Introduction
Glioblastoma is one of the most fatal human cancers
and is characterized by a high proliferative rate and aggressive
invasiveness. Despite advances in surgery, chemotherapy and
radiotherapy, no significant increase in survival has been achieved
for patients with glioblastoma over the last 20 years. The survival
of most glioblastoma patients is less than 2 years (1,2). To
develop novel treatments to improve the prognosis of glioblastoma
patients, it will be important to further elucidate the molecular
mechanisms that support invasion and proliferation of these cancer
cells.
Homeobox transcriptional factors are a family of
proteins with a homeobox domain that binds to conserved DNA
sequences. Homeobox proteins are known to play pivotal roles in the
developmental processes of multicellular organisms, and
accumulating evidence has revealed that certain homeobox genes
regulate the progression of numerous types of cancers (3). For example, SIX homeobox 1 (SIX1) is
overexpressed in human breast cancers, and transgenic mice that
overexpressed SIX1 in mammary epithelial cells exhibited increased
tumor development (4,5). A recent study reported that
aristaless-like homeobox 1 (ALX1) promoted the invasion of ovarian
cancer cells by inducing the epithelial-to-mesenchymal transition
(EMT), which is a morphological conversion of epithelial to
mesenchymal cells (6). In
glioblastoma, HOXA9 expression increased cell proliferation and
inhibited apoptosis, and its expression was associated with poor
prognosis (7). These studies have
shown the important functions of homeobox proteins in tumor
progression and support the notion that the inhibition of
tumor-associated homeobox proteins can be a novel therapeutic
strategy.
Paired related homeobox 1 (PRRX1) is a member of the
paired-type family of homeobox transcription factors, which have
important functions in the regulation of developmental
morphogenetic processes (8,9). PRRX1 is highly expressed in cardiac
and skeletal muscle of mouse embryos as well as in adults, and mice
lacking PRRX1 die perinatally due to craniofacial and limb
malformations (10). PRRX1 is also
associated with tumor progression. A high level of PRRX1 expression
is correlated with metastasis and poor prognosis of colon cancer
cells (11). In contrast, Ocaña
et al reported that PRRX1 expression suppressed metastasis
and that strong PRRX1 expression could be a marker for good
prognosis of breast and lung cancer (12). Although their conclusions are
contradictory, both groups reported that PRRX1 expression promotes
the EMT in cancer cells. A recent report showed that PRRX1
associates with sex determining region-Y box 2 (SOX2) and is
required to maintain the stemness of neuronal stem cells (13). SOX2 has been reported to be
overexpressed in numerous types of tumors and to promote invasion
of glio-blastoma cells (14–17).
In the present study, we examined the role of PRRX1 in glioblastoma
cells and showed that PRRX1 plays an important role in the invasion
of glioblastoma cells.
Materials and methods
Cells and antibodies
The human glioblastoma cell lines T98, U251MG,
U251sp, U87, SKMG1, U251nu/nu and AO2 and the human embryonic
kidney 293T (HEK293T) cell line were maintained in Dulbecco’s
modified Eagle’s medium (DMEM) supplemented with 10% FBS
(Equitech-Bio, Kerrville, TX, USA) at 37°C in a humidified
atmosphere of 5% CO2. U251sp and U251nu/nu cells are
derived from U251MG cells (18).
Anti-E-cadherin, anti-N-cadherin and anti-vimentin antibodies were
obtained from BD Biosciences (San Jose, CA, USA) and the
anti-β-actin antibody was from Sigma-Aldrich (St. Louis, MO, USA).
The anti-HES1 antibody was purchased from Cell Signaling
Technology, Inc. (Danvers, MA, USA).
siRNA transfection
The sequences of the siRNAs specific for PRRX1
consisted of 5′-GCACUAAAGUCUACAUCUA-3′ (siPRRX-1) and
5′-CCACUGUUCUUAUCUCUAU-3′ (siPRRX-2). The sequence of the control
siRNA that targeted luciferase was 5′-CUUACGCUGAGUACUUCGATT-3′.
siRNAs were obtained from Sigma-Aldrich. Cells were transfected
with 20 nM of siRNA using Lipofectamine RNAiMAX (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer’s
instructions.
Quantitative RT-PCR
RNA was extracted from glioma samples and cells
using the RNeasy Mini kit (Qiagen, Venlo, The Netherlands), and
cDNA was generated using PrimeScript reverse transcriptase (Takara,
Tokyo, Japan). Glioma samples were obtained from patients at Nagoya
University Hospital with informed consent. Normal human brain RNAs
were obtained from Takara and BioChain (Newark, CA, USA). Catalog
nos. of the normal samples are 636530 (Takara) and R1234035-50
(BioChain). PCR was performed using the SYBR Premix Ex Taq™
II, and the Thermal Cycler Dice™ Real-time System TP800 (both from
Takara) was used for analysis. The relative mRNA expression levels
were normalized to GAPDH. The sequences of primers used to amplify
each gene were: 5′-AGGTGGAGGAGTGGGTGTCGCTG TT-3′ and 5′-CCGGGA A
ACTGTGGCGTGATGG-3′ (GAPDH); 5′-AGTTCCGCAGGAATGAGAGA-3′ and 5′-AT
GGCGCTTTTCAGTGTCTT-3′ (PRRX1A); 5′-CATCGTACC TCGTCCTGCTC-3′ and
5′-GCCCCTCGTGTAAACAAC AT-3′ (PRRX1B); and
5′-AGGCGGACATTCTGGAAATG-3′ and 5′-TCGTTCATGCACTCGCTGA-3′
(HES1).
Generation of stable cell lines
To generate T98 and U251MG cells that constitutively
expressed GFP, GFP-PRRX1A and GFP-PRRX1B, 293T cells were
transfected with a pQCXIP vector (Clontech Laboratories, Inc.,
Mountain View, CA, USA) encoding each gene as well as the pVPack-GP
and pVPack-Ampho vectors (Stratagene, Tokyo, Japan). The culture
supernatant was collected 48 h later and applied to T98 or U251MG
cells with 2 μg/ml of Polybrene (Sigma-Aldrich). The cells were
cultured for 24 h, and then 1 μg/ml of puromycin (Sigma-Aldrich)
was added to select for infected cells. T98 and U251MG cells that
constitutively expressed dominant-negative RBPJ (DN-RBPJ) with
PRRX1 were produced by infecting PRRX1-expressing cells with
recombinant retrovirus that encoded DN-RBPJ. The infected cells
were selected with neomycin for 5 days. To generate U87 and U251sp
cells that constitutively expressed the shRNAs, oligonucleotides
encoding shRNAs specific for human PRRX1 and luciferase were cloned
into the pSIREN-RetroQ vector (Clontech Laboratories, Inc.). The
sequences of the shRNAs were: 5′-GCTTGAAGCTACAGATTAT-3′ (shPRRX1),
5′-CCACTG TTCTTATCTCTAT-3′ (shPRRX2) and 5′-CTTACGCTGAG TACTTCGA-3′
(control). Recombinant retrovirus was produced, and infected U87
and U251sp cells were selected with 1 μg/ml puromycin for 3
days.
Invasion assay
To measure cell invasion using Boyden chambers, a
filter was pre-coated with Matrigel, and 4.5×104 cells
(T98, U251sp, U87) or 1.2×104 cells (U251MG) were seeded
into the upper surface of the chamber. Twenty hours after seeding,
the cells were fixed with 70% methanol and stained with 0.5%
crystal violet. Cells that invaded the lower surface of the filters
were counted in five randomly selected fields. Three independent
experiments were performed. To evaluate cell invasion in the
presence of the Notch inhibitor, DAPT, cells were treated with DMSO
or DAPT (10 μM) for 12 h and then subjected to an invasion assay in
the presence of DMSO or DAPT.
Neurosphere assay
Cells were cultured in a 6-well, ultra-low
attachment plate (Iwaki, Tokyo, Japan) with DMEM/F-12 medium
supplemented with 10 ng/ml bFGF and 20 ng/ml EGF (both from
PeproTech, Rocky Hill, NJ, USA). Two weeks later, the number of
spheres that were >50 μm in diameter was counted in 10
different, randomly selected fields.
Reporter assay
Cells were transfected with a reporter construct
with pRTK-Luc to normalize to the transfection efficiency. Reporter
constructs for Notch (19) and
Hedgehog signaling (20) were
obtained from RIKEN BioResource Center, and a construct for Wnt
signaling (21) was obtained from
Addgene (Addgene plasmid 12456). Twenty-four hours after
transfection, the activities of firefly and Renilla
lucif-erase were measured using the dual-luciferase reporter assay
system (Promega, Madison, WI, USA). Luciferase activity was
measured in triplicate, and three independent experiments were
performed.
Animal experiments
Animal experiments were conducted in accordance with
the Faculty of Medicine of Nagoya University. U87 cells expressing
control or PRRX1 shRNAs (3.5×105 cells/5 μl) were
stereotactically injected into the brains of Balb-c nu/nu nude mice
(female, 5 weeks old) under anesthesia using a Hamilton syringe
(Hamilton, Reno, NV, USA). The coordination was 1.4 mm posterior
from the bregma, 3.0 mm to the right and at a 4.0-mm depth from the
brain surface.
Results
PRRX1 is expressed in the glioma
samples
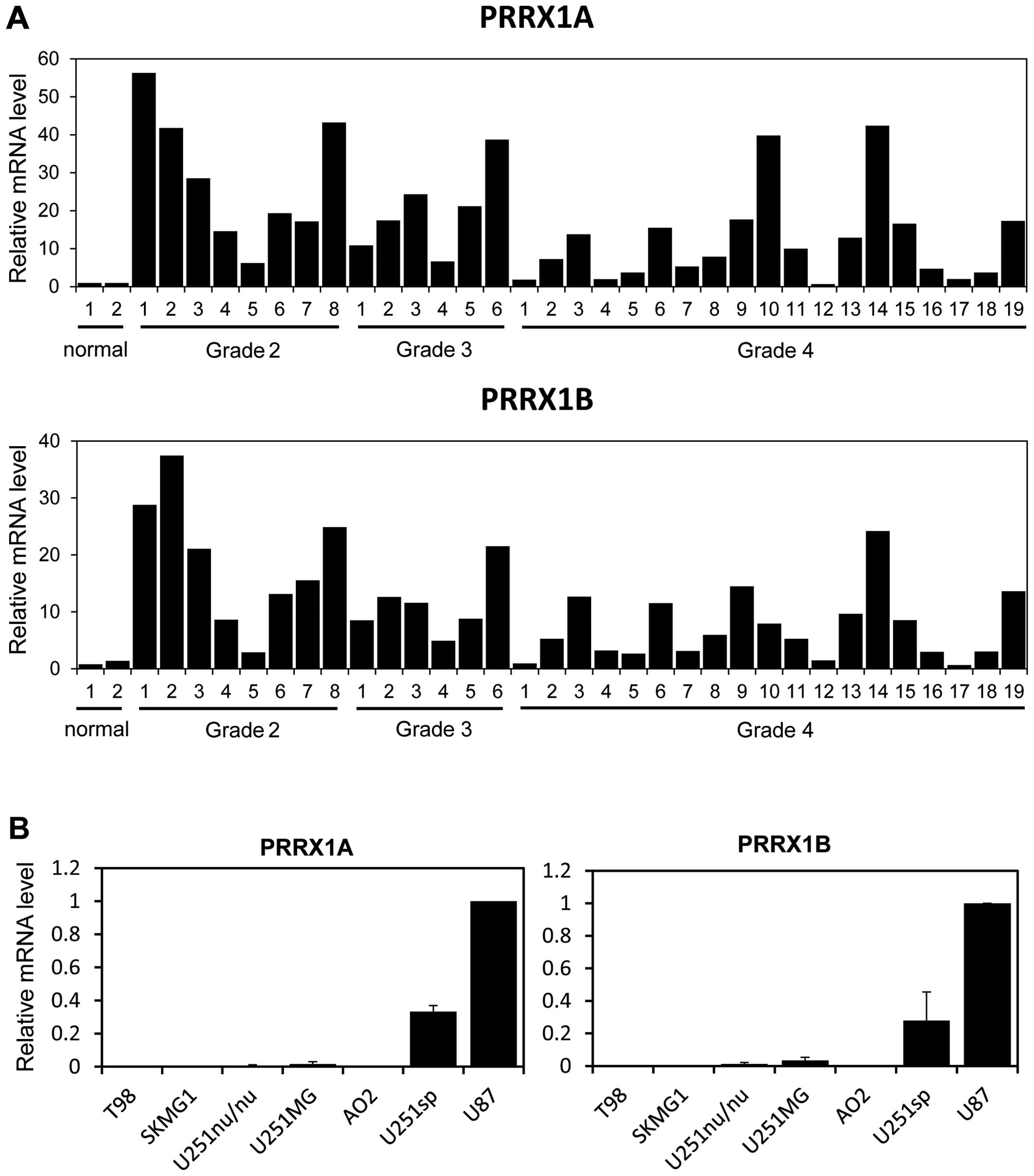
We first evaluated the mRNA levels of PRRX1 in
multiple low-grade and high-grade glioma samples. Two alternative
forms of PRRX1, PRRX1A and PRRX1B, exist; both isoforms have a
homeobox domain in their central regions, yet they differ at their
C-termini. PRRX1B has a so-called otp, aristaless and rax (OAR)
domain at its C-terminus, whereas the C-terminal end of PRRX1A
lacks this domain. Real-time PCR analysis revealed that the mRNA
levels of both isoforms were increased in most of the low-grade and
high-grade glioma samples when compared to the levels in the normal
brain tissue (Fig. 1A). We next
examined expression of PRRX1 in several glioblastoma cell lines.
PRRX1A and PRRX1B were highly expressed in the U251sp and U87 cells
compared with that in the other glioblastoma cell lines (Fig. 1B).
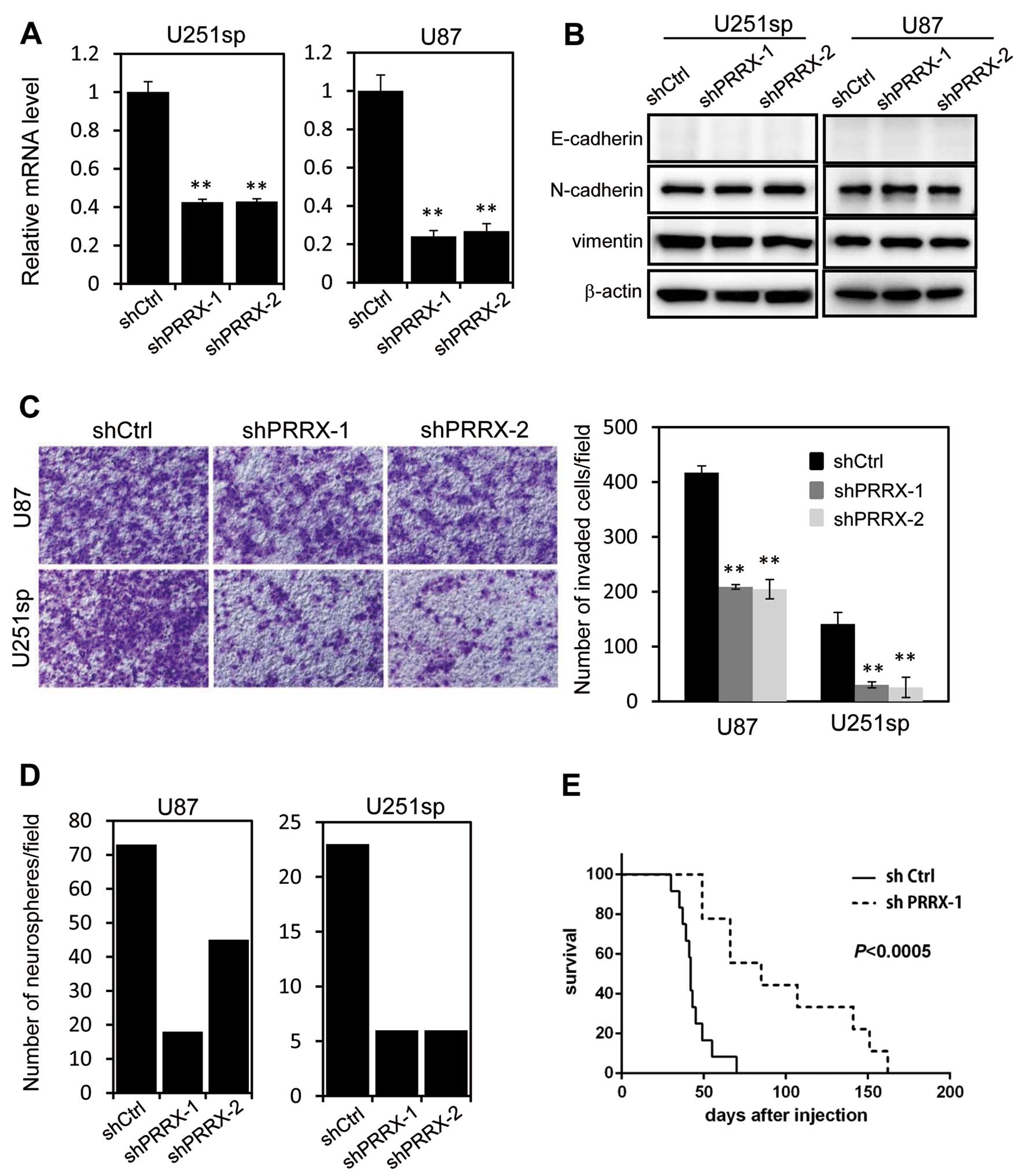
Silencing of PRRX1 inhibits invasion and
neurosphere formation
To examine the role of PRRX1 in glioblastoma cells,
we used two different shRNAs that targeted both isoforms of PRRX1.
U251sp and U87 cells that constitutively expressed the control
(shCtrl) or PRRX1 shRNA (shPRRX-1 and shPRRX-2) were established by
retrovirus infection. RT-PCR analysis confirmed a significant
reduction in the PRRX1 mRNA in the shPRRX-1 and shPRRX-2 cells
(Fig. 2A). Although a previous
study reported that PRRX1 is associated with EMT, we did not
observe any changes in the expression of EMT markers, such as
E-cadherin and vimentin, following PRRX1 knockdown (Fig. 2B). We examined the invasion of
PRRX1-knockdown cells using Matrigel-coated Boyden chambers. The
invasion of U251sp and U87 cells was significantly reduced by PRRX1
knockdown (Fig. 2C). We performed
in vitro neurosphere forming assays to assess the
tumorigenicity of the PRRX1-depleted cells. Cells were cultured in
suspension with EGF and FGF in the absence of serum for 2 weeks,
and then the number of neurospheres was determined. As shown in
Fig. 2D, neurosphere formation was
suppressed by PRRX1 depletion. We then examined the effect of PRRX1
suppression on the proliferation of glioblastoma cells in
vivo. Both shCtrl and shPRRX-1 U87 cells were implanted in the
brains of mice by intracranial injection, and the survival of the
mice was observed. Mice implanted with the PRRX1-knockdown cells
exhibited a significantly longer survival when compared with the
survival in the shCtrl cell-injected mice (Fig. 2E). These results indicate that PRRX1
is associated with invasion and proliferative properties of
glioblastoma cells.
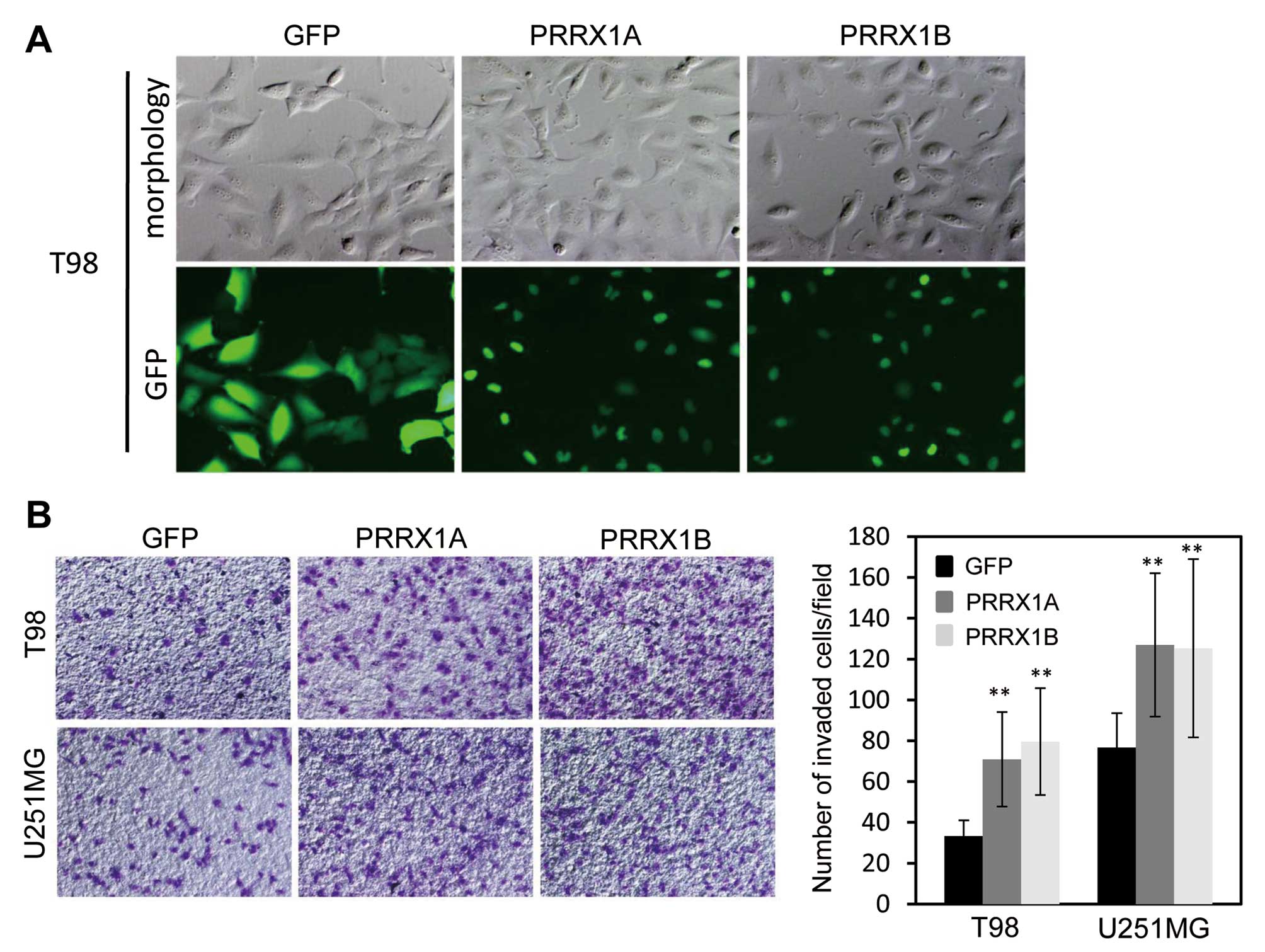
Exogenous expression of PRRX1 promotes
invasiveness in glioblastoma cells
To further confirm the effects of PRRX1 expression
on the invasion of glioblastoma cells, we expressed both isoforms
of PRRX1 in T98 and U251MG cells, which showed lower expression of
PRRX1 compared with U87 and U251sp cells. T98 and U251MG cells were
infected with recombinant virus that encoded GFP-tagged PRRX1A or
PRRX1B and selected with puromycin for 2 days. Most of the selected
cells clearly expressed GFP-PRRX1, and both isoforms localized to
the nucleus (Fig. 3A). We then
examined the invasion of PRRX1-expressing cells using
Matrigel-coated Boyden chambers. The expression of either isoform
of PRRX1 promoted the invasion of T98 and U251MG cells (Fig. 3B). Neurosphere formation assays were
also performed to assess tumorigenicity, yet neither GFP- nor
GFP-PRRX1-expressing cells formed neurospheres (data not
shown).
PRRX1 induces Notch activation
We next investigated the molecular mechanisms by
which PRRX1 promotes the invasion of glioblastoma cells. A number
of pathways, including the Notch, Wnt and Hedgehog pathways, are
activated in glioblastoma cells and are known to be associated with
the progression of glioblastoma (22–24).
We tested whether these pathways were activated by PRRX1 expression
using a reporter assay. GFP- or GFP-PRRX1-expressing T98 cells were
transfected with a reporter plasmid encoding luciferase that could
be activated by either pathway, and the luciferase activity was
measured 24 h later. In this analysis, we found specific activation
of the Notch pathway in PRRX1-expressing cells (Fig. 4A). To further confirm Notch
activation by PRRX1 expression, we examined the level of HES1,
which is a target gene of Notch signaling. Consistent with the
activation of Notch signaling, increased levels of HES1 mRNA and
protein by PRRX1 expression were observed (Fig. 4B and C). Conversely, depletion of
PRRX1 expression in U251MG cells by siRNA transfection suppressed
HES1 mRNA levels (Fig. 4D). We also
tested whether suppression of PRRX1 affected Notch signaling in U87
and U251sp cells, but we did not observe any changes in Notch
activation after PRRX1 knockdown (data not shown). It appears that
PRRX1-mediated activation of Notch signaling is dependent upon the
cellular context. We next examined whether PRRX1 expression was
related with HES1 expression in the glioma samples. The expression
levels of the PRRX1A and HES1 mRNAs were evaluated in glioma
samples using RT-PCR analysis. As shown in Fig. 4E, we observed a correlation between
PRRX1A and HES1 expression.
 | Figure 4PRRX1 activates Notch signaling. (A)
GFP- or GFP-PRRX1-expressing T98 cells were transfected with a
reporter plasmid for each signaling pathway together with pRK-Luc
to normalize for transfection efficiency. Twenty-four hours later,
the cells were lysed, and luciferase activities were measured. The
graphs indicate the relative activities of luciferase. Three
independent experiments were performed, and the data are shown as
the means ± SD (**P<0.01, n.s., not significant). (B)
Level of HES1 mRNA in each cell line was determined by quantitative
RT-PCR and normalized to GAPDH. The graphs show the relative levels
of the HES1 mRNA. Three independent experiments were performed, and
the data are shown as the means ± SD (**P<0.01). (C)
GFP- or GFP-PRRX1-expressing cells were lysed, and expression of
HES1 was examined by immunoblotting. (D) U251MG cells were
transfected with two different PRRX1 siRNAs and 3 days later, the
level of HES1 mRNA in each cell line was determined by quantitative
RT-PCR. Three independent experiments were performed, and the data
are shown as the means ± SD (**P<0.01). (E) Relative
expression of the PRRX1A and HES1 mRNAs normalized to GAPDH mRNA in
glioma tissues was determined by quantitative RT-PCR. The Pearson’s
correlation coefficients (r) are shown. PRRX1, paired related
homeobox 1. |
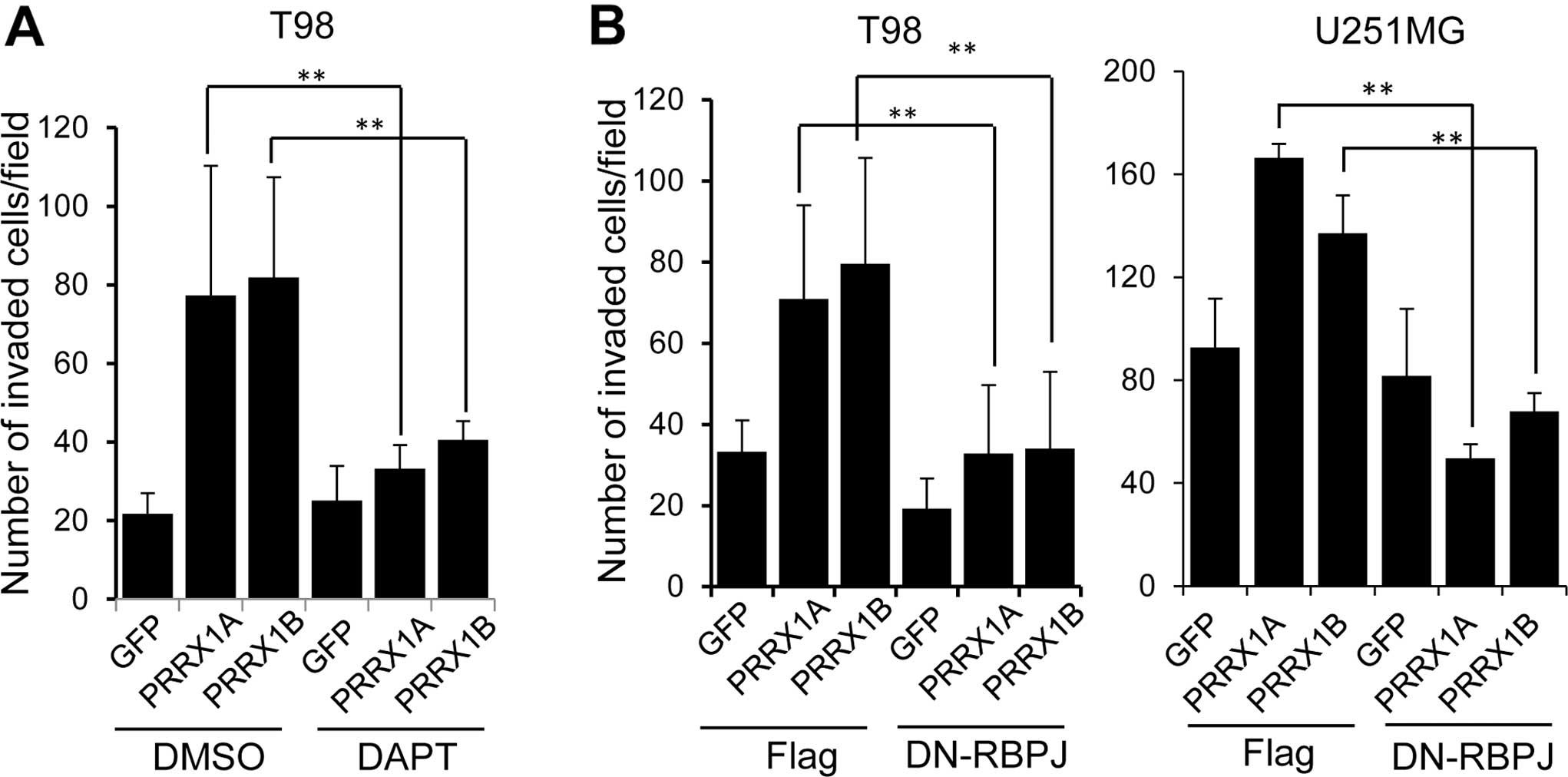
Promotion of invasion by PRRX1 is
dependent on Notch pathway activation
We tested whether Notch activation was responsible
for the PRRX1-mediated promotion of cell invasion. To inhibit Notch
activation, we used a chemical inhibitor, DAPT. Cells were
incubated with DAPT for 12 h and subjected to the invasion assay in
the presence of DAPT. Addition of DAPT clearly suppressed the
invasion of PRRX1-expressing T98 cells (Fig. 5A). We also used a dominant negative
form of the recombination signal binding protein for immunoglobulin
kappa J region (RBPJ), which is a transcriptional regulator that is
critical for the Notch signaling pathway (25). We established T98 and U251MG cells
that constitutively expressed PRRX1 and a dominant-negative form of
RBPJ (DN-RBPJ) and examined cell invasion. As shown in Fig. 5B, expression of the DN-RBPJ
suppressed the invasion of PRRX1-expressing cells.
Discussion
In the present study, we studied the role of PRRX1
in glioblastoma cells. Depletion of PRRX1 in U87 and U251sp cells
reduced the invasive potential of cells. In addition, tumor
implantation experiments showed longer survival of mice injected
with PRRX1-knockdown cells when compared with the survival of mice
injected with the control cells. Conversely, exogenous expression
of PRRX1 in the T98 and U251MG cells clearly promoted cellular
invasion. These results clearly indicate that PRRX1 has the
potential to promote invasion of glioblastoma. A previous study
reported that PRRX1 is an EMT inducer that promotes cancer cell
invasion (12). During EMT,
non-motile epithelial cells dissolve cell-cell junctions and become
motile, invasive mesenchymal cells; therefore, EMT is an important
step for cancer cells to acquire invasive potential (26). Although we examined whether
PRRX1-mediated invasion was associated with EMT, we did not observe
clear changes in cellular morphology or in the expression of marker
proteins, such as vimentin and E-cadherin. These results indicate
that the induction of EMT by PRRX1 is dependent on the cell
type.
PRRX1 has two isoforms. PRRX1B has an OAR domain at
its C-terminal end, while PRRX1A lacks this domain. The exact role
of the OAR domain is not yet clear, yet some studies have reported
that it is associated with its transcriptional activities (27). PRRX1A deleted of the OAR domain was
capable of promoting transcriptional activity, which indicates that
the OAR domain has repressive function (28). The OAR domain was also found to
attenuate the activity of other transcription factors (29). A previous study showed that PRRX1A
and PRRX1B induced different sets of genes, and the expression of
PRRX1B is more effective in promoting sphere formation and invasion
of pancreatic cells (30). We
expressed PRRX1A or PRRX1B in the T98 and U251 cells and examined
cell invasion; however, we did not observe any clear differences
between them. It has been suggested that the difference in activity
of PRRX1A and PRRX1B is mediated by undetermined co-factors; thus,
the function of the OAR may differ depending on the cell
context.
The Notch signaling pathway is an evolutionarily
conserved pathway that regulates differentiation, proliferation and
survival. Recent studies have revealed that the Notch signaling
pathway is activated in numerous types of cancer. For example, in
ovarian cancers, the expression of Notch3 is increased, and
inhibition of the Notch pathway inhibits cancer progression
(31). In glioblastoma cells, the
Notch pathway is important for proliferation, stem cell maintenance
and tumorigenesis (32). These
studies have clearly shown the important functions of Notch
activation for the progression of numerous cancers, including
glioblastoma. We showed that PRRX1 expression promoted the
activation of Notch signaling, and inhibition of the Notch pathway
abolished PRRX1-mediated tumor invasion. In addition, expression
levels of PRRX1 and HES1, which is a target gene of the Notch
pathway, were correlated in numerous glioma samples. These results
suggest a potential role of PRRX1 in the activation of Notch
signaling in glioblastoma. PRRX1 is widely expressed, and the Notch
pathway is activated in numerous tumors; thus, PRRX1 may regulate
Notch activation in various types of tumors.
In summary, we demonstrated that PRRX1 is expressed
in glioma and that its inhibition suppresses tumor invasion. We
also showed that PRRX1-mediated tumor invasion was dependent on
activation of the Notch pathway. Accumulating evidence has clearly
shown that PRRX1 is associated with the progression of several
types of cancer. Detailed analyses of the functions of PRRX1 may
provide novel molecular mechanisms for tumor progression and
therapeutic strategies.
Acknowledgements
We would like to thank Dr Honjo, Dr Sasaki and Dr
Moon for the Notch, Hedgehog and Wnt signaling reporter constructs.
This study was funded by a grant from the Ministry of Education,
Culture, Sports, Science and Technology of Japan (Nanomedicine
Molecular Science, 23107010), and from the Takeda Science
Foundation.
References
|
1
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bello L, Giussani C, Carrabba G, Pluderi
M, Costa F and Bikfalvi A: Angiogenesis and invasion in gliomas.
Cancer Treat Res. 117:263–284. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shah N and Sukumar S: The Hox genes and
their roles in onco-genesis. Nat Rev Cancer. 10:361–371. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Coletta RD, Christensen KL, Micalizzi DS,
Jedlicka P, Varella-Garcia M and Ford HL: Six1 overexpression in
mammary cells induces genomic instability and is sufficient for
malignant transformation. Cancer Res. 68:2204–2213. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
McCoy EL, Iwanaga R, Jedlicka P, et al:
Six1 expands the mouse mammary epithelial stem/progenitor cell pool
and induces mammary tumors that undergo epithelial-mesenchymal
transition. J Clin Invest. 119:2663–2677. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yuan H, Kajiyama H, Ito S, et al: ALX1
induces snail expression to promote epithelial-to-mesenchymal
transition and invasion of ovarian cancer cells. Cancer Res.
73:1581–1590. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Costa BM, Smith JS, Chen Y, et al:
Reversing HOXA9 oncogene activation by PI3K inhibition: epigenetic
mechanism and prognostic significance in human glioblastoma. Cancer
Res. 70:453–462. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kern MJ, Argao EA, Birkenmeier EH, Rowe LB
and Potter SS: Genomic organization and chromosome localization of
the murine homeobox gene Pmx. Genomics. 19:334–340. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Norris RA, Scott KK, Moore CS, et al:
Human PRRX1 and PRRX2 genes: cloning, expression, genomic
localization, and exclusion as disease genes for Nager syndrome.
Mamm Genome. 11:1000–1005. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Martin JF, Bradley A and Olson EN: The
paired-like homeo box gene MHox is required for early events of
skeletogenesis in multiple lineages. Genes Dev. 9:1237–1249. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Takahashi Y, Sawada G, Kurashige J, et al:
Paired related homoeobox 1, a new EMT inducer, is involved in
metastasis and poor prognosis in colorectal cancer. Br J Cancer.
109:307–311. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ocaña OH, Córcoles R, Fabra A, et al:
Metastatic colonization requires the repression of the
epithelial-mesenchymal transition inducer Prrx1. Cancer Cell.
22:709–724. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shimozaki K, Clemenson GD Jr and Gage FH:
Paired related homeobox protein 1 is a regulator of stemness in
adult neural stem/progenitor cells. J Neurosci. 33:4066–4075. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bass AJ, Watanabe H, Mermel CH, et al:
SOX2 is an amplified lineage-survival oncogene in lung and
esophageal squamous cell carcinomas. Nat Genet. 41:1238–1242. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gangemi RM, Griffero F, Marubbi D, et al:
SOX2 silencing in glioblastoma tumor-initiating cells causes stop
of proliferation and loss of tumorigenicity. Stem Cells. 27:40–48.
2009. View Article : Google Scholar
|
|
16
|
Fang X, Yu W, Li L, et al: ChIP-seq and
functional analysis of the SOX2 gene in colorectal cancers. OMICS.
14:369–384. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Alonso MM, Diez-Valle R, Manterola L, et
al: Genetic and epigenetic modifications of Sox2 contribute to the
invasive phenotype of malignant gliomas. PLoS One. 6:e267402011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ohta S, Mizuno M, Takaoka T and Yoshida J:
Augmentation of anti-Fas antibody-mediated apoptosis on human
glioma cells by liposomes associated with the antibody. J
Neurooncol. 35:7–11. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kurooka H, Kuroda K and Honjo T: Roles of
the ankyrin repeats and C-terminal region of the mouse Notch1
intracellular region. Nucleic Acids Res. 26:5448–5455. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sasaki H, Hui C, Nakafuku M and Kondoh H:
A binding site for Gli proteins is essential for HNF-3β floor plate
enhancer activity in transgenics and can respond to Shh in vitro.
Development. 124:1313–1322. 1997.PubMed/NCBI
|
|
21
|
Kaykas A, Yang-Snyder J, Héroux M, Shah
KV, Bouvier M and Moon RT: Mutant Frizzled 4 associated with
vitreoretinopathy traps wild-type Frizzled in the endoplasmic
reticulum by oligo-merization. Nat Cell Biol. 6:52–58. 2004.
View Article : Google Scholar
|
|
22
|
Lino MM, Merlo A and Boulay JL: Notch
signaling in glioblastoma: a developmental drug target? BMC Med.
8:722010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mimeault M and Batra SK: Complex oncogenic
signaling networks regulate brain tumor-initiating cells and their
progenies: pivotal roles of wild-type EGFR, EGFRvIII mutant and
hedgehog cascades and novel multitargeted therapies. Brain Pathol.
21:479–500. 2011.PubMed/NCBI
|
|
24
|
Paul I, Bhattacharya S, Chatterjee A and
Ghosh MK: Current understanding on EGFR and Wnt/β-catenin signaling
in glioma and their possible crosstalk. Genes Cancer. 4:427–446.
2013. View Article : Google Scholar
|
|
25
|
Tanigaki K and Honjo T: Two opposing roles
of RBP-J in Notch signaling. Curr Top Dev Biol. 92:231–252. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Leussink B, Brouwer A, el Khattabi M,
Poelmann RE, Gittenberger-de Groot AC and Meijlink F: Expression
patterns of the paired-related homeobox genes MHox/Prx1 and S8/Prx2
suggest roles in development of the heart and the forebrain. Mech
Dev. 52:51–64. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Norris RA and Kern MJ: The identification
of Prx1 transcription regulatory domains provides a mechanism for
unequal compensation by the Prx1 and Prx2 loci. J Biol Chem.
276:26829–26837. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Brouwer A, ten Berge D, Wiegerinck R and
Meijlink F: The OAR/aristaless domain of the homeodomain protein
Cart1 has an attenuating role in vivo. Mech Dev. 120:241–252. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Reichert M, Takano S, von Burstin J, et
al: The Prrx1 homeodomain transcription factor plays a central role
in pancreatic regeneration and carcinogenesis. Genes Dev.
27:288–300. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen X, Thiaville MM, Chen L, et al:
Defining NOTCH3 target genes in ovarian cancer. Cancer Res.
72:2294–2303. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Saito N, Fu J, Zheng S, et al: A high
Notch pathway activation predicts response to γ secretase
inhibitors in proneural subtype of glioma tumor-initiating cells.
Stem Cells. 32:301–312. 2014. View Article : Google Scholar :
|