Globally, hepatocellular carcinoma (HCC) is the
fifth most common human cancer and the third leading cause of
cancer-associated mortalities (1).
In spite of the great achievements of novel therapies and
diagnostic techniques, the early detection of HCC is difficult,
resulting in a poor 5-year survival for HCC patients (ranging from
0 to 14%) (2,3). Therefore, the identification of the
most specific and sensitive biomarkers for HCC is crucial.
MicroRNAs (miRNAs) are a large set of small
non-coding RNAs, ~22 nucleotides in length, that mainly bind to the
seed sequences located within the 3′ untranslated region (3′UTR) of
target mRNAs. miRNAs can promote the degradation or suppress the
translation of target mRNAs, eventually inhibiting the biological
functions of their target genes. Different genes can be regulated
by the same miRNA, while different miRNAs can be regulated by the
same gene (4,5). Over 2,500 human miRNAs have been found
to play important roles in various physiological and pathological
processes such as embryonic development, cell proliferation,
differentiation, cell cycle progression, apoptosis, autophagy,
angiogenesis and metabolism (6,7).
Recently, it has been demonstrated that miRNAs exhibit tissue- and
disease-specific patterns in human cancers, indicating that miRNAs
may be novel biomarkers for the early diagnosis and prognosis
prediction of HCC (8). In the
present review, we summarized the known alterations of miRNAs and
their biological roles in the develop ment of HCC.
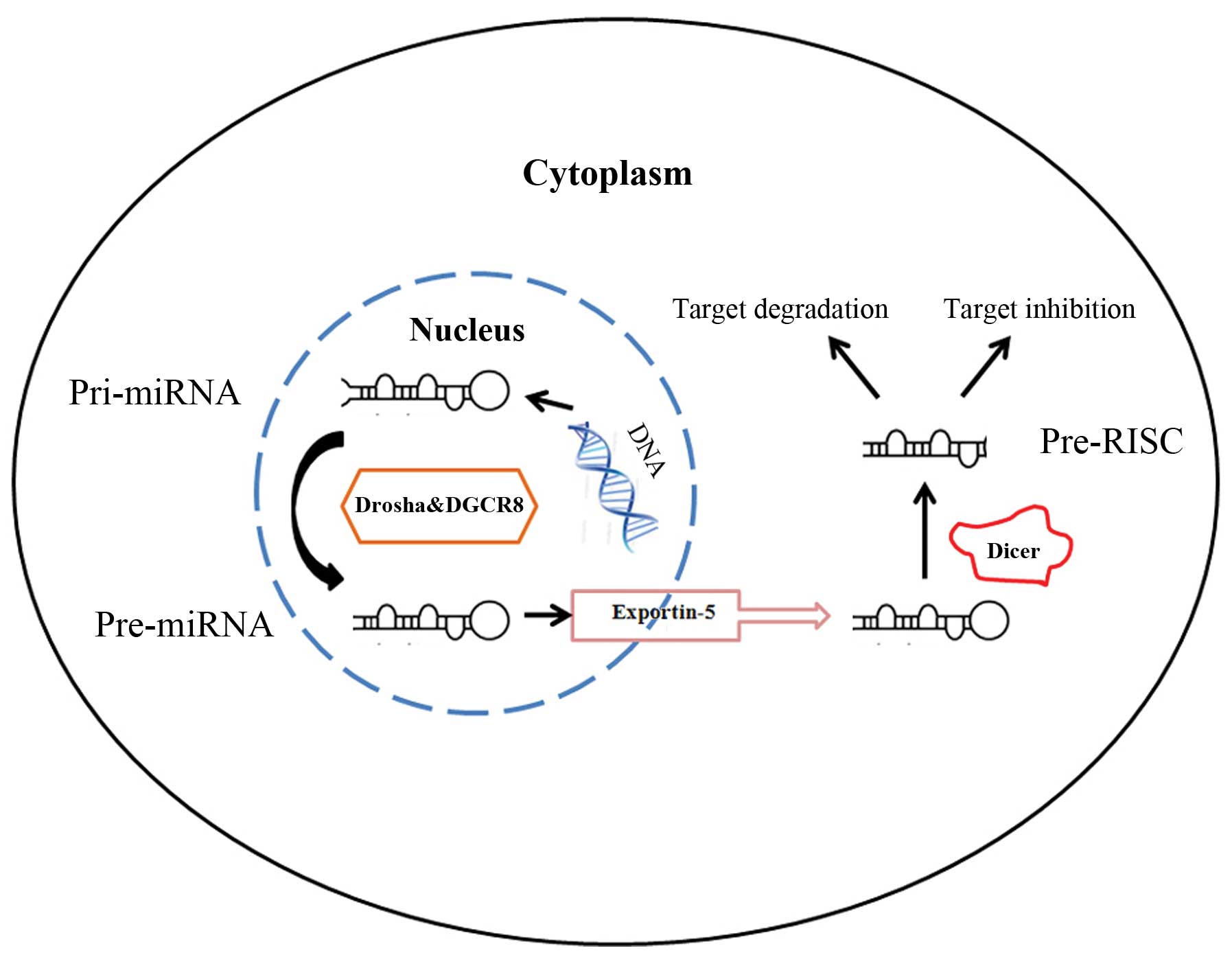
Primary miRNAs (pri-miRNAs, containing stem-loop
structures) are first transcribed by RNA polymerase II and then
processed into the hair-shaped precursor miRNA (pre-miRNA, 70–90
nucleotides in length) by the complex comprising RNAase III (also
known as Drosha) and DGCR8/Pasha in the nucleus (Fig. 1). Pre-miRNAs are transported into
the cytoplasm by the exportin-5 complex and cleaved into mature
miRNAs by Dicer (9–13). Earlier studies have identified
enhancers and silencers of miRNA transcription (11,14,15).
Recently, other mechanisms including DNA methylation and histone
modification have been shown to regulate the production of miRNAs
(16,17).
During the past decade, the multiple roles of miRNAs
in the initiation and progression of human cancer have been well
established. miRNAs are commonly dysregulated in tumor tissues and
act as oncogenes or tumor suppressors, respectively. miRNAs are
able to manipulate tumor proliferation, migration, invasion,
metastasis, angiogenesis, cell cycle progression, apoptosis and
autophagy. Consistently, the crucial roles of miRNAs in
tumorigenesis and development have been further demonstrated in
several animal models. For example, miRNA-15a/miRNA-16-1 knockout
mice were predisposed to develop chronic lymphocytic leukemia
(18). In addition, Eμ-miRNA-155
transgenic mice were prone to develop a proliferative B-cell
malignancy in lymph nodes (19).
Collectively, these results indicated that miRNAs may be valid
therapeutic targets for treating human cancers.
Numerous studies have focused on the ectopic
expression (up- or downregulation) of miRNAs in HCC. A panel of
miRNAs have been identified to be candidate tumor suppressors or
oncogene inducers and further proven to be critical factors in the
regulation of malignant tumor behaviors. A brief description of
these miRNAs is provided below.
The miRNA let-7g is a member of the large let-7
family. It is significantly downregulated in human HCC tissues and
is closely associated with the metastasis and poor overall survival
of HCC. In vitro, restoration of let-7g markedly inhibited
tumor proliferation and migration, suppressed the
epithelial-mesenchymal transition (EMT), and induced cell apoptosis
and cell cycle arrest by blocking the K-Ras/HMGA2/Snail signaling
pathway (20). Let-7g also targets
Bcl-xL and collagen type I α2, thus promoting apoptosis and
inhibiting migration in HCC cells.
The liver-specific miRNA-122 is significantly
downregulated in a large number of HCC patients and is often
inversely associated with a poor prognosis and metastasis.
Restoration of miR-122 inhibits proliferation and migration, and
increases apoptosis by targeting AKT3 in HCC cell lines (21). miR-122 was able to directly bind to
the 3′UTR of the DLX4 gene (Distal-less 4) and downregulate
its expression, which markedly suppressed HCC cell proliferation.
Considering that miR-122 is associated with tumor invasion and
metastasis in HCC, Wang et al (22) performed a panel of experiments and
demonstrated that miR-122 was capable of triggering the EMT, induce
disruption of the cellular cytoskeleton, block the RhoA/Rock
signaling pathway, enhance adhesion and suppress invasion in HCC
cells. Recently, it has been suggested that cell morphology and
mitochondrial functions can be markedly regulated by miR-122,
proposing that miR-122 is critical for hepatocarcinogenesis
(23). Thus, miR-122 knockout mice
eventually developed spontaneous tumors resembling human HCC. In
particular, in the Huh7, HepG2 and QSG-7701 HCC cell lines, the
expression of miR-122 has been shown to be significantly inhibited
by the methylation of its promoter, which was restored by treatment
with the demethylation agent 5-aza-dC. Additionally, the
overexpression of miR-122 induced further cell apoptosis in these
HCC cells (24).
miR-199a-1, miR-199a-2 and miR-199b belong to the
miR-199 family. In HCC tissues, miR-199a was significantly
downregulated, correlating with a higher recurrence rate and a poor
prognosis. In HCC cell lines, miR-199a was able to suppress tumor
proliferation and induce apoptosis and cell cycle arrest by
regulating the expression of matrix metallo-proteinase-9 (MMP-9),
frizzled type 7 receptor (FZD7) and hypoxia-inducible factor-1α. In
a similar manner, miR-199a-3p expression was significantly reduced
in a panel of human HCC cell lines. In addition, it was shown that
miR-199a-3p directly targeted CD44 and inactivated the c-Met
signaling pathway. The other known targets of miR-199a-3p include
the mammalian target of rapamycin (mTOR) and c-Met, which play
important roles for the biological functions of miR-199a-3p as a
tumor suppressor. Downregulation of miR-199a-5p was observed in
more than two-thirds of HCC samples and notably associated with an
advanced tumor stage. In vitro experiments suggested that
miR-199a-5p directly inhibits the expression of discoidin domain
receptor-1 (DDR1) and that the loss of miR-199a-5p leads to the
upregulation of DDR1 and enhances the invasion of HCC cells. In
another study, it was found that upregulated miR-199a-5p directly
inhibited the activity of E2F3 and sensitized HCC cells to routine
chemotherapies (25). Of note,
miR-199a-5p significantly enhanced the suppressive effects of
cisplatin on cell proliferation (26), while cisplatin downregulated the
level of miR-199a-5p and then induced autophagy by activating
autophagy-associated gene 7, a direct target of miR-199a-5p. These
controversial results remain to be clarified in the future using
more powerful experimental approaches (25–34).
In addition, the marked reduction of miR-20a,
miR-138, miR-503, miR-218 and miR-376a expression was detected in
HCC tissues. These miRNAs have been shown to be involved in tumor
proliferation, apoptosis and cell cycle regulation (35–37).
Collectively, a large number of miRNAs are downregulated in human
HCC and have been found to be closely associated with tumor
progression and prognosis.
miRNAs can also serve as oncogenes in human cancers.
In HCC, a group of onco-miRNAs (listed in Table II) has been identified, and their
functions have been well defined. Of these, miR-21, miR-221 and
miR-224 are the best studied. Consequently, the roles in HCC of
these miRNAs are reviewed.
miR-21 is one of the most common dysregulated miRNAs
in human cancers and is involved in the regulation of cell
proliferation, differentiation, apoptosis, angiogenesis, migration
and invasion (38–40). Recent studies have suggested that
miR-21 functions as a pro-metastatic miRNA in HCC (41) and can promote the invasion and
metastasis of HCC by targeting phosphatase and tensin homolog
(PTEN) and heparin-degrading endosulfatase-1 (hSulf-1) and
activating the AKT/ERK signaling pathways (42). The HEPN1 gene is another
target of miR-21, and its silence induced by miR-21 significantly
accelerated the tumor growth of HCC. In addition, mitogen-activated
protein kinase-kinase 3 (MAP2K3), reversion-inducing-cysteine-rich
protein with kazal motifs (RECK), and programmed cell death 4
(PDCD4) were the direct targets of miR-21, and their expression and
functions were notably suppressed in HCC (41,43,44).
Another overexpressed miRNA in human HCC is miR-221,
and its expression is significantly correlated with a poor overall
survival and recurrence-free survival. It was found that miR-221
causes rapid S-phase entry and enhances tumor growth by targeting
p27, p57 and aryl hydrocarbon nuclear translocator (Arnt) in HCC.
Additionally, the upregulation of miR-221 has been associated with
a more aggressive phenotype of HCC and can suppress cell apoptosis
by targeting the Bmf gene (45).
Recently, miR-221 was shown to be able to silence histone
deacetylase 6 (HDAC6) and enhance the malignant progression of
HCC.
miR-224 is upregulated in HCC, and recent studies
have shown that miR-224 can act as an onco-miRNA in HCC through
activating the AKT signaling pathway (46). In a previous study, we demonstrated
that miR-224 can promote migration and invasion in HCC cells by
targeting the homeobox D10 (HOXD10) gene (47).
Thus, dysregulated miRNAs are frequently involved in
almost every step of the initiation and progression of HCC,
indicating that these miRNAs are potential targets for the
diagnosis and prognosis prediction for HCC patients.
Since a large number of HCC patients are diagnosed
at an advanced stage of disease, the development of novel valid
approaches to detect HCC earlier is crucial. Currently,
α-fetoprotein is the only marker commonly used for HCC detection in
the clinic. However, its reliability is questionable and its
accuracy is not satisfactory (6).
Based on the amount of evidence from clinical and basic research,
it has been suggested that miRNAs have potential characteristics as
diagnostic markers for HCC. Technically, miRNAs in circulation and
in the cytoplasm are abundant and stable enough for detection using
commercial kits. For example, a recent study has shown that miR-127
is significantly downregulated in HCC and that it is a potential
diagnostic biomarker for HCC (48).
It has been previously demonstrated that miR-21, miR-26a, miR-27a,
miR-122, miR-192, miR-223 and miR-801 can discriminate between HCC
and healthy, chronic hepatitis B and cirrhosis groups (6). miR-126, miR-141 and miR-200c are also
able to differentiate HCC from metastatic liver cancer with a high
accuracy (49). In addition,
aberrant DNA methylation of miRNA is potentially a useful parameter
for the early diagnosis of HCC, and it has been found that single
locus hypermethylation of miR-129-2 functions as a highly
specific marker to distinguish HCC from chronic hepatitis and
healthy liver tissues (50).
Circulating miRNAs are also valuable for the early detection and
prognosis prediction of HCC. The circulating miRNAs present in the
serum and plasma of HCC patients are provided in Table III.
As previously reported, miRNAs are a powerful
predictor of prognosis for cancer patients. Recently, several
studies have shown the validity of miRNAs as prognostic markers in
HCC (35,51–70).
Upregulation of miR-9, miR-17-5p, miR-18, miR-25 and miR-590-5p/3p
presented a high metastatic potential and a shorter survival in
patients with HCC (35,67–70).
Another study has indicated that concordant DNA methylation at
certain miRNA loci correlated with a poor survival for HCC
patients. Therefore, methylation may be used as a biomarker to
predict the prognosis for HCC patients (50). In other studies, either a single
miRNA or a panel of several miRNAs was proven to be a good
predictor of prognosis for HCC patients (71,72).
Taken together, all of the abovementioned studies
suggest that miRNAs can act as valuable biomarkers for the early
diagnosis and prognosis prediction of HCC. However, controversies
regarding the application of these markers in the clinic remain,
thus further investigations are required.
miRNAs function as tumor suppressors or oncogenes in
HCC. Therefore, targeting these miRNAs may be a novel approach to
treat HCC (24,35,60,61,64,73–102).
Currently, the therapeutic application of miRNAs mainly consists of
two strategies: miRNA inhibition and miRNA replacement.
The aim of miRNA inhibition is to suppress oncogenic
miRNAs using miRNA antagonists that usually involve some chemical
changes to heighten binding, reduce nuclease resistance and promote
cellular intake (6). Evidence has
suggested that miR-146a, miR-182, miR-184, and miR-190b can act as
therapeutic targets for HCC (98–102).
The aim of miRNA replacement is to restore the level
of tumor suppressor miRNAs. In HCC cell lines, restoration of
miR-26a/b was able to increase their chemosensitivity, which was
favorable for the targeted molecular therapy of HCC (75–77). A
previous study has shown that miR-26a replacement using an
adeno-associated (AAV8) delivery system decreased the tumori
genicity in a mouse model of HCC (49). Another example is that miR-122 can
be used as a tumor suppressor through AAV-mediated delivery in a
mouse model of HCC (49). Of these
miRNAs, miR-34a is particularly noteworthy as it is the first miRNA
mimic to reach the clinic (49) and
is capable of inducing sensitivity to the antitumor effect of
sorafenib in the treatment of HCC (79,80).
Furthermore, miR-425-3p is a promising prognostic marker in HCC
treated with sorafenib (93).
The therapeutic application of miRNAs is a promising
strategy for HCC treatment. However, it is well known that one
miRNA regulates multiple target genes and that artificially up- or
downregulating the level of miRNAs may result in undesirable
off-target effects. Thus, the application of miRNAs for HCC
treatment remains to be examined in clinical trials.
The rapid development of radiation techniques has
led to radiotherapy becoming a major treatment for HCC. However, a
subgroup of HCC patients present intrinsic or acquired resistance
to routine radiotherapy, which significantly hinders the
therapeutic effects and patient outcomes. Therefore, determining
methods to improve the effects of radiotherapy is of interest to
oncologists and radiologists. Recent findings have shown that
miRNAs are closely associated with radiotherapy outcomes and are
involved in radiosensitivity (103). Some miRNAs can act as biomarkers
to predict the cellular sensitivities to radiotherapy, while others
enhance or reduce radiosensitivity in vitro and in
vivo. For example, miRNA-381 promoted the radiosensitivity of
esophageal squamous cell carcinoma (ESCC), and its expression
played a vital role in the radiosensitivity of ESCC (104). In addition, miRNA-25 is
overexpressed in radio-resistant non-small cell lung cancer (NSCLC)
patients, and miRNA-25 affected radiosensitivity by regulating BTG2
directly in NSCLC cells. Overexpression of miRNA-145 promoted the
radiosensitivity of cervical cancer and is a potential new
biomarker of radio-sensitizing treatment for cervical cancer
(105). Based on the
abovementioned data, miRNAs play a crucial role in radiosensitivity
for cancer. However, the relationship between miRNAs and HCC
radio-sensitivity has yet to be reported. Therefore, in view of the
importance of miRNAs on radiosensitivity, their mechanisms in HCC
should be elucidated.
In summary, miRNAs are widely used in many areas of
cancer, especially in HCC, including the early diagnosis, prognosis
prediction, follow-up monitoring and target therapies. Undoubtedly,
miRNAs have important effects on HCC; therefore, miRNA-based
therapies pose a significant challenge for HCC treatment. However,
investigations regarding miRNAs cannot yet be applied in the
clinic. Thus, more well-designed studies are required to focus on
their translational values in the future. The accuracy of miRNA
detection needs to be further improved to avoid variations in the
technical procedures. Additionally, more large randomized
prospective clinical trials are required for the application of
miRNAs to assess their potential efficacy in HCC treatment,
especially for the radiotherapy of HCC.
The present review was supported by the National
Natural Science Foundation of China (grant nos. 81272498, 30973457
and 30901764).
|
1
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schwartz M, Roayaie S and Konstadoulakis
M: Strategies for the management of hepatocellular carcinoma. Nat
Clin Pract Oncol. 4:424–432. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Poon RT and Fan ST: Hepatectomy for
hepatocellular carcinoma: Patient selection and postoperative
outcome. Liver Transpl. 10(2 Suppl 1): S39–S45. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
D'Anzeo M, Faloppi L, Scartozzi M,
Giampieri R, Bianconi M, Del Prete M, Silvestris N and Cascinu S:
The role of micro-RNAs in hepatocellular carcinoma: From molecular
biology to treatment. Molecules. 19:6393–6406. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Garajová I, Le Large TY, Frampton AE,
Rolfo C, Voortman J and Giovannetti E: Molecular mechanisms
underlying the role of microRNAs in the chemoresistance of
pancreatic cancer. Biomed Res Int. 2014:6784012014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han J, Lee Y, Yeom KH, Kim YK, Jin H and
Kim VN: The Drosha-DGCR8 complex in primary microRNA processing.
Genes Dev. 18:3016–3027. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S, et al: The nuclear RNase III
Drosha initiates microRNA processing. Nature. 425:415–419. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yi R, Qin Y, Macara IG and Cullen BR:
Exportin-5 mediates the nuclear export of pre-microRNAs and short
hairpin RNAs. Genes Dev. 17:3011–3016. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Denli AM, Tops BB, Plasterk RH, Ketting RF
and Hannon GJ: Processing of primary microRNAs by the
microprocessor complex. Nature. 432:231–235. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Borchert GM, Lanier W and Davidson BL: RNA
polymerase III transcribes human microRNAs. Nat Struct Mol Biol.
13:1097–1101. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cai X, Hagedorn CH and Cullen BR: Human
microRNAs are processed from capped, polyadenylated transcripts
that can also function as mRNAs. RNA. 10:1957–1966. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Weber B, Stresemann C, Brueckner B and
Lyko F: Methylation of human microRNA genes in normal and
neoplastic cells. Cell Cycle. 6:1001–1005. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Scott GK, Mattie MD, Berger CE, Benz SC
and Benz CC: Rapid alteration of microRNA levels by histone
deacetylase inhibition. Cancer Res. 66:1277–1281. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Klein U, Lia M, Crespo M, Siegel R, Shen
Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, et
al: The DLEU2/miR-15a/16-1 cluster controls B cell proliferation
and its deletion leads to chronic lymphocytic leukemia. Cancer
Cell. 17:28–40. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Costinean S, Zanesi N, Pekarsky Y, Tili E,
Volinia S, Heerema N and Croce CM: Pre-B cell proliferation and
lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155
transgenic mice. Proc Natl Acad Sci USA. 103:7024–7029. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen KJ, Hou Y, Wang K, Li J, Xia Y, Yang
XY, Lv G, Xing XL and Shen F: Reexpression of Let-7g microRNA
inhibits the proliferation and migration via K-Ras/HMGA2/snail axis
in hepatocellular carcinoma. Biomed Res Int.
2014:7424172014.PubMed/NCBI
|
|
21
|
Nassirpour R, Mehta PP and Yin MJ: miR-122
regulates tumori-genesis in hepatocellular carcinoma by targeting
AKT3. PLoS One. 8:e796552013. View Article : Google Scholar
|
|
22
|
Wang SC, Lin XL, Li J, Zhang TT, Wang HY,
Shi JW, Yang S, Zhao WT, Xie RY, Wei F, et al: MicroRNA-122
triggers mesenchymal-epithelial transition and suppresses
hepatocellular carcinoma cell motility and invasion by targeting
RhoA. PLoS One. 9:e1013302014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jin JC, Zhang X, Jin XL, Qian CS, Jiang H
and Ruan Y: Micro-RNA-122 regulation of the morphology and
cytoarchitecture of hepatoma carcinoma cells. Mol Med Rep.
9:1376–1380. 2014.PubMed/NCBI
|
|
24
|
Xing TJ, Xu HT, Yu WQ and Jiang DF:
Methylation regulation of liver-specific microRNA-122 expression
and its effects on the proliferation and apoptosis of
hepatocellular carcinoma cells. Genet Mol Res. 12:3588–3597. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee JM, Heo MJ, Lee CG, Yang YM and Kim
SG: Increase of miR-199a-5p by protoporphyrin IX, a photocatalyzer,
directly inhibits E2F3, sensitizing mesenchymal tumor cells to
anticancer agents. Oncotarget. 6:3918–3931. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu N, Zhang J, Shen C, Luo Y, Xia L, Xue F
and Xia Q: Cisplatin-induced downregulation of miR-199a-5p
increases drug resistance by activating autophagy in HCC cell.
Biochem Biophys Res Commun. 423:826–831. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Peveling-Oberhag J, Seiz A, Döring C,
Hartmann S, Köberle V, Liese J, Zeuzem S, Hansmann ML and Piiper A:
MicroRNA profiling of laser-microdissected hepatocellular carcinoma
reveals an oncogenic phenotype of the tumor capsule. Transl Oncol.
7:672–680. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang CX, Song W, Li ZJ, Song B, Wu DH, Sun
AM and Chen LH: MicroRNA profiling in patients with hepatocellular
carcinoma. Nan Fang Yi Ke Da Xue Xue Bao. 30:976–977. 2010.In
Chinese. PubMed/NCBI
|
|
29
|
Zhang J, Zhang D, Wu GQ, Feng ZY and Zhu
SM: Propofol inhibits the adhesion of hepatocellular carcinoma
cells by upreg-ulating microRNA-199a and downregulating MMP-9
expression. Hepatobiliary Pancreat Dis Int. 12:305–309. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fornari F, Milazzo M, Chieco P, Negrini M,
Calin GA, Grazi GL, Pollutri D, Croce CM, Bolondi L and Gramantieri
L: MiR-199a-3p regulates mTOR and c-Met to influence the
doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res.
70:5184–5193. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Henry JC, Park JK, Jiang J, Kim JH,
Nagorney DM, Roberts LR, Banerjee S and Schmittgen TD: miR-199a-3p
targets CD44 and reduces proliferation of CD44 positive
hepatocellular carcinoma cell lines. Biochem Biophys Res Commun.
403:120–125. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jia XQ, Cheng HQ, Qian X, Bian CX, Shi ZM,
Zhang JP, Jiang BH and Feng ZQ: Lentivirus-mediated overexpression
of microRNA-199a inhibits cell proliferation of human
hepatocellular carcinoma. Cell Biochem Biophys. 62:237–244. 2012.
View Article : Google Scholar
|
|
33
|
Shen Q, Cicinnati VR, Zhang X, Iacob S,
Weber F, Sotiropoulos GC, Radtke A, Lu M, Paul A, Gerken G, et al:
Role of microRNA-199a-5p and discoidin domain receptor 1 in human
hepatocellular carcinoma invasion. Mol Cancer. 9:2272010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Song J, Gao L, Yang G, Tang S, Xie H, Wang
Y, Wang J, Zhang Y, Jin J, Gou Y, et al: MiR-199a regulates cell
proliferation and survival by targeting FZD7. PLoS One.
9:e1100742014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Otsuka M, Kishikawa T, Yoshikawa T, Ohno
M, Takata A, Shibata C and Koike K: The role of microRNAs in
hepato-carcinogenesis: Current knowledge and future prospects. J
Gastroenterol. 49:173–184. 2014. View Article : Google Scholar
|
|
36
|
Fan MQ, Huang CB, Gu Y, Xiao Y, Sheng JX
and Zhong L: Decrease expression of microRNA-20a promotes cancer
cell proliferation and predicts poor survival of hepatocellular
carcinoma. J Exp Clin Cancer Res. 32:212013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xiao F, Zhang W, Chen L, Chen F, Xie H,
Xing C, Yu X, Ding S, Chen K, Guo H, et al: MicroRNA-503 inhibits
the G1/S transition by downregulating cyclin D3 and E2F3 in
hepatocellular carcinoma. J Transl Med. 11:1952013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen J and Wang X: MicroRNA-21 in breast
cancer: Diagnostic and prognostic potential. Clin Transl Oncol.
16:225–233. 2014. View Article : Google Scholar
|
|
39
|
Huang Y, Yang YB, Zhang XH, Yu XL, Wang ZB
and Cheng XC: MicroRNA-21 gene and cancer. Med Oncol. 30:3762013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pan X, Wang ZX and Wang R: MicroRNA-21: A
novel therapeutic target in human cancer. Cancer Biol Ther.
10:1224–1232. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhou L, Yang ZX, Song WJ, Li QJ, Yang F,
Wang DS, Zhang N and Dou KF: MicroRNA-21 regulates the migration
and invasion of a stem-like population in hepatocellular carcinoma.
Int J Oncol. 43:661–669. 2013.PubMed/NCBI
|
|
42
|
Bao L, Yan Y, Xu C, Ji W, Shen S, Xu G,
Zeng Y, Sun B, Qian H, Chen L, et al: MicroRNA-21 suppresses PTEN
and hSulf-1 expression and promotes hepatocellular carcinoma
progression through AKT/ERK pathways. Cancer Lett. 337:226–236.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hu S, Tao R, Wang S, Wang C, Zhao X, Zhao
H, Li L, Zhu S, He Y, Jiang X, et al: MicroRNA-21 promotes cell
proliferation in human hepatocellular carcinoma partly by targeting
HEPN1. Tumour Biol. Feb 17–2015.Epub ahead of print. View Article : Google Scholar
|
|
44
|
Xu G, Zhang Y, Wei J, Jia W, Ge Z, Zhang Z
and Liu X: MicroRNA-21 promotes hepatocellular carcinoma HepG2 cell
proliferation through repression of mitogen-activated protein
kinase-kinase 3. BMC Cancer. 13:4692013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gramantieri L, Fornari F, Ferracin M,
Veronese A, Sabbioni S, Calin GA, Grazi GL, Croce CM, Bolondi L and
Negrini M: MicroRNA-221 targets Bmf in hepatocellular carcinoma and
correlates with tumor multifocality. Clin Cancer Res. 15:5073–5081.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ma D, Tao X, Gao F, Fan C and Wu D:
miR-224 functions as an onco-miRNA in hepatocellular carcinoma
cells by activating AKT signaling. Oncol Lett. 4:483–488. 2012.
|
|
47
|
Li Q, Ding C, Chen C, Zhang Z, Xiao H, Xie
F, Lei L, Chen Y, Mao B, Jiang M, et al: miR-224 promotion of cell
migration and invasion by targeting Homeobox D 10 gene in human
hepatocel-lular carcinoma. J Gastroenterol Hepatol. 29:835–842.
2014. View Article : Google Scholar
|
|
48
|
Zhou J, Lu S, Yang S, Chen H, Shi H, Miao
M and Jiao B: MicroRNA-127 post-transcriptionally downregulates
Sept7 and suppresses cell growth in hepatocellular carcinoma cells.
Cell Physiol Biochem. 33:1537–1546. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Callegari E, Elamin BK, Sabbioni S,
Gramantieri L and Negrini M: Role of microRNAs in hepatocellular
carcinoma: A clinical perspective. Onco Targets Ther. 6:1167–1178.
2013.PubMed/NCBI
|
|
50
|
Anwar SL and Lehmann U: DNA methylation,
microRNAs, and their crosstalk as potential biomarkers in
hepatocellular carcinoma. World J Gastroenterol. 20:7894–7913.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhou L, He J and Zhang Y: MicroRNA-22
expression in hepatocellular carcinoma and its correlation with
ezrin protein. J Int Med Res. 41:1009–1016. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bae HJ, Noh JH, Kim JK, Eun JW, Jung KH,
Kim MG, Chang YG, Shen Q, Kim SJ, Park WS, et al: MicroRNA-29c
functions as a tumor suppressor by direct targeting oncogenic SIRT1
in hepato-cellular carcinoma. Oncogene. 33:2557–2567. 2014.
View Article : Google Scholar
|
|
53
|
Xie K, Liu J, Chen J, Dong J, Ma H, Liu Y
and Hu Z: Methylation-associated silencing of microRNA-34b in
hepatocellular carcinoma cancer. Gene. 543:101–107. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Shen Q, Bae HJ, Eun JW, Kim HS, Park SJ,
Shin WC, Lee EK, Park S, Park WS, Lee JY, et al: MiR-101 functions
as a tumor suppressor by directly targeting nemo-like kinase in
liver cancer. Cancer Lett. 344:204–211. 2014. View Article : Google Scholar
|
|
55
|
Lu Y, Yue X, Cui Y, Zhang J and Wang K:
MicroRNA-124 suppresses growth of human hepatocellular carcinoma by
targeting STAT3. Biochem Biophys Res Commun. 441:873–879. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tsang FH, Au V, Lu WJ, Shek FH, Liu AM,
Luk JM, Fan ST, Poon RT and Lee NP: Prognostic marker microRNA-125b
inhibits tumorigenic properties of hepatocellular carcinoma cells
via suppressing tumorigenic molecule eIF5A2. Dig Dis Sci.
59:2477–2487. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Liu LL, Lu SX, Li M, Li LZ, Fu J, Hu W,
Yang YZ, Luo RZ, Zhang CZ and Yun JP: FoxD3-regulated microRNA-137
suppresses tumour growth and metastasis in human hepatocellular
carcinoma by targeting AKT2. Oncotarget. 5:5113–5124. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Long XR, He Y, Huang C and Li J:
MicroRNA-148a is silenced by hypermethylation and interacts with
DNA methyltransferase 1 in hepatocellular carcinogenesis. Int J
Oncol. 44:1915–1922. 2014.PubMed/NCBI
|
|
59
|
Zhang Z, Zheng W and Hai J: MicroRNA-148b
expression is decreased in hepatocellular carcinoma and associated
with prognosis. Med Oncol. 31:9842014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhi Q, Zhu J, Guo X, He S, Xue X, Zhou J,
Hu B, Li H, Chen S, Zhao H, et al: MMetastasis-related miR-185 is a
potential prognostic biomarker for hepatocellular carcinoma in
early stage. Biomed Pharmacother. 67:393–398. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Qadir XV, Han C, Lu D, Zhang J and Wu T:
miR-185 inhibits hepatocellular carcinoma growth by targeting the
DNMT1/PTEN/Akt pathway. Am J Pathol. 184:2355–2364. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Xiao F, Zhang W, Zhou L, Xie H, Xing C,
Ding S, Chen K and Zheng S: microRNA-200a is an independent
prognostic factor of hepatocellular carcinoma and induces cell
cycle arrest by targeting CDK6. Oncol Rep. 30:2203–2210.
2013.PubMed/NCBI
|
|
63
|
Zhang Y, Zheng D, Xiong Y, Xue C, Chen G,
Yan B and Ye Q: miR-202 suppresses cell proliferation in human
hepatocellular carcinoma by downregulating LRP6
post-transcriptionally. FEBS Lett. 588:1913–1920. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang J, Li J, Wang X, Zheng C and Ma W:
Downregulation of microRNA-214 and overexpression of FGFR-1
contribute to hepatocellular carcinoma metastasis. Biochem Biophys
Res Commun. 439:47–53. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Yu L, Ding GF, He C, Sun L, Jiang Y and
Zhu L: MicroRNA-424 is down-regulated in hepatocellular carcinoma
and suppresses cell migration and invasion through c-Myb. PLoS One.
9:e916612014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Miao HL, Lei CJ, Qiu ZD, Liu ZK, Li R, Bao
ST and Li MY: MicroRNA-520c-3p inhibits hepatocellular carcinoma
cell proliferation and invasion through induction of cell apoptosis
by targeting glypican-3. Hepatol Res. 44:338–348. 2014. View Article : Google Scholar
|
|
67
|
Sun Z, Han Q, Zhou N, Wang S, Lu S, Bai C
and Zhao RC: MicroRNA-9 enhances migration and invasion through
KLF17 in hepatocellular carcinoma. Mol Oncol. 7:884–894. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Murakami Y, Tamori A, Itami S, Tanahashi
T, Toyoda H, Tanaka M, Wu W, Brojigin N, Kaneoka Y, Maeda A, et al:
The expression level of miR-18b in hepatocellular carcinoma is
associated with the grade of malignancy and prognosis. BMC Cancer.
13:992013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Su ZX, Zhao J, Rong ZH, Geng WM, Wu YG and
Qin CK: Upregulation of microRNA-25 associates with prognosis in
hepatocellular carcinoma. Diagn Pathol. 9:472014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yang H, Zheng W, Zhao W, Guan C and An J:
Roles of miR-590-5p and miR-590-3p in the development of
hepatocellular carcinoma. Nan Fang Yi Ke Da Xue Xue Bao.
33:804–811. 2013.In Chinese. PubMed/NCBI
|
|
71
|
Jiang L, Cheng Q, Zhang BH and Zhang MZ:
Circulating microRNAs as biomarkers in hepatocellular carcinoma
screening: A validation set from China. Medicine. 94:e6032015.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wen Y, Han J, Chen J, Dong J, Xia Y, Liu
J, Jiang Y, Dai J, Lu J, Jin G, et al: Plasma miRNAs as early
biomarkers for detecting hepatocellular carcinoma. Int J Cancer.
37:1679–1690. 2015. View Article : Google Scholar
|
|
73
|
Zhang X, Hu S, Zhang X, Wang L, Zhang X,
Yan B, Zhao J, Yang A and Zhang R: MicroRNA-7 arrests cell cycle in
G1 phase by directly targeting CCNE1 in human hepatocellular
carcinoma cells. Biochem Biophys Res Commun. 443:1078–1084. 2014.
View Article : Google Scholar
|
|
74
|
Wang N, Zhu M, Tsao SW, Man K, Zhang Z and
Feng Y: MiR-23a-mediated inhibition of topoisomerase 1 expression
potentiates cell response to etoposide in human hepatocellular
carcinoma. Mol Cancer. 12:1192013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Chai ZT, Kong J, Zhu XD, Zhang YY, Lu L,
Zhou JM, Wang LR, Zhang KZ, Zhang QB, Ao JY, et al: MicroRNA-26a
inhibits angiogenesis by down-regulating VEGFA through the
PIK3C2α/Akt/HIF-1α pathway in hepatocellular carcinoma. PLoS One.
8:e779572013. View Article : Google Scholar
|
|
76
|
Shen G, Lin Y, Yang X, Zhang J, Xu Z and
Jia H: MicroRNA-26b inhibits epithelial-mesenchymal transition in
hepatocellular carcinoma by targeting USP9X. BMC Cancer.
14:3932014. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zhao N, Wang R, Zhou L, Zhu Y, Gong J and
Zhuang SM: MicroRNA-26b suppresses the NF-κB signaling and enhances
the chemosensitivity of hepatocellular carcinoma cells by targeting
TAK1 and TAB3. Mol Cancer. 13:352014. View Article : Google Scholar
|
|
78
|
Chen Z, Ma T, Huang C, Zhang L, Lv X, Xu
T, Hu T and Li J: MiR-27a modulates the MDR1/P-glycoprotein
expression by inhibiting FZD7/β-catenin pathway in hepatocellular
carcinoma cells. Cell Signal. 25:2693–2701. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Yang F, Li QJ, Gong ZB, Zhou L, You N,
Wang S, Li XL, Li JJ, An JZ, Wang DS, et al: MicroRNA-34a targets
Bcl-2 and sensitizes human hepatocellular carcinoma cells to
sorafenib treatment. Technol Cancer Res Treat. 13:77–86. 2014.
|
|
80
|
Dang Y, Luo D, Rong M and Chen G:
Underexpression of miR-34a in hepatocellular carcinoma and its
contribution towards enhancement of proliferating inhibitory
effects of agents targeting c-MET. PLoS One. 8:e610542013.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhang J, Jin H, Liu H, Lv S, Wang B, Wang
R, Liu H, Ding M, Yang Y, Li L, et al: MiRNA-99a directly regulates
AGO2 through translational repression in hepatocellular carcinoma.
Oncogenesis. 3:e972014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Chen P, Zhao X and Ma L: Downregulation of
microRNA-100 correlates with tumor progression and poor prognosis
in hepatocellular carcinoma. Mol Cell Biochem. 383:49–58. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zha R, Guo W, Zhang Z, Qiu Z, Wang Q, Ding
J, Huang S, Chen T, Gu J, Yao M, et al: Genome-wide screening
identified that miR-134 acts as a metastasis suppressor by
targeting integrin β1 in hepatocellular carcinoma. PLoS One.
9:e876652014. View Article : Google Scholar
|
|
84
|
Duan X, Hu J, Wang Y, Gao J, Peng D and
Xia L: MicroRNA-145: A promising biomarker for hepatocellular
carcinoma (HCC). Gene. 541:67–68. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Liu Y, Wu C, Wang Y, Wen S, Wang J, Chen
Z, He Q and Feng D: MicroRNA-145 inhibits cell proliferation by
directly targeting ADAM17 in hepatocellular carcinoma. Oncol Rep.
32:1923–1930. 2014.PubMed/NCBI
|
|
86
|
Wang Y, Hu C, Cheng J, Chen B, Ke Q, Lv Z,
Wu J and Zhou Y: MicroRNA-145 suppresses hepatocellular carcinoma
by targeting IRS1 and its downstream Akt signaling. Biochem Biophys
Res Commun. 446:1255–1260. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Noh JH, Chang YG, Kim MG, Jung KH, Kim JK,
Bae HJ, Eun JW, Shen Q, Kim SJ, Kwon SH, et al: MiR-145 functions
as a tumor suppressor by directly targeting histone deacetylase 2
in liver cancer. Cancer Lett. 335:455–462. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Amer M, Elhefnawi M, El-Ahwany E, Awad AF,
Gawad NA, Zada S and Tawab FM: Hsa-miR-195 targets PCMT1 in
hepa-tocellular carcinoma that increases tumor life span. Tumour
Biol. 35:11301–11309. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Yang Y, Li M, Chang S, Wang L, Song T, Gao
L, Hu L, Li Z, Liu L, Yao J, et al: MicroRNA-195 acts as a tumor
suppressor by directly targeting Wnt3a in HepG2 hepatocellular
carcinoma cells. Mol Med Rep. 10:2643–2648. 2014.PubMed/NCBI
|
|
90
|
Yang T, Zheng ZM, Li XN, Li ZF, Wang Y,
Geng YF, Bai L and Zhang XB: MiR-223 modulates multidrug resistance
via down-regulation of ABCB1 in hepatocellular carcinoma cells. Exp
Biol Med. 238:1024–1032. 2013. View Article : Google Scholar
|
|
91
|
Yao J, Liang LH, Zhang Y, Ding J, Tian Q,
Li JJ and He XH: GNAI1 suppresses tumor cell migration and invasion
and is post-transcriptionally regulated by mir-320a/c/d in
hepatocellular carcinoma. Cancer Biol Med. 9:234–241. 2012.
|
|
92
|
Zhou P, Huang G, Zhao Y, Zhong D, Xu Z,
Zeng Y, Zhang Y, Li S and He F: MicroRNA-363-mediated
downregulation of S1PR1 suppresses the proliferation of
hepatocellular carcinoma cells. Cell Signal. 26:1347–1354. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Vaira V, Roncalli M, Carnaghi C, Faversani
A, Maggioni M, Augello C, Rimassa L, Pressiani T, Spagnuolo G, Di
Tommaso L, et al: MicroRNA-425-3p predicts response to sorafenib
therapy in patients with hepatocellular carcinoma. Liver Int.
35:1077–1086. 2015. View Article : Google Scholar
|
|
94
|
Zhang H, Feng Z, Huang R, Xia Z, Xiang G
and Zhang J: MicroRNA-449 suppresses proliferation of hepatoma cell
lines through blockade lipid metabolic pathway related to SIRT1.
Int J Oncol. 45:2143–2152. 2014.PubMed/NCBI
|
|
95
|
Zhou Y, Li Y, Ye J, Jiang R, Yan H, Yang
X, Liu Q and Zhang J: MicroRNA-491 is involved in metastasis of
hepatocellular carcinoma by inhibitions of matrix metalloproteinase
and epithelial to mesenchymal transition. Liver Int. 33:1271–1280.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Tang J, Tao ZH, Wen D, Wan JL, Liu DL,
Zhang S, Cui JF, Sun HC, Wang L, Zhou J, et al: MiR-612 suppresses
the stemness of liver cancer via Wnt/β-catenin signaling. Biochem
Biophys Res Commun. 447:210–215. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Lin F, Ding R, Zheng S, Xing D, Hong W,
Zhou Z and Shen J: Decrease expression of microRNA-744 promotes
cell proliferation by targeting c-Myc in human hepatocellular
carcinoma. Cancer Cell Int. 14:582014. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Zhu K, Pan Q, Zhang X, Kong LQ, Fan J, Dai
Z, Wang L, Yang XR, Hu J, Wan JL, et al: MiR-146a enhances
angiogenic activity of endothelial cells in hepatocellular
carcinoma by promoting PDGFRA expression. Carcinogenesis.
34:2071–2079. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Qin J, Luo M, Qian H and Chen W:
Upregulated miR-182 increases drug resistance in cisplatin-treated
HCC cell by regulating TP53INP1. Gene. 538:342–347. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Gao B, Gao K, Li L, Huang Z and Lin L:
miR-184 functions as an oncogenic regulator in hepatocellular
carcinoma (HCC). Biomed Pharmacother. 68:143–148. 2014. View Article : Google Scholar
|
|
101
|
Wu GG, Li WH, He WG, Jiang N, Zhang GX,
Chen W, Yang HF, Liu QL, Huang YN, Zhang L, et al: Mir-184
post-transcriptionally regulates SOX7 expression and promotes cell
proliferation in human hepatocellular carcinoma. PLoS One.
9:e887962014. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Hung TM, Ho CM, Liu YC, Lee JL, Liao YR,
Wu YM, Ho MC, Chen CH, Lai HS and Lee PH: Up-regulation of
microRNA-190b plays a role for decreased IGF-1 that induces insulin
resistance in human hepatocellular carcinoma. PLoS One.
9:e894462014. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Ren J, Chu Y, Ma H, Zhang Y, Zhang X, Zhao
D, Li Z, Wang J, Gao YE, Xiao L, et al: Epigenetic interventions
increase the radiation sensitivity of cancer cells. Curr Pharm Des.
20:1857–1865. 2014. View Article : Google Scholar
|
|
104
|
Zhou S, Ye W, Ren J, Shao Q, Qi Y, Liang J
and Zhang M: MicroRNA-381 increases radiosensitivity in esophageal
squamous cell carcinoma. Am J Cancer Res. 5:267–277.
2015.PubMed/NCBI
|
|
105
|
Ye C, Sun NX, Ma Y, Zhao Q, Zhang Q, Xu C,
Wang SB, Sun SH, Wang F and Li W: MicroRNA-145 contributes to
enhancing radiosensitivity of cervical cancer cells. FEBS Lett.
589:702–709. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Li T, Yin J, Yuan L, Wang S, Yang L, Du X
and Lu J: Downregulation of microRNA-139 is associated with
hepato-cellular carcinoma risk and short-term survival. Oncol Rep.
31:1699–1706. 2014.PubMed/NCBI
|
|
107
|
Gu W, Li X and Wang J: miR-139 regulates
the proliferation and invasion of hepatocellular carcinoma through
the WNT/TCF-4 pathway. Oncol Rep. 31:397–404. 2014.
|
|
108
|
Liang X, Zeng J, Wang L, Fang M, Wang Q,
Zhao M, Xu X, Liu Z, Li W, Liu S, et al: Histone demethylase
retinoblastoma binding protein 2 is overexpressed in hepatocellular
carcinoma and negatively regulated by hsa-miR-212. PLoS One.
8:e697842013. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Ma Y, She XG, Ming YZ and Wan QQ: miR-24
promotes the proliferation and invasion of HCC cells by targeting
SOX7. Tumour Biol. 35:10731–10736. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Dai W, Wang C, Wang F, Wang Y, Shen M,
Chen K, Cheng P, Zhang Y, Yang J, Zhu R, et al: Anti-miR-197
inhibits migration in HCC cells by targeting KAI 1/CD82. Biochem
Biophys Res Commun. 446:541–548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Yang YF, Wang F, Xiao JJ, Song Y, Zhao YY,
Cao Y, Bei YH and Yang CQ: MiR-222 overexpression promotes
proliferation of human hepatocellular carcinoma HepG2 cells by
downregulating p27. Int J Clin Exp Med. 7:893–902. 2014.PubMed/NCBI
|
|
112
|
Pang F, Zha R, Zhao Y, Wang Q, Chen D,
Zhang Z, Chen T, Yao M, Gu J and He X: MiR-525-3p enhances the
migration and invasion of liver cancer cells by downregulating
ZNF395. PLoS One. 9:e908672014. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Sun Z, Meng C, Wang S, Zhou N, Guan M, Bai
C, Lu S, Han Q and Zhao RC: MicroRNA-1246 enhances migration and
invasion through CADM1 in hepatocellular carcinoma. BMC Cancer.
14:6162014. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Köberle V, Kronenberger B, Pleli T, Trojan
J, Imelmann E, Peveling-Oberhag J, Welker MW, Elhendawy M, Zeuzem
S, Piiper A, et al: Serum microRNA-1 and microRNA-122 are
prognostic markers in patients with hepatocellular carcinoma. Eur J
Cancer. 49:3442–3449. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Ge W, Yu DC, Li QG, Chen X, Zhang CY and
Ding YT: Expression of serum miR-16, let-7f, and miR-21 in patients
with hepatocellular carcinoma and their clinical significances.
Clin Lab. 60:427–434. 2014.PubMed/NCBI
|
|
116
|
Meng FL, Wang W and Jia WD: Diagnostic and
prognostic significance of serum miR-24-3p in HBV-related
hepatocellular carcinoma. Med Oncol. 31:1772014. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Luo J, Chen M, Huang H, Yuan T, Zhang M,
Zhang K and Deng S: Circulating microRNA-122a as a diagnostic
marker for hepatocellular carcinoma. Onco Targets Ther. 6:577–583.
2013.PubMed/NCBI
|
|
118
|
Zhang ZQ, Meng H, Wang N, Liang LN, Liu
LN, Lu SM and Luan Y: Serum microRNA 143 and microRNA 215 as
potential biomarkers for the diagnosis of chronic hepatitis and
hepatocel-lular carcinoma. Diagn Pathol. 9:1352014. View Article : Google Scholar
|
|
119
|
Li J, Wang Y, Yu W, Chen J and Luo J:
Expression of serum miR-221 in human hepatocellular carcinoma and
its prognostic significance. Biochem Biophys Res Commun. 406:70–73.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Zhan MX, Li Y, Hu BS, Shao PJ, Meng QW, He
X, Huang JW and Lu LG: Expression of serum microRNAs (miR-222,
miR-181, miR-216) in human hepatocellular carcinoma and its
clinical significance. Zhonghua Yi Xue Za Zhi. 93:1830–1832.
2013.In Chinese. PubMed/NCBI
|