Introduction
Gastric carcinoma is the second leading cause of
cancer-related mortality and the fourth most common type of cancer
worldwide (1,2). There are over 750,000 new cases
diagnosed annually worldwide (3),
and the overall 5-year survival rate remains low (4). Although progress has been made, the
pathophysiological mechanisms and reliable molecular biomarkers of
gastric cancer (GC) remain to be clarified. Therefore, relevant
molecular and genetic studies to establish effective control of the
initiation and progression of GC are imperative.
Interferon-induced transmembrane protein 3 (IFITM3),
also known as 1-8U, is a member of the IFN-inducible transmembrane
protein family. The IFITM3 gene was initially isolated from
a genetic screen to identify the genes involved in the acquisition
of germ-cell competence (5).
Previously, it was shown that IFITM3 belongs to a family of murine
genes (6), which consists of short,
two transmembrane domain proteins (5–18 kDa) with a high core
sequence similarity but divergent N- and C-termini. The human
homologues including IFITM1, IFITM2 and IFITM3 are clustered on
chromosome 11 within an 18-kb genomic sequence (7–9).
Moreover, the IFITMs play an important role in different cell
processes, including cell adhesion, immune-cell regulation,
germ-cell homing and maturation, and bone mineralization (10–14).
The function of the IFITMs in tumorigenesis was
previously confirmed. For example, the expression of IFITM1 and
IFITM3 was significantly upregulated in astrocytoma cells compared
to normal astrocytes in mice (11,15,16)
and IFITM1 overexpression was promoted, with IFITM1 knockdown
suppressing the invasion of HNSCC cells (17). Furthermore, IFITM2 as an independent
pro-apoptotic p53 gene has played a vital role in regulating the
tumor cell death pathway (18).
However, the concrete function and potential mechanisms of IFITM3
in GC pathogenesis are unclear. In the present study, we aimed to
determine the IFITM3 expression level and its regulation mechanism
in GC.
Materials and methods
Human tissue specimens
Forty-eight specimens of GC tissues and adjacent
non-cancer tissues were surgically obtained between June 2012 and
December 2013 at the First Affiliated Hospital of Nanjing Medical
University (Jiangsu, China). The corresponding normal gastric
tissue samples were extracted >5 cm from the edge of the tumor,
while no evident organization of the tumor cells was detectable.
Resected tissue samples were immediately frozen in liquid nitrogen,
and stored at −80°C until the extraction of total RNA. TNM disease
stage was classified according to The American Joint Committee on
Cancer (AJCC), 7th edition. In the present study, patients that had
undergone any preoperative treatments were not included.
Cell lines and cell culture
The GES-1 human gastric epithelial mucosa cell line,
and SGC7901, MKN28, MKN45, AGS and BGC823 GC cell lines were
obtained in our general laboratory. The cells were cultured in
RPMI-1640 containing 10% fetal bovine serum (FBS), penicillin (100
U/ml) and streptomycin (100 μg/ml) under cell culture conditions
(5% CO2, 95% relative humidity and 37°C).
Quantitative real-time PCR (RT-qPCR)
Total RNA was isolated from cell cultures or tissues
using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). First-strand
complementary DNA (cDNA) was reverse-transcribed using the
PrimeScript RT kit (Takara, Dalian, China). RT-qPCR was performed
with FastStart Universal SYBR-Green Master (Rox) (Roche
Diagnostics, Indianapolis, IN, USA) with an ABI 7500 (Applied
Biosystems, Life Technologies Corporation, Carlsbad, CA, USA). The
specific oligonucleotide primers are listed in Table I. PCR was performed using the
following cycles: 95°C for 30 sec, 40 cycles of 95°C for 5 sec,
60°C for 30 sec; and the dissociation stage: 95°C for 15 sec, 60°C
for 1 min and 95°C for 15 sec. Each sample was analyzed in
triplicate.
 | Table IOligonucleotide primer sequences used
in the RT-qPCR. |
Table I
Oligonucleotide primer sequences used
in the RT-qPCR.
| Product primer | Sequence | RT-qPCR length
(bp) |
|---|
| IFITM3 | F:
5′-CGAAACTACTGGGGAAAGGGA-3′
R: 5′-ATTCATGGTGTCCAGCGAAGA-3′ | 83 |
| β-catenin | F:
5′-AAAGCGGCTGTTAGTCACTGG-3′
R: 5′-CGAGTCATTGCATACTGTCCAT-3′ | 215 |
| E-cadherin | F:
5′-GACCGAGAGAGTTTCCCTACG-3′
R: 5′-TCAGGCACCTGACCCTTGTA-3′ | 158 |
| Vimentin | F:
5′-ATGACCGCTTCGCCAACTAC-3′
R: 5′-CGGGCTTTGTCGTTGGTTAG-3′ | 178 |
| MMP-2 | F:
5′-TACAGGATCATTGGCTACACACC-3′
R: 5′-GGTCACATCGCTCCAGACT-3′ | 90 |
| MMP-9 | F:
5′-TGTACCGCTATGGTTACACTCG-3′
R: 5′-RGGCAGGGACAGTTGCTTCT-3′ | 97 |
Lentivirus packaging and stable
transfection cell line generation
To investigate the overexpression of IFITM3, we
modified LV-IFITM3 vector lentiviral constructs (Shanghai
GenePharma Co., Ltd., Shanghai, China) to knock down IFITM3 in GC
cells. The empty lentiviral (LV-NC) with enhanced green fluorescent
protein (EGFP) served as a negative control. The constructed
vectors were verified by DNA sequencing. The GC cells were infected
with LV-IFITM3 and LV-NC. The supernatant was removed after 24 h
and replaced with fresh culture medium. The cells were selected
with 2 μg/ml puromycin for 2 weeks.
Wound-healing assay
Cells (5×105) were seeded in 6-well plate
and incubated for 24 h. When the cells were grown to 90–100%
confluency, a linear wound was generated by scratching the adherent
cells using a sterile plastic pipette tip. The medium was displaced
with serum-free medium. Immediately and 24 h after incubation at
37°C, migrating cells at the wound front were photographed using an
inverted microscope. Each experiment was performed at least three
times in triplicate.
Transwell migration and invasion
assay
In vitro invasion was determined using
24-well Transwell chamber. Cells (2×104) were suspended
in 100 μl of serum-free medium and placed in the upper compartment
of a Transwell chamber with 8-mm membrane filter inserts coated
with Matrigel (BD Biosciences). Subsequently, the medium with 10%
FBS was added to the lower chamber as a chemoattractant. After
incubation for 24 h, the cells on the upper surface of the membrane
filter were removed, and the cells that had penetrated to the lower
surface of the membrane were fixed and stained with crystal violet.
Five visual fields of each insert were randomly selected and
counted under a light microscope. Migration assays were performed
using a Transwell compartment, and with the exception of Matrigel,
all other steps were identical. For each experimental condition,
the assay was performed at least in triplicate.
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was measured by the CCK-8
(Beyotime Institute of Biotechnology, Shanghai, China) according to
the manufacturer’s instructions. Cells were seeded in a 96-well
plate at 5×103 cells/well with 100 μl of complete
culture medium. After adhesion for 24 h, 10 μl CCK-8 was added to
each well and incubated at 37°C for 2 h. The absorbance was
measured at 450 nm using a microplate reader. Experiments were
performed in triplicate and repeated three times.
Cell cycle assay
Cells (3×105 cells/well) were seeded in
6-well plates and cultured for 24 h. After adhesion, the cells were
harvested and centrifuged at 1,500 rpm for 5 min. Pellets were
washed with cold phosphate-buffered saline (PBS) and fixed with
cold 70% ethanol at 4°C for 30 min and stored at −20°C overnight.
The following day, the cells were centrifuged at 1,500 rpm for 5
min to remove ethanol and washed with PBS. The supernatant was
discarded and propidium iodide (PI) containing RNase A was added at
4°C for 30 min in the dark. Cell suspension was filtered through a
mesh filter and analyzed by flow cytometry.
Western blot analysis
Cultured cells were lysed using RIPA buffer
(Beyotime Institute of Biotechnology), and equivalent amounts of
protein were subjected to SDS-PAGE separation and subsequently
diverted to polyvinylidene fluoride (PVDF) membranes (Millipore,
Bedford, MA, USA). The membranes were blocked with 5% non-fat milk
in Tris-buffered saline solution containing 0.05% Tween-20 and then
incubated with antibody-specific IFITM3 (1:1,000; ab109429, Abcam),
E-cadherin, vimentin, β-catenin, MMP-2, MMP-9 (1:1,000), GAPDH
(1:10,000) (both from Cell Signaling Technology). The membranes
were washed three times with TBST, and incubated with horseradish
peroxidase (HRP)-conjugated secondary antibody (1:1,000; Beijing
Biosynthesis Biotechnology) at 37°C for 2 h. Bound proteins were
visualized using ECL (Pierce) after three TBST washes and detected
using a Bio-Imaging System. GAPDH was used as an internal loading
control.
Immunohistochemistry (IHC)
For immunohistochemistry, the tissue samples were
fixed in 10% formalin solution and embedded in paraffin. Previous
sections of 3 mm were prepared, then deparaffinized in xylene, and
rehydrated through a graded series of ethanol. Endogenous
peroxidase was blocked with 0.3% hydrogen peroxide in methanol for
20 min at room temperature. For pretreatment, antigen retrieval
sections were performed in citrate buffer (pH 6.0) using a
microwave for 10 min. Overnight incubation at 4°C was carried out
for the binding of primary antibodies (IFITM3, 1:400; ab109429,
Abcam). The secondary anti-mouse antibody was subsequently applied
to the sections for 30 min at room temperature. After rinsing with
TBST, the slides were treated with diaminobenzidine solution and
counterstained with hematoxylin. Immunostaining intensity (0, no
staining; 1, weak staining; 2, moderate staining; and 3, strong
staining) as well as the percentage of cells stained (0, no cells;
1, <10% of cells; 2, 11–50% of cells; 3, 51–80% of cells; and 4,
>80% of cells) were evaluated.
Statistical analysis
The Statistical Program for Social Sciences (SPSS)
20.0 software (IBM, SPSS, Inc., Chicago, IL, USA) was used for the
statistical analysis. The data were expressed as means ± standard
deviation (SD). Differences were analyzed with the unpaired
Student’s t-test and analysis of variance (ANOVA). A two-tailed
value of P<0.05 was considered to indicate a statistically
significant result.
Results
IFITM3 overexpression is identified in GC
tissues and cell lines
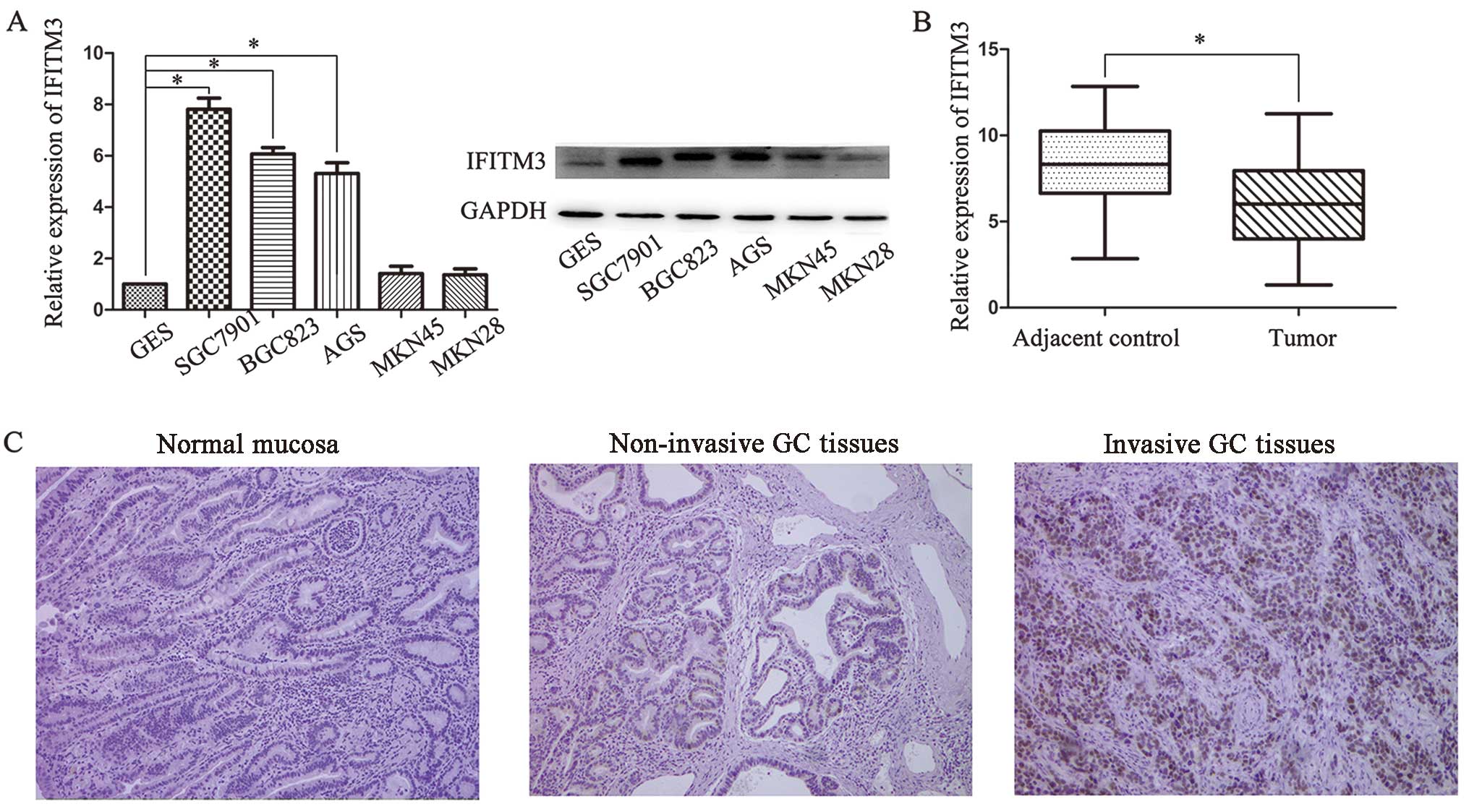
To examine the role of IFITM3 in GC progression, the
mRNA and protein expression of IFITM3 in the AGS, SGC7901, BGC823,
MKN45 and MKN28 human GC cell lines and the GES-1 human gastric
epithelial mucosa cell line were analyzed by RT-qPCR and western
blot analysis, respectively. As shown in Fig. 1A, the expression levels of IFITM3
mRNA and protein were significantly higher in SGC7901 and BGC823
cell lines than in normal cell lines. However, there was no
significant difference for MKN28 and MKN45. Thus, SGC7901 and
BGC823 were selected as the optimal experimental groups.
Furthermore, we examined the expression in 48 pairs of GC tissues
compared with adjacent normal tissues. IFITM3 levels were markedly
upregulated in cancer tissues compared with corresponding
non-cancer tissues (Fig. 1B). The
potential related factors affecting IFITM3 expression were
determined and it was confirmed that IFITM3 expression was
significantly associated with tumor differentiation, lymph node and
distant metastasis, and TNM stages. Nevertheless, there was no
significant correlation between IFITM3 expression and other
clinicopathologic characteristics, such as age, gender, diameter,
tumor location and CEA values (Table
II). We also examined the expression of IFITM3 by
immunohistochemistry and it was found that IFITM3 was positively
stained in the cytoplasm of the majority of invasive cancer
specimens, whereas IFITM3 was negatively or weakly stained in the
adjacent normal tissues. Furthermore, IFITM3 expression levels were
increased with the pathological stages of GC (Fig. 1C).
 | Table IIAssociations between IFITN3
expression (ΔCT) and clinicopathological characteristics of 48 GC
patients. |
Table II
Associations between IFITN3
expression (ΔCT) and clinicopathological characteristics of 48 GC
patients.
|
Characteristics | No. of patients
(%) | Mean ± SD | P-value |
|---|
| Age (years) | | | 0.539 |
| ≥60 | 32 | 5.84±2.31 | |
| <60 | 16 | 6.24±1.72 | |
| Gender | | | 0.157 |
| Male | 33 | 5.68±2.02 | |
| Female | 15 | 6.62±2.25 | |
| Diameter (cm) | | | 0.685 |
| ≥5 (large) | 19 | 5.82±2.50 | |
| <5 (small) | 29 | 6.08±1.87 | |
| Location | | | 0.685 |
| Cardia or
body | 30 | 6.07±2.20 | |
| Antrum | 18 | 5.81±2.03 | |
|
Differentiation | | | 0.009 |
| Poor or not | 32 | 5.42±2.15 | |
| Well or
moderate | 16 | 7.09±1.59 | |
| Lymphatic
metastasis | | | 0.002 |
| Present | 38 | 5.50±2.00 | |
| Absent | 10 | 7.77±1.60 | |
| Distal
metastasis | | | 0.024 |
| Present | 44 | 6.18±2.08 | |
| Absent | 4 | 3.71±1.00 | |
| AJCC clinical
stage | | | 0.005 |
| I+II | 13 | 7.35±2.18 | |
| III+IV | 35 | 5.47±1.88 | |
| Serum CEA
value | | | 0.752 |
| ≥5 (μg/l) | 20 | 5.86±2.52 | |
| <5 (μg/l) | 28 | 6.06±1.83 | |
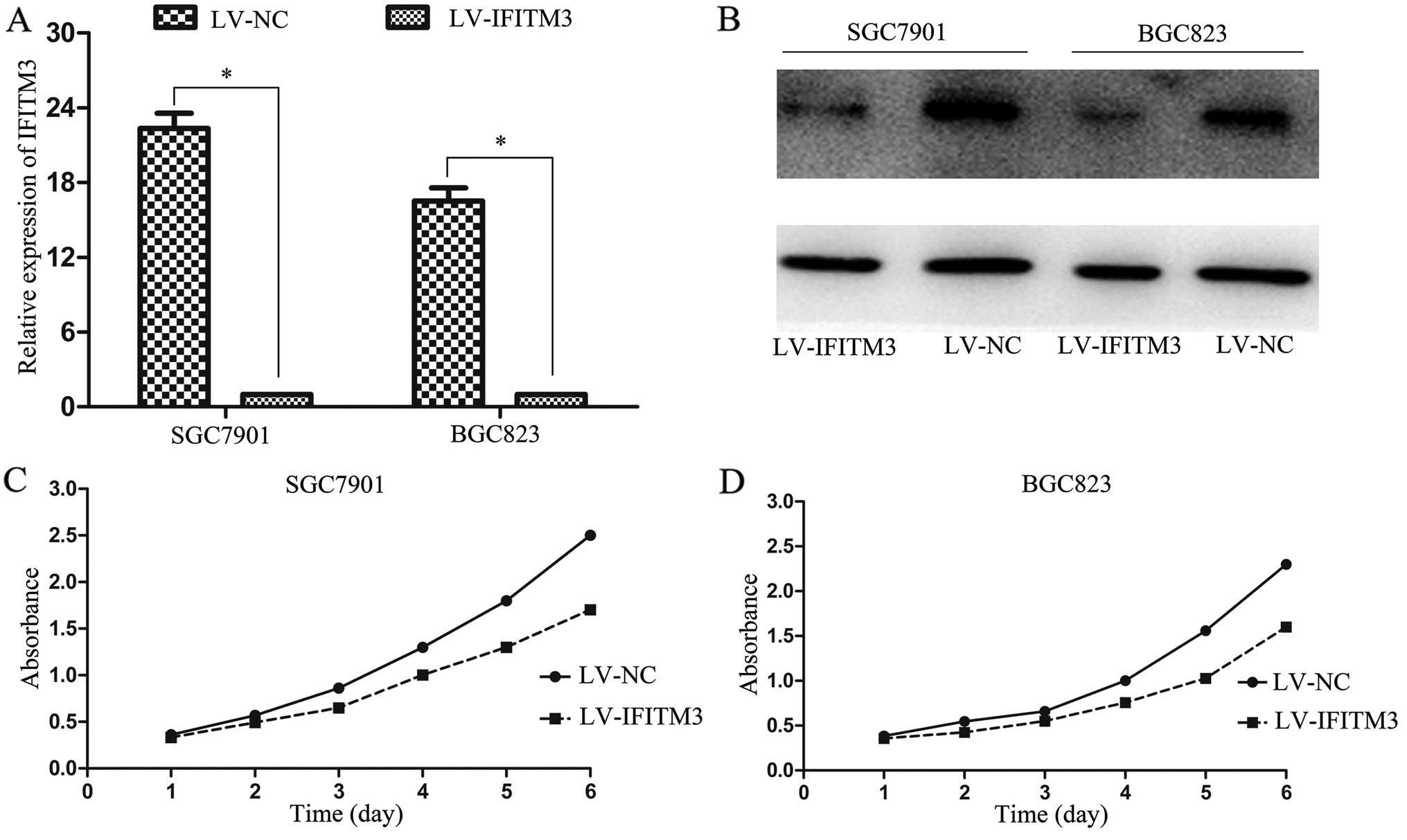
IFITM3 knockdown is negatively regulates
GC cell proliferation
To demonstrate the functional role of IFITM3 in GC
tumorigenesis, we infected IFITM3 shRNA into SGC7901 and BGC823
cells and examined the transfection efficiency by RT-qPCR and
western blot analysis, respectively. We found that the levels of
IFITM3 expression in the LV-NC were markedly higher than that of
LV-IFITM3 (Fig. 2A and B).
Furthermore, obvious inhibitory effects on cell proliferation were
observed in IFITM3-silenced cells at 4–6 days (Fig. 2C and D). These results suggested
that IFITM3 overexpression contributes to the proliferation of
colon cancer cells.
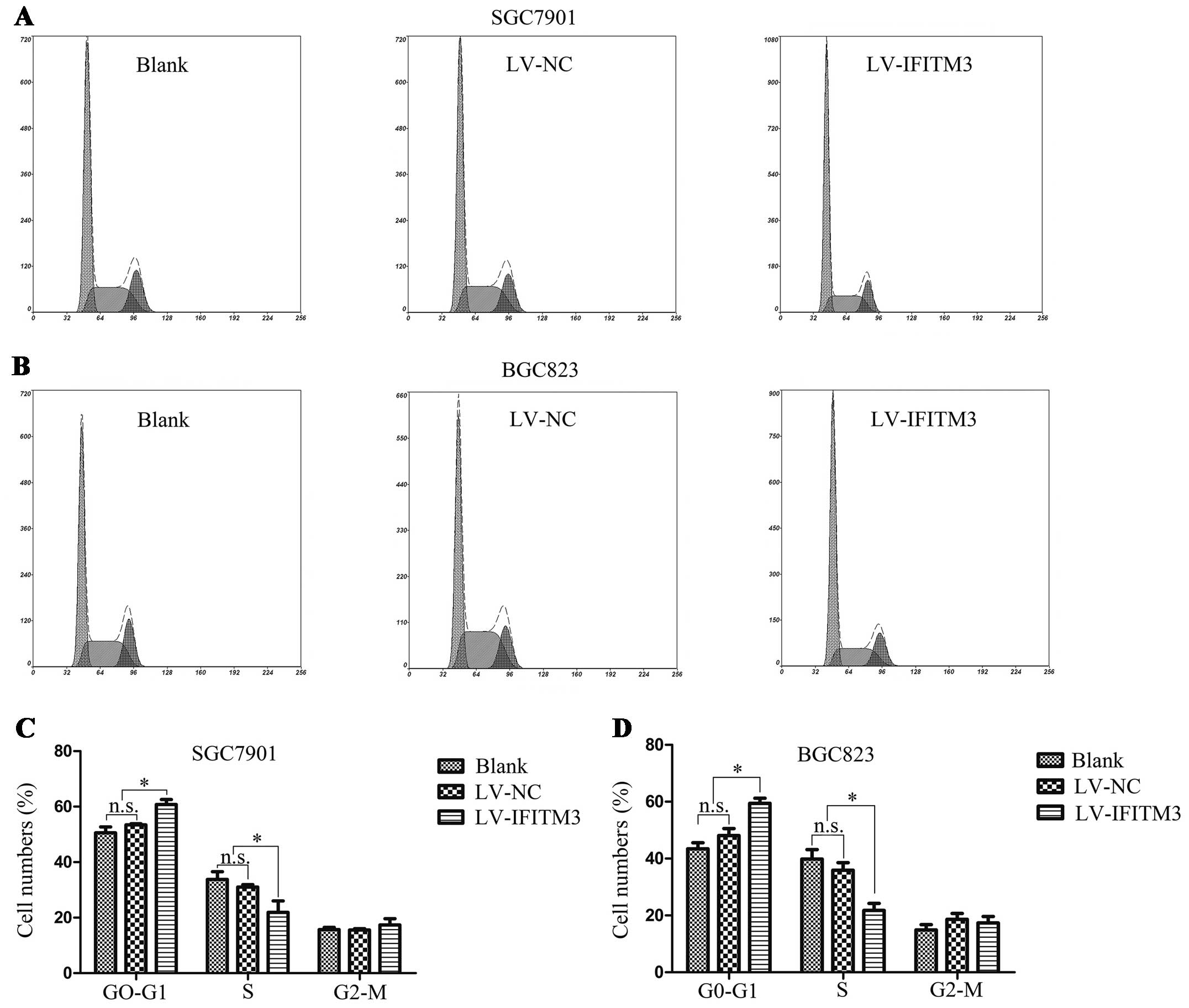
Depletion of IFITM3 arrests GC cells in
the G0/G1 phase
To elucidate the effect of IFITM3 downregulation on
cell cycle distribution, the flow cytometry instrument analysis was
performed to show the increasing proportion of G0/G1 phase in GC
cells. As shown in Fig. 3A and B,
there was a significant increase in the cell population at the
G0/G1 phase, associated with a decrease in the cell population at
the G2/M and S phases following IFITM3 depletion (P<0.05;
Fig. 3C and D). These data
suggested that depletion of IFITM3 arrests GC cells in the G0/G1
phase.
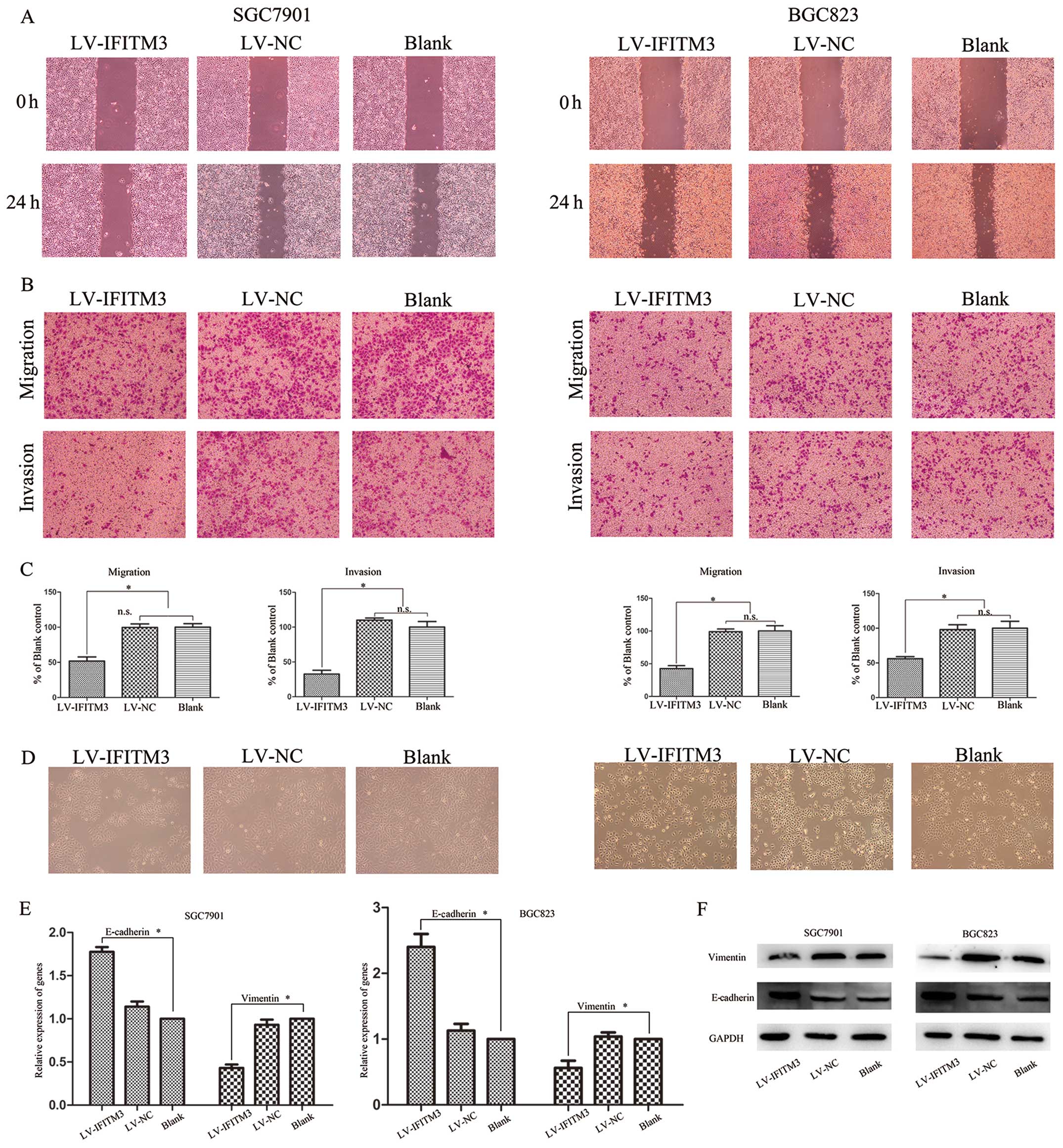
IFITM3 silencing inhibits cell migration
and invasion through epithelial-to-mesenchymal transition (EMT)
change
To exhibit the involvement of IFITM3 in the invasion
of gastric cells, wound-healing and Transwell assays were performed
using SGC7901 and BGC823 cells transfected with LV-IFITM1 or LV-NC.
The cell migration effect identified by the wound-healing assay
showed that compared with negative control vector-transfected
cells, IFITM3 knockdown significantly inhibits GC cell migration
(Fig. 4A). Similar results were
obtained from the Transwell assay, while transfection with
LV-IFITM3 significantly suppressed the migration and invasion of
SGC7901 and BGC823 cells (P<0.05; Fig. 4B and C). EMT, an essential cell
biological program during embryonic development, contributes to
cancer invasion and metastasis (19,20).
To confirm the role of IFITM3 in the process of GC cells, we
transfected the LV-IFITM3 vector and evaluated the expression of
epithelial marker E-cadherin and mesenchymal marker vimentin at
mRNA and protein levels. As shown in Fig. 4D, IFITM3 knockdown cells induced
round spheroids with no or few protrusions. The increased
expression of E-cadherin and decreased expression of vimentin was
detected in IFITM3 knockdown cells in the mRNA and protein levels
(Fig. 4E and F). This suggested
that IFITM3 may regulate the migration and invasion potential of
the gastric cell lines by inducing EMT.
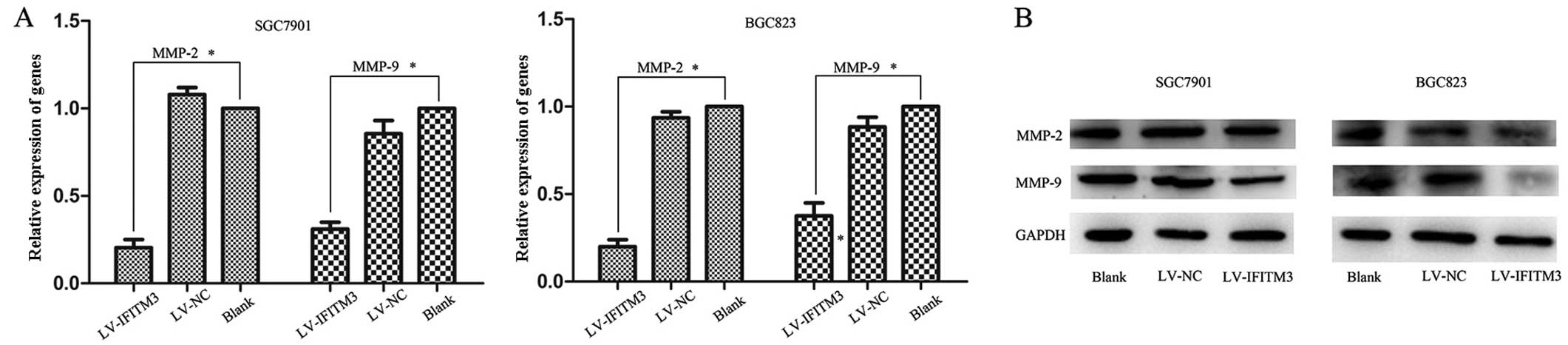
Knockdown of IFITM3 induces
downregulation of MMPs
MMPs are matrix metalloproteinases that were
previously shown to play a crucial role in the tumor
microenvironment by enhancing cancer cell invasion, proliferation
and cancer matastasis (21,22). The results showed that IFITM3 is
able to promote the migration and invasion of GC cancer cells.
However, whether IFITM3 mediates MMP expression remains to be
determined. Therefore, we examined MMP-2 and MMP-9 expression in
transfected cells and negative control by qPCR and western
blotting, respectively. A significant downregulation of MMP-2 and
MMP-9 expression was observed in IFITM3-silencing cells, as
compared with the empty lentiviral cells (P<0.05; Fig. 5A and B). These results suggested
that MMPs are involved in IFITM3-induced cell migration and
invasion.
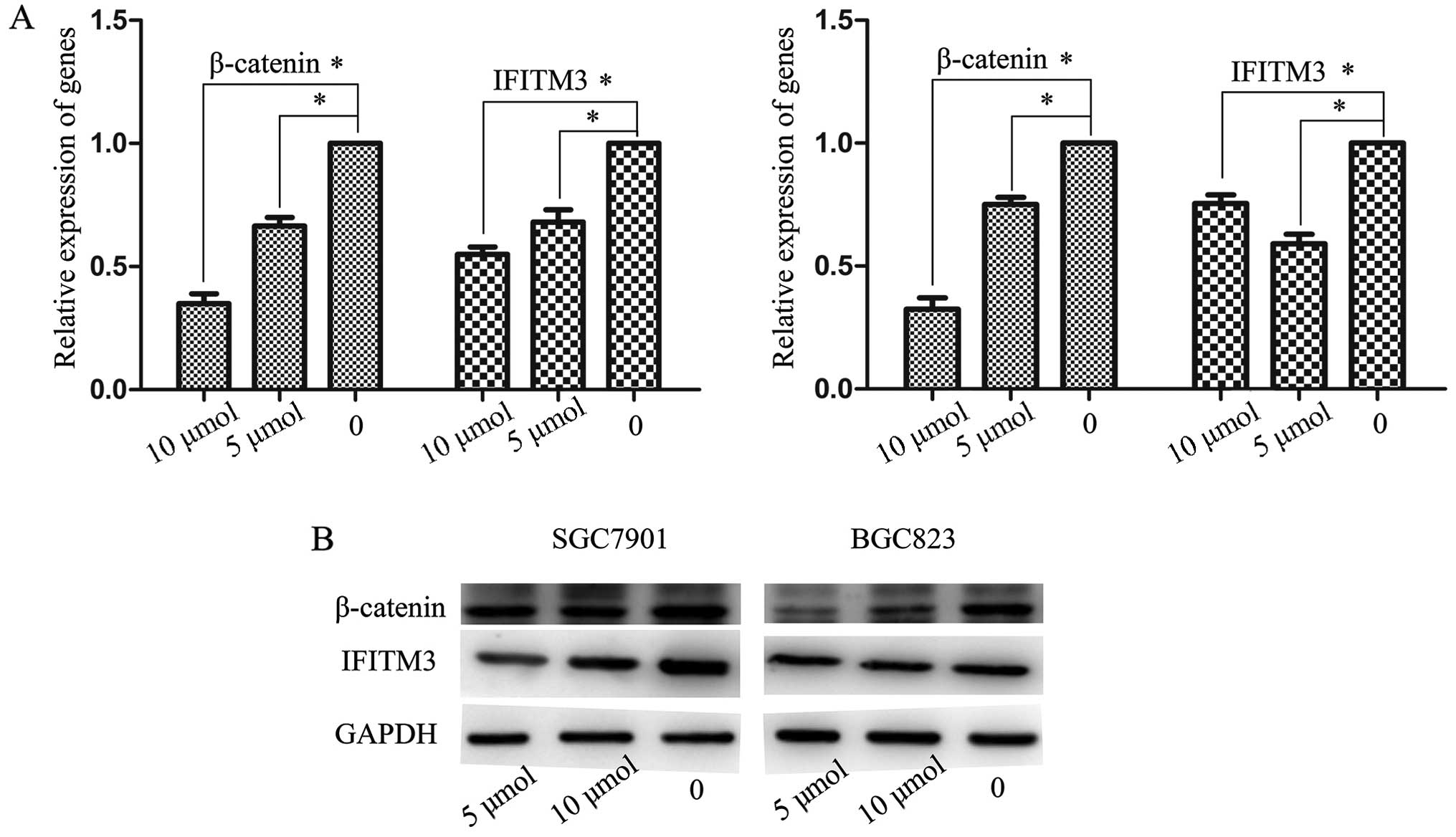
Involvement of the Wnt/β-catenin pathway
in the expression of IFITM3
The canonical Wnt/β-catenin pathway plays an
important role in embryonic development and tumorigenesis. It has
been shown that Wnt/β-catenin signaling participates in the
regulation of IFITM genes during intestinal tumorigenesis
(23). Wnt signaling and stable
axin protein leading to β-catenin Therefore, we investigated
whether the upregulation of IFITM3 destruction (24). Concentrations of 5 and 10 μmol/l,
and an in GC cells was induced by activation of Wnt/β-catenin
optimal processing time of 48 h were selected. GC cells were
signaling. XAV939, a new molecular inhibitor, blocked subsequently
treated with various concentrations of XAV939 and IFITM3 expression
was analyzed using RT-qPCR and western blot analysis. The results
showed that the expression of β-catenin and IFITM3 maintained a
high level in normal GC cells. After incubating with XAV939 with 5
and 10 μ/mol, IFITM3 expression markedly decreased together with
the reduction of β-catenin, both of which were significantly
inferior to the untreated controls (P<0.05; Fig. 6A and B).
Discussion
IFITM3 is known to function as a cancer-promoting
gene and is overexpressed in the oncogenesis of several
malignancies, including glioma, colorectal and breast cancer
(25–27). However, the mechanism and
significance of IFITM3 is unclear. Thus, in the present study, we
concretely investigated the expression of IFITM3 in GC tissues and
cell lines. RT-qPCR and western blot analysis revealed that IFITM3
expression was significantly higher in SGC7901 and BGC823 cells
than in normal cells at the mRNA and protein expression level. In
the tissues, the level of IFITM3 expression was upregulated in GC
tumor tissues, which is consistent with the adjacent normal tissues
by RT-qPCR analysis. We also found that IFITM3 was significantly
correlated with tumor differentiation, lymph node and distant
metastasis, and TNM stages. Consistent with the abovementioned
results, the results of the immunohistochemical staining indicated
that IFITM3 expression was closely associated with an advanced
cancer biology. IFITM3-positive staining was significantly higher
in advanced GC than early GC. Alteration of IFITM3 expression
offered further functional evidence that it was involved in the
enhancement of aggressive biological behavior of cancer cells.
There were also certain limitations in the present study. First,
all of the patients were enlisted from a single institution and the
number of samples was not adequate. For instance, there were only
four patients at M1 stage. Additionally, since the tissues used
were resected in the previous two years, the follow-up period after
surgery was insufficient to investigate the relationship between
IFITM3 expression and overall survival of GC patients.
IFITM3 is a double trans-membrane protein that can
be upregulated by IFNs (10,28).
It is involved in signal transduction of antiviral,
anti-inflammation, immune cell regulation and somitogenesis
(11,12,15,29).
However, the precise role of IFITM3 in GC pathogenesis remains
unknown. Thus, we used lentivirus-mediated RNAi to knock down
IFITM3 in GC cells. shRNA targeting IFITM3 induced obvious and
efficient inhibition of cell proliferation, migration and invasion.
The flow cytometry data showed that the growth inhibitory effect of
LV-IFITM3 was mediated by cell cycle arrest at the G0/G1 phase, and
reduced the number of cells in the S phase. Concordant with our
findings, it has previously been confirmed that IFITM1 expression
promotes invasion in head and neck tumor in the early stages of
disease progression (17).
El-Tanani et al reported that IFITM3 reduces osteopontin
mRNA expression by affecting OPN mRNA stability to modulate
anchorage-independent growth, cell adhesion and invasion (30). Thus, the present study confirms that
IFITM3 is important in GC tumorigenesis, suggesting IFITM3 as a
potential oncogene in human GC.
Metastasis remains the leading cause of treatment
failure and poor prognosis in patients with malignant tumors. It is
a complex and multistage process including proteolysis, motility
and migration of cells, proliferation in a new site and
neoangiogenesis (31). EMT in tumor
progression and metastasis may be induced by autonomous oncogenic
activation or inactivation of signaling molecules with or without
additional stimulation (32).
During this progression, epithelial markers including E-cadherin,
ZO-1 and MUC1 are downregulated and molecules such as transcription
factors Snail, Slug, Twist as well as N-cadherin and vimentin are
upregulated (33). To understand
whether the mechanism of IFITM3-regulated invasion and metastasis
is associated with the EMT, we tested the expression of the EMT
representative markers E-cadherin and vimentin treated with the
control or IFITM3 knockdown. The results showed a tendency of
E-cadherin to decrease and of vimentin and the morphological change
(growth pattern, decreased formation of lamellipodia) to increase,
suggesting that IFITM3 is probably an EMT-like phenotype. These
results indicate that the alterative expression of IFITM3 had a
significant impact on the process of EMT by mediating the
E-cadherin and vimentin expression in GC cells. In addition, it is
well known that an increased expression of MMPs is required in
tumor invasion and metastasis. Among the MMPs, MMP-2 and MMP-9 are
considered to be crucial enzymes in this process with degrading
type IV collagen regulating various cell behaviors with relevance
for cancer biology (34). Enhanced
levels of MMP-2 and MMP-9 are important factors associated with
metastasis and invasion in the process of GC (35). To validate the effect of IFITM3 on
cell invasion and metastasis, we examined MMP-2 and MMP-9
expression in transfected cells and the negative control by qPCR
and western blot analysis, respectively. Our results suggested that
depletion of IFITM3 decreased the expression of MMP-2 and MMP-9.
Furthermore, we confirmed that the downregulation of IFITM3
expression decreased the invasion and migration ability of GC
cells. The results confirmed that changes in the expression of
MMP-2 and MMP-9 were consistent with IFITM3. However, the concrete
mechanisms of how IFITM3 regulates invasion and metastasis in GC
cells remain to be clarified in future studies.
The Wnt/β-catenin signaling pathway is a fundamental
mechanism that regulates cell proliferation, polarity and
differentiation during embryonic development. Stability of
β-catenin in the cytoplasm was assessed to determine activation of
the Wnt/β-catenin signaling pathway (36). Findings of a previous study showed
the expression of the IFITM family members is induced and that
their transcription is dependent on the activation of β-catenin
signaling in intestinal tumorigenesis (23). Thus, we hypothesize that a similar
event occurs in GC. In the present study, XAV939, a specific
inhibitor of β-catenin, was used to treat gastric cells at various
concentrations. Our results suggest that IFITM3 expression was
positively correlated with the β-catenin level, indicating that the
Wnt/β-catenin signaling pathway may play an important role in
IFITM3 expression in GC cells and activated β-catenin signaling
including IFITM3 may be involved in gastric carcinoma.
Nevertheless, the molecular mechanism of IFITM3 overexpression
underlying the regulating process of the Wnt/β-catenin signaling
pathway in GC remains to be elucidated. However, more studies are
required to elucidate the molecular regulation for this
possibility.
In conclusion, the present study has offered insight
into the role of the IFITM3 gene in the pathogenesis of GC. An
increased expression of IFITM3 was observed in GC tissues and cell
lines, which is closely associated with the deregulation of Wnt
signaling. Additionally, IFITM3 exhibits tumor promoter activity
that facilitates the growth, migration, invasion and metastatic
potential of tumor cells via the regulation of EMT and MMPs. These
findings provide new and important information on the progression
of GC and suggest that IFITM3 may be beneficial as a novel
molecular target for the treatment of GC patients.
Acknowledgements
The present study was supported by the Department of
Health of the Jiangsu Province Fund.
Abbreviations:
|
AJCC
|
American Joint Committee on Cancer
|
|
EMT
|
epithelial-to-mesenchymal
transition
|
|
mRNA
|
messenger RNA
|
|
MMPs
|
matrix metalloproteinases
|
References
|
1
|
Bertuccio P, Chatenoud L, Levi F, et al:
Recent patterns in gastric cancer: a global overview. Int J Cancer.
125:666–673. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
3
|
Cheng Y, Jin Z, Agarwal R, et al: LARP7 is
a potential tumor suppressor gene in gastric cancer. Lab Invest.
92:1013–1019. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Khosravi Shahi P, Díaz Muñoz de la Espada
VM, García Alfonso P, et al: Management of gastric adenocarcinoma.
Clin Transl Oncol. 9:438–442. 2007.
|
|
5
|
Saitou M, Payer B, Lange UC, Erhardt S,
Barton SC and Surani MA: Specification of germ cell fate in mice.
Philos Trans R Soc Lond B Biol Sci. 358:1363–1370. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lange UC, Saitou M, Western PS, Barton SC
and Surani MA: The fragilis interferon-inducible gene family of
transmembrane proteins is associated with germ cell specification
in mice. BMC Dev Biol. 3:12003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lewin AR, Reid LE, McMahon M, Stark GR and
Kerr IM: Molecular analysis of a human interferon-inducible gene
family. Eur J Biochem. 199:417–423. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Evans SS, Collea RP, Appenheimer MM and
Gollnick SO: Interferon-α induces the expression of the L-selectin
homing receptor in human B lymphoid cells. J Cell Biol.
123:1889–1898. 1993.
|
|
9
|
Tanaka SS, Yamaguchi YL, Tsoi B, Lickert H
and Tam PP: IFITM/Mil/fragilis family proteins IFITM1 and IFITM3
play distinct roles in mouse primordial germ cell homing and
repulsion. Dev Cell. 9:745–756. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Friedman RL, Manly SP, McMahon M, Kerr IM
and Stark GR: Transcriptional and posttranscriptional regulation of
interferon-induced gene expression in human cells. Cell.
38:745–755. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tanaka SS, Nagamatsu G, Tokitake Y, Kasa
M, Tam PP and Matsui Y: Regulation of expression of mouse
interferon-induced transmembrane protein like gene-3, Ifitm3
(mil-1, fragilis), in germ cells. Dev Dyn.
230:651–659. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Smith RA, Young J, Weis JJ and Weis JH:
Expression of the mouse fragilis gene products in immune cells and
association with receptor signaling complexes. Genes Immun.
7:113–121. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Saitou M, Barton SC and Surani MA: A
molecular programme for the specification of germ cell fate in
mice. Nature. 418:293–300. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nibbe RK, Markowitz S, Myeroff L, Ewing R
and Chance MR: Discovery and scoring of protein interaction
subnetworks discriminative of late stage human colon cancer. Mol
Cell Proteomics. 8:827–845. 2009. View Article : Google Scholar
|
|
15
|
Seyfried NT, Huysentruyt LC, Atwood JA
III, Xia Q, Seyfried TN and Orlando R: Up-regulation of NG2
proteoglycan and interferon-induced transmembrane proteins 1 and 3
in mouse astrocytoma: a membrane proteomics approach. Cancer Lett.
263:243–252. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wylie C: IFITM1-mediated cell repulsion
controls the initial steps of germ cell migration in the mouse. Dev
Cell. 9:723–724. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hatano H, Kudo Y, Ogawa I, et al:
IFN-induced transmembrane protein 1 promotes invasion at early
stage of head and neck cancer progression. Clin Cancer Res.
14:6097–6105. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Daniel-Carmi V, Makovitzki-Avraham E,
Reuven EM, et al: The human 1-8D gene (IFITM2) is a
novel p53 independent proapoptotic gene. Int J Cancer.
125:2810–2819. 2009.
|
|
19
|
Li J, Yang B, Zhou Q, et al: Autophagy
promotes hepatocellular carcinoma cell invasion through activation
of epithelial-mesenchymal transition. Carcinogenesis. 34:1343–1351.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ledford H: Cancer theory faces doubts.
Nature. 472:2732011. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gialeli C, Theocharis AD and Karamanos NK:
Roles of matrix metalloproteinases in cancer progression and their
pharmacological targeting. FEBS J. 278:16–27. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Roy R, Yang J and Moses MA: Matrix
metalloproteinases as novel biomarkers and potential therapeutic
targets in human cancer. J Clin Oncol. 27:5287–5297. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Andreu P, Colnot S, Godard C, et al:
Identification of the IFITM family as a new molecular marker
in human colorectal tumors. Cancer Res. 66:1949–1955. 2006.
|
|
24
|
Tian XH, Hou WJ, Fang Y, et al: XAV939, a
tankyrase 1 inhibitor, promotes cell apoptosis in neuroblastoma
cell lines by inhibiting Wnt/β-catenin signaling pathway. J Exp
Clin Cancer Res. 32:1002013.PubMed/NCBI
|
|
25
|
Zhao B, Wang H, Zong G and Li P: The role
of IFITM3 in the growth and migration of human glioma cells.
BMC Neurol. 13:2102013.
|
|
26
|
Li D, Peng Z, Tang H, et al: KLF4-mediated
negative regulation of IFITM3 expression plays a critical role in
colon cancer pathogenesis. Clin Cancer Res. 17:3558–3568. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang M, Gao H, Chen P, Jia J and Wu S:
Knockdown of interferon-induced transmembrane protein 3 expression
suppresses breast cancer cell growth and colony formation and
affects the cell cycle. Oncol Rep. 30:171–178. 2013.PubMed/NCBI
|
|
28
|
Kelly JM, Gilbert CS, Stark GR and Kerr
IM: Differential regulation of interferon-induced mRNAs and c-myc
mRNA by α- and γ-interferons. Eur J Biochem. 153:367–371. 1985.
|
|
29
|
Jiang D, Weidner JM, Qing M, et al:
Identification of five interferon-induced cellular proteins that
inhibit West Nile virus and dengue virus infections. J Virol.
84:8332–8341. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
El-Tanani MK, Jin D, Campbell FC and
Johnston PG: Interferon-induced transmembrane 3 binds osteopontin
in vitro: expressed in vivo IFITM3 reduced OPN expression.
Oncogene. 29:752–762. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang YY, Chen B and Ding YQ:
Metastasis-associated factors facilitating the progression of
colorectal cancer. Asian Pac J Cancer Prev. 13:2437–2444. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Scheel C, Eaton EN, Li SH, et al:
Paracrine and autocrine signals induce and maintain mesenchymal and
stem cell states in the breast. Cell. 145:926–940. 2011. View Article : Google Scholar
|
|
33
|
Zhao Y, Guo Q, Chen J, Hu J, Wang S and
Sun Y: Role of long non-coding RNA HULC in cell proliferation,
apoptosis and tumor metastasis of gastric cancer: A clinical and
in vitro investigation. Oncol Rep. 31:358–364.
2014.PubMed/NCBI
|
|
34
|
Egeblad M and Werb Z: New functions for
the matrix metalloproteinases in cancer progression. Nat Rev
Cancer. 2:161–174. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kabashima A, Maehara Y, Kakeji Y, Baba H,
Koga T and Sugimachi K: Clinicopathological features and
overexpression of matrix metalloproteinases in intramucosal gastric
carcinoma with lymph node metastasis. Clin Cancer Res. 6:3581–3584.
2000.PubMed/NCBI
|
|
36
|
Logan CY and Nusse R: The Wnt signaling
pathway in development and disease. Annu Rev Cell Dev Biol.
20:781–810. 2004. View Article : Google Scholar : PubMed/NCBI
|