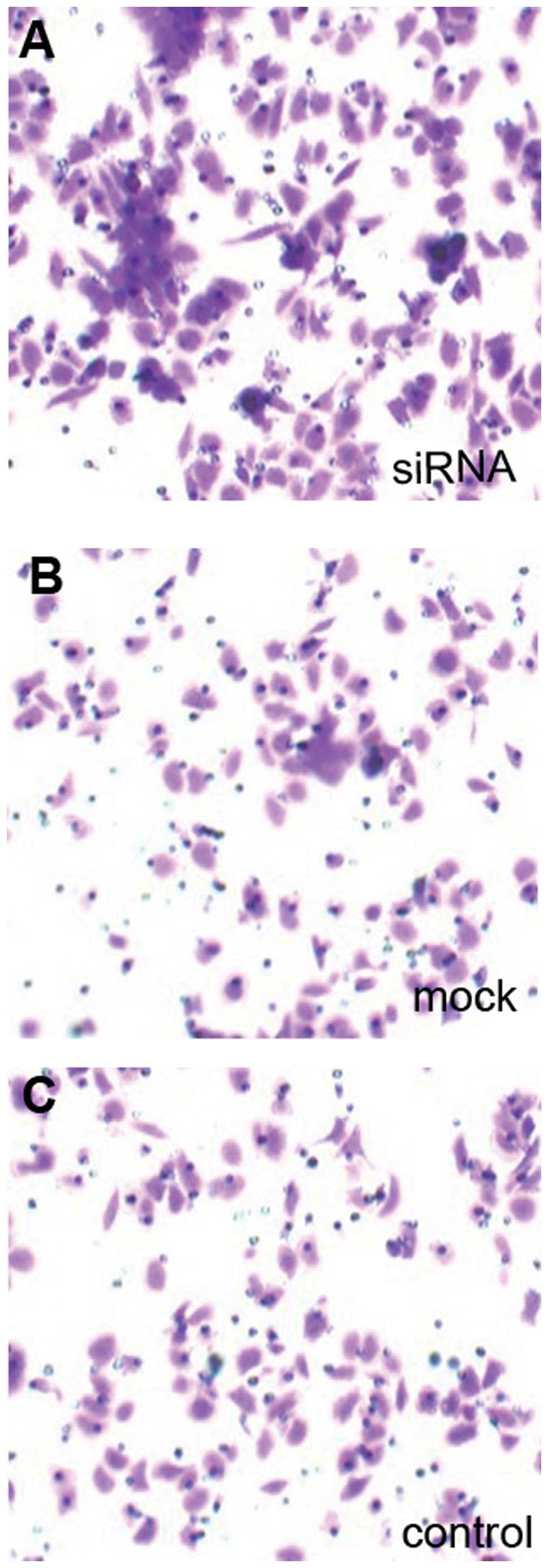
7 and 9A). Furthermore,
Dicer-siRNA transfection increased Transwell migration to
167.20±6.38 cells from 92.40±13.22 cells and 91.60±11.61 cells in
negative-control siRNA cells and non-transfected cells,
respectively (P=0.000, P=0.000; Figs.
8 and 9B). Therefore,
transfecting Dicer-siRNA into Tca-8113 cells increased cell
migration and invasion in vitro to levels significantly
greater than in cells transfected with the negative-control siRNA
and in non-transfected cells. Collectively, our results suggest
that knocking down Dicer expression in Tca-8113 cells increases the
invasive and proliferative capacities of these cells.
Discussion
Dicer, a member of RNase III family essential for
miRNA processing, is known to be associated with cancer. Dicer is
upregulated in prostate, colorectal cancer and lung squamous cell
carcinoma (1,10–14),
and is downregulated in hepatocellular carcinoma, gastric, breast
and ovarian cancer (15–18). Dicer is mis-expressed in different
types of cancer, and Dicer expression changes are biomarkers of
poor prognostics for cancer patients.
Cancer can be promoted or suppressed by miRNAs
(19). In certain types of cancer,
miRNAs are upregulated (20), but
in most cancers the miRNAs are globally downregulated, and this
downregulation plays a key role in the phenotypic transformation of
cancer (4). Since Dicer is an
upstream regulator of miRNAs, the mis-expression of Dicer in cancer
can partly explain the abnormal expression of miRNAs in the cancer
cells, and several studies have demonstrated changes in cancer
behavior after silencing Dicer both in vivo and in
vitro. For instance, knocking down Dicer in breast cancer can
lead to a more malignant and invasive phenotype in vitro
(21). Moreover, in animal studies,
silencing Dicer in the mouse model of lung cancer using K-Ras
promoted the development of the cancer, and suppressing the
expression of Dicer contributed to liver cancer in mice (22,23).
However, no evidence thus far has suggested a role of Dicer in
tongue squamous cell carcinoma.
We found that Dicer was expressed at lower levels in
2 cell lines of tongue squamous cell carcinoma (Tca-8113 and UM-1
cells) compared to normal gingival epithelial cells, which supports
the theory that miRNAs are downregulated in cancer due to the
suppression of Dicer expression. Our results further link Dicer
downregulation and the potential impairment of miRNA processing
with the transformation of tumor cells; we also demonstrated that
depletion of Dicer, a key component in the miRNA processing
machinery in cells, enhanced malignant transformation of tongue
squamous carcinoma cells. Specifically, knocking down Dicer in
Tca-8113 cells significantly increased the proliferative ability,
G1 arrest and invasiveness of the cells.
Collectively, our results suggest that Dicer
functions as an indirect tumor suppressor, as silencing Dicer
expression makes cancer cells more malignant. Although the
mechanism of this transformation remains elusive, the present study
suggests that rectifying the aberrant expression of Dicer could be
one approach in treating tongue squamous cell carcinoma.
Acknowledgements
This study was supported by a grant from Guangdong
Provincial International Science and Technology Cooperation Project
(2012B050300028). The authors thank Dr AnXun Wang for providing the
tongue squamous carcinoma cell lines (Tca-8113 and UM-1), and
Professor Jun Xia from the Hong Kong University of Science and
Technology for collaboration and guidance in the research.
References
|
1
|
Macrae IJ, Zhou K, Li F, et al: Structural
basis for double-stranded RNA processing by Dicer. Science.
211:195–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Si ML, Zhu S, Wu H, et al: miR-21-mediated
tumor growth. Oncogene. 26:2799–2803. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shi B, Sepp-Lorenzino L, Prisco M, et al:
MicroRNA-145 targets the insulin receptor substrate-1 and inhibits
the growth of colon cancer cells. J Biol Chem. 282:32582–32590.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lu J, Getz G, Miska EK, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bu Y, Lu C, Bian C, et al: Knockdown of
Dicer in MCF-7 human breast carcinoma cells results in G1 arrest
and increased sensitivity to cisplatin. Oncol Rep. 21:13–17.
2009.PubMed/NCBI
|
|
6
|
Han L, Zhang A, Zhou X, et al:
Downregulation of Dicer enhances tumor cell proliferation and
invasion. Int J Oncol. 37:299–305. 2010.PubMed/NCBI
|
|
7
|
Liu Tian-Run, Xu Li-Hua, Yang An-Kui, et
al: Decreased expression of SATB2: a novel independent prognostic
marker of worse outcome in laryngeal carcinoma patients. PLoS One.
7:e407042012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jiang Lu, Dai Yang, Liu Xiqiang, et al:
Identification and experimental validation of G protein alpha
inhibiting activity polypeptide 2 (GNAI2) as a microRNA-138 target
in tongue squamous cell carcinoma. Hum Genet. 129:189–197. 2011.
View Article : Google Scholar
|
|
9
|
Nakayama S, Sasaki A, Mese H, Alcalde RE
and Matsumura T: Establishment of high and low metastasis cell
lines derived from a human tongue squamous cell carcinoma. Invasion
Metastasis. 18:219–228. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chiosea S, Jelezcova E, Chandran U, et al:
Up-regulation of Dicer, a component of the microRNA machinery, in
prostate adenocarcinoma. Am J Pathol. 169:1812–1820. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Faber C, Horst D, Hlubek F, et al:
Overexpression of Dicer predicts poor survival in colorectal
cancer. Eur J Cancer. 47:1414–1419. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Papachristou DJ, Korpetinou A,
Giannopoulou E, et al: Expression of the ribonucleases Drosha,
Dicer, and Ago2 in colorectal cancinomas. Virchows Arch.
459:431–440. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Stratmann J, Wang CJ, Gnosa S, et al:
Dicer and miRNA in relation to clinicopathological variables in
colorectal cancer patients. BMC Cancer. 11:3452011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chiosea S, Jelezcova E, Chandran U, et al:
Overexpression of Dicer in precursor lesions of lung
adenocarcinoma. Cancer Res. 67:2345–2450. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu JF, Shen W, Liu NZ, et al:
Down-regulation of Dicer in hepotocellular carcinoma. Med Oncol.
28:804–809. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Grelier G, Voirin N, Ay AS, et al:
Prognostic value of Dicer expression in human breast cancers and
association with the mesenchymal phenotype. Br J Cancer.
101:673–683. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan M, Huang HY, Wang T, et al:
Dysregulated expression of dicer and drosha in breast cancer.
Pathol Oncol Res. 18:343–348. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pampalakis G, Diamandis EP, Katsaros D, et
al: Down-regulation of dicer expression in ovarian cancer tissues.
Clin Biochem. 43:324–327. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kumar MS, Lu J, Mercer KL, et al: Impaired
microRNA processing enhances cellular transformation and
tumorigenesis. Nat Genet. 39:673–677. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martello G, Rosato A, Ferrari F, et al: A
MicroRNA targeting dicer for metastasis control. Cell.
141:1195–1207. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Iwanaga K, Yang Y, Raso MG, et al: Pten
inactivation accelerates oncogenic K-ras-initiated
tumorigenesis in a mouse model of lung cancer. Cancer Ras.
68:1119–1127. 2008.PubMed/NCBI
|
|
23
|
Sekine S, Ogawa R, Ito R, et al:
Disruption of Dicer1 induces dysregulated fetal gene expression and
promotes hepatocarcinogenesis. Gastroenterology. 136:2304–2315.
2009. View Article : Google Scholar : PubMed/NCBI
|