Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignant tumors with increasing morbidity and mortality all
over the world (1). Although
current therapeutic strategies have improved over the past decades,
the treatment outcome and survival rate remain poor because of
frequent recurrence and high invasion and metastasis capacity
(2–4). One of the obstacles that hampers the
development of efficient therapies is the lack of understanding of
the precise molecular mechanism underlying HCC development and
progression. Thus, a better understanding of the pathological
mechanism is important to provide theoretical basis for developing
potential and promising therapeutics for HCC.
In recent years, a class of small and non-coding
RNAs called microRNAs (miRs) have emerged as prospective tools for
cancer therapy, including HCC therapy (5). miRs are negative regulators of gene
expression that regulate protein translation by targeting the
3′-untranslated region (UTR) of mRNA (6,7).
Studies have indicated that miRs act as potential and useful
biomarkers and targets for disease diagnosis, prognosis and
treatment (8,9). Other studies have also reported that
miRs such as miR-302b (10),
miR-1285-3p (11), mR-520g
(12) and miR-139 (13) serve as oncogenes or tumor
suppressors in HCC. miR-381 is extensively associated with various
cancer types (14,15), but its role in HCC has not been
comprehensively investigated.
The liver receptor homolog-1 (LRH-1) has been
reported as an important regulator in many different cancer types
(16). Various studies have
uncovered that LRH-1 is involved in regulating embryonic
development, ovulation, pregnancy and metabolism (17–22).
LRH-1 deficiency blunts intestinal tumorigenesis (23). LRH-1 is elucidated as an oncogene in
pancreatic cancer (24), colon
cancer (25), gastric cancer
(26), breast cancer (27) and ovarian cancer (28). Moreover, LRH-1 is associated with
the activation of Wnt signaling pathways (29), which were frequently aberrantly
activated in cancers, such as HCC (30). These findings indicate that LRH-1
can serve as a potential molecular target for cancer therapy.
Several studies have also attempted to develop an effective
antagonist for LRH-1 to treat cancers (31).
The suppression of LRH-1 in HCC has been
demonstrated to induce cell cycle arrest and apoptosis (32), thus indicating its important role in
HCC. However, the precise role of LRH-1 in HCC remains largely
unknown. In the present report, we demonstrate a targeted
relationship between LRH-1 and miR-381. We found that miR-381 was
significantly downregulated in HCC cells and tissues. miR-381
overexpression inhibited HCC cell growth and invasion ability,
whereas miR-381 suppression displayed an opposite effect.
Bioinformatics analysis and dual-luciferase reporter assay results
showed that miR-381 directly targeted the 3′-UTR of LRH-1, and
quantitative polymerase chain reaction (qPCR) and western blot
analysis results showed that miR-381 negatively modulated LRH-1
expression. Data elucidated that miR-381 regulated HCC cell growth
and invasion through LRH-1-mediated Wnt signaling. Clinical tissue
detection data revealed that an inverse correlation existed between
miR-381 and LRH-1 expression, thus further indicating the
functional significance of miR-381-LRH-1 in regulating HCC
tumorigenesis. The present study provides evidence that the use of
miR-381 to target LRH-1 may be a potential therapeutic strategy for
HCC treatment.
Materials and methods
Tissue specimens
A total of 20 paired malignant HCC and adjacent
non-tumorous liver tissues were obtained from HCC patients
undergoing surgical resection in the First Hospital of Jilin
University. These patients were first diagnosed without local or
systemic treatment before surgical resection. The tissue samples
were removed and immediately snap-frozen in liquid nitrogen for
further use. Informed consent was obtained from each patient, and
the experimental protocol was reviewed and approved by the
Institutional Human Experiment and Ethic Committee of the First
Hospital of Jilin University.
Cell cultures
Human HCC cell lines including HepG2, SMMC-7721,
HuH7, BEL-7402, human normal liver cell line HL-7702 and human
embryonic kidney (HEK) 293T cell line were purchased from the
American Type Culture Collection (ATCC; Manassas, VA, USA) and were
grown in Dulbecco's modified Eagle's medium (DMEM) in a supplement
with 10% heat-inactivated fetal bovine serum (FBS) supplemented
with 1% penicillin-streptomycin (all from Gibco, Grand Island, NY,
USA). The cells were cultured in a humidified incubator containing
5% CO2 atmosphere at 37°C.
RNA isolation and quantitative polymerase
chain reaction (qPCR)
Total RNA was extracted by using the miRNeasy Mini
kit (Qiagen, Dusseldorf, Germany) according to the manufacturer's
recommended method. For mRNA expression analysis, the first-strand
cDNA was synthesised by using Moloney murine leukaemia virus
(M-MLV) reverse transcriptase (Invitrogen, Carlsbad, CA, USA). The
cDNA was then used as templates to amplify LRH-1, cyclin D1, cyclin
E1 and MMP9 with specific primers by using SYBR Green Master Mix
(Bio-Rad, Hercules, CA, USA). Glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) was used as the internal normalisation
control. For miRNA expression analysis, the cDNA was obtained using
miScript reverse transcription kit and amplified using miScript
SYBR Green PCR kit (both from Qiagen) according to the
manufacturer's instructions. U6 small nuclear RNA (U6) was used as
internal normalization control. The specific primers used in this
study were as follows: LRH-1 (forward, 5′-CTGATAGTGGAACTTTTGAA-3′
and reverse, 5′-CTTCATTTGGTCATCAACCTT-3′); cyclin D1 (forward,
5′-GTTCGTGGCCTCTAAGATGAAG-3′ and reverse,
5′-GTGTTTGCGGATGATCTGTTTG-3′); cyclin E1 (forward,
5′-GCTTCGGCCTTGTATCATTTC-3′ and reverse,
5′-CTGTCTCTGTGGGTCTGTATG-3′); MMP9 (forward,
5′-GCCCGACCCGAGCTGACTC-3′ and reverse,
5′-TTCAGGGCGAGGACCATAGAGG-3′); GAPDH (forward,
5′-GAAATCCCATCACCATCTTCCAGG-3′ and reverse,
5′-GAGCCCCAGCCTTCTCCATG-3′); miR-381 (forward,
5′-TAATCTGACTATACAAGGGCAAGCT-3′ and reverse,
5′-TATGGTTGTTCTGCTCTCTGTCTC-3′); and U6 (forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′). Relative gene expression was
analyzed by using the 2−ΔΔCt method.
Transfection of HCC cell lines
miR-381 mimics (sense,
5′-UAUACAAGGGCAAGCUCUCUGUTT-3′ and antisense,
5′-ACAGAGAGCUUGCCCUUGUCGCTT-3′), scrambled mimics (sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′), miR-381 LNA oligonucleotide
(inhibitor; 5′-LNAACAGAGAGCUUGCCCUUGUAUA-3′) and scrambled LNA
(5′-LNACAGUACUUUUGUGUAGUACAA-3′) were purchased from Shanghai
GenePharma Co., Ltd. (Shanghai, China). Cells were plated onto
6-well plates and cultured until reaching 80% confluence. At that
time, cells were transiently transfected with miRNA mimics, miR-LNA
or their scrambled controls for a final concentration of 20 nM
using Lipofectamine 2000 (Invitrogen) according to the supplier's
recommended protocol. For LRH-1 overexpression, 4 µg of
pcDNA3.0/LRH-1 expressing vectors were transiently transfected into
HCC cells using Lipofectamine 2000 (Invitrogen) for 48 h. The
transfection efficiency was then detected by using western blot
analysis.
MTT assay
Cell growth was evaluated by using the MTT method.
The HCC cells were briefly seeded in 96-well plates at
1×104 cells/well and cultured overnight. The cells were
then transfected with miRNA-381 mimics, miR-381 LNA or their
scrambled controls and were incubated for 24, 48 and 72 h followed
by discarding the old medium and adding fresh medium containing MTT
(5 mg/ml in PBS; Sigma, St. Louis, MO, USA). After culturing for
another 4 h at 37°C, the formed formazan products were solubilised
by adding dimethyl sulfoxide (200 µl/well). The optical
density (OD) value was detected at a wavelength of 490 nm by using
a multi-well spectrophotometer (Bio-Tek Instruments, Winooski, VT,
USA).
Cell cycle analysis
Cell cycle distribution was detected by using flow
cytometry. HCC cells were briefly transfected with miRNA-381 mimics
or miR-381 LNA for 48 h. Cells were harvested by trypsinization,
washed with PBS and fixed with 70% ethanol overnight at 4°C.
Approximately 1×106 cells were stained with propidium
iodide (PI, 100 µg/ml; Sigma) solution containing 10
µg/ml of RNase A for 30 min in the dark. The percentage of
cells in each cell cycle phase was then measured by using a FACScan
flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) and
analyzed by using ModFit software.
Colony formation assay
HCC cells were cultured in a 6-well plate and
transfected with miRNA-381 mimics or miR-381 LNA for 48 h.
Thereafter, the transfected cells were re-plated into a 6-well
plate in a growth medium containing 0.3% noble agar at 200
cells/well to form natural colonies. After 2 weeks, cells were
washed with PBS, fixed with 4% paraformaldehyde and stained with
Giemsa (Sigma). The total number of colonies was counted under a
microscope.
Cell invasion assay
HCC cells were transfected with miRNA-381 mimics or
miR-381 LNA for 48 h and then starved overnight in serum-free
medium. Thereafter, the transfected cells (1×105 cells)
were seeded into the upper chamber of a 24-well Transwell (Corning
Incorporated, Toledo, NY, USA) containing 200 µl of
serum-free medium. Transwell membrane filter inserts were precoated
with Matrigel (BD Biosciences, San Jose, CA, USA). A medium
containing 10% FBS added in the lower chamber was used as a
chemoattractant. After incubation for 24 h, the non-invasive cells
in the upper chamber were gently removed by a cotton swab. The
invasive cells on the lower surface were then fixed, stained and
observed under the microscope. The mean number of invaded cells in
5 random fields per membrane was counted and averaged.
Luciferase reporter assay
The targeting relationship between miR-381 and LRH-1
3′-UTR was detected by using a dual-luciferase reporter assay. The
cDNA fragments from the 3′-UTR of LRH-1 containing miR-381 binding
site were briefly amplified and subcloned into pmirGLO vector
(Promega, Madison, WI, USA). HEK293T cells were seeded into 6-well
plates at 2×105 cells/well for 24 h and then
co-transfected with the 10 ng of pmirGLO-LRH-1 recombinant vectors
and 20 nM of miR-381 mimics or scrambled mimics by using
Lipofectamine 2000 (Invitrogen). Cells were lysed after 48 h of
transfection, and the luciferase activity was measured by using the
dual-luciferase reporter system (Promega). Wnt signaling activity
was determined by using Tcf luciferase assays. Cells were briefly
co-transfected with TOPFlash firefly luciferase reporter vector
(100 ng; Addgene, Cambridge, MA, USA) and phRL-TK Renilla
luciferase vectors (5 ng; Promega) in the presence of miRNA-381
mimics or miR-381 LNA for 48 h. Cells were lysed, and firefly and
Renilla luciferase activities were detected by using the
dual-luciferase reporter system (Promega). The data were expressed
as firefly/Renilla luciferase activity.
Western blot analysis
Cells were lysed by using RIPA lysis buffer, and
protein concentrations were measured by using a BCA kit (both from
Beyotime Biotechnology, Haimen, China). A total of 25 µg
proteins were separated by sodium dodecyl sulphate-polyacrylamide
gel electrophoresis and then electro-transferred onto a
nitrocellulose membrane (Bio-Rad). The membrane was then blocked by
3% fat-free milk and blotted with primary antibodies (rabbit
polyclonal anti-LRH-1, 1:500; rabbit polyclonal anti-GAPDH, 1:800;
Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) at 4°C
overnight. Thereafter, the membrane was incubated with goat
anti-rabbit horseradish peroxidase-conjugated secondary antibodies
(1:2,000; Santa Cruz Biotechnology Inc.) for 1 h at room
temperature. The signals of the protein bands were detected by
using the enhanced chemiluminescence method (Amersham, Little
Chalfont, UK). The signal intensity on the membrane was analysed by
Image-Pro Plus 6.0 (Media Cybernetics, Inc., Rockville, MD, USA)
and normalized to the control group.
Data analysis
All data were reported as means ± standard
deviation. Statistical analyses were performed with SPSS version
11.5 (SPSS Inc., Chicago, IL, USA) using Student's t-test or
one-way analysis of variance. Correlations were assessed by
Pearson's correlation test. A p-value of <0.05 was set as the
level of statistical difference.
Results
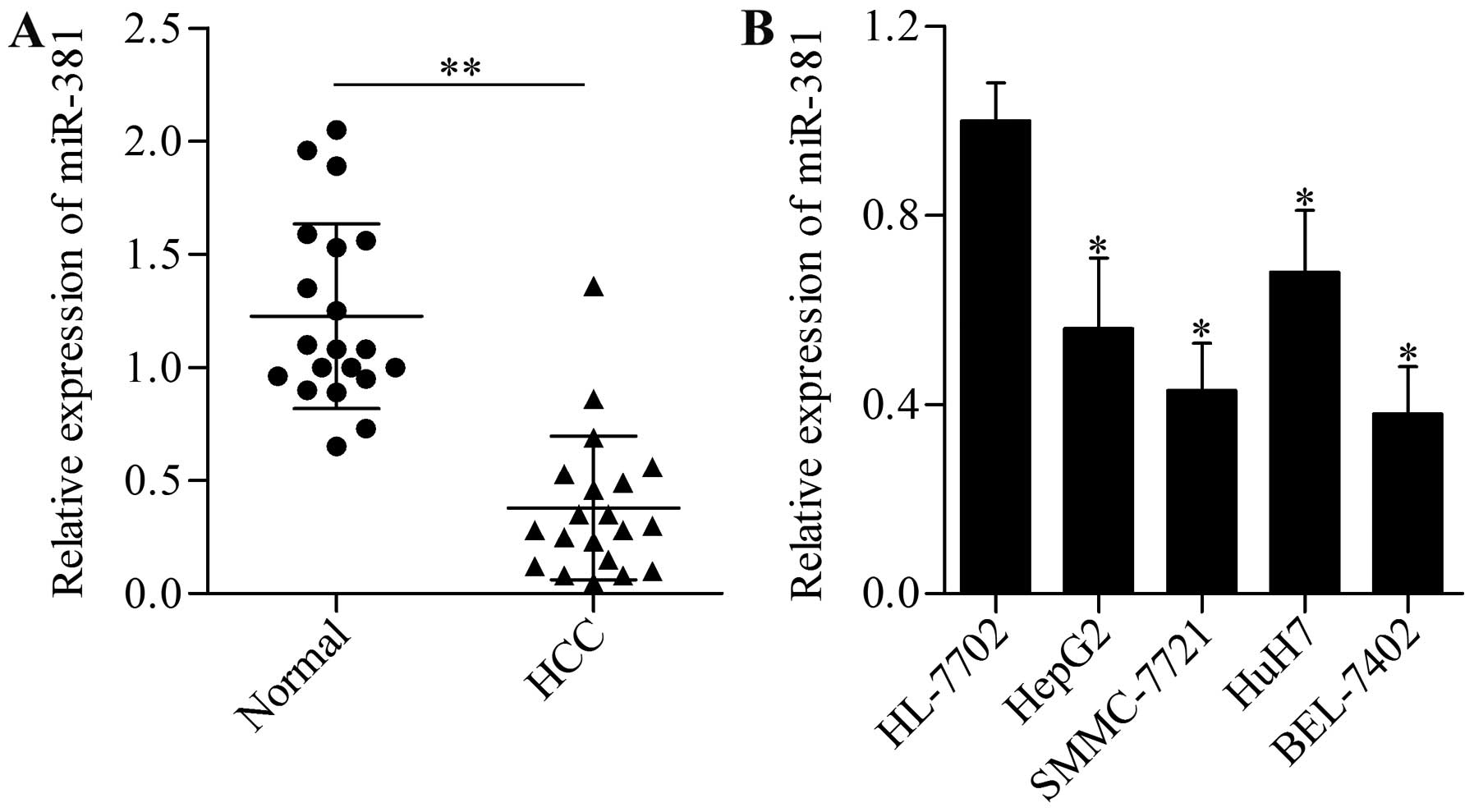
miR-381 downregulation frequently
occurred in HCC tissues and cell lines
To investigate the potential function of miR-381 in
HCC, we detected first the expression pattern of miR-381 in 20
paired malignant HCC and adjacent non-tumorous liver tissues by
using qPCR. The results showed that miR-381 was frequently and
significantly decreased in live HCC samples compared with their
peritumor counterparts (Fig. 1A).
Furthermore, we detected the expression levels of miR-381 in
several HCC cell lines (HepG2, SMMC-7721, HuH7 and BEL-7402) and
normal liver cells. This observation was consistent with the
results of miR-381 in HCC tissues wherein miR-381 expression was
prevalently downregulated compared with human normal liver HL-7702
cells (Fig. 1B). These results
indicated that miR-381 may play an important role in HCC
development and progression.
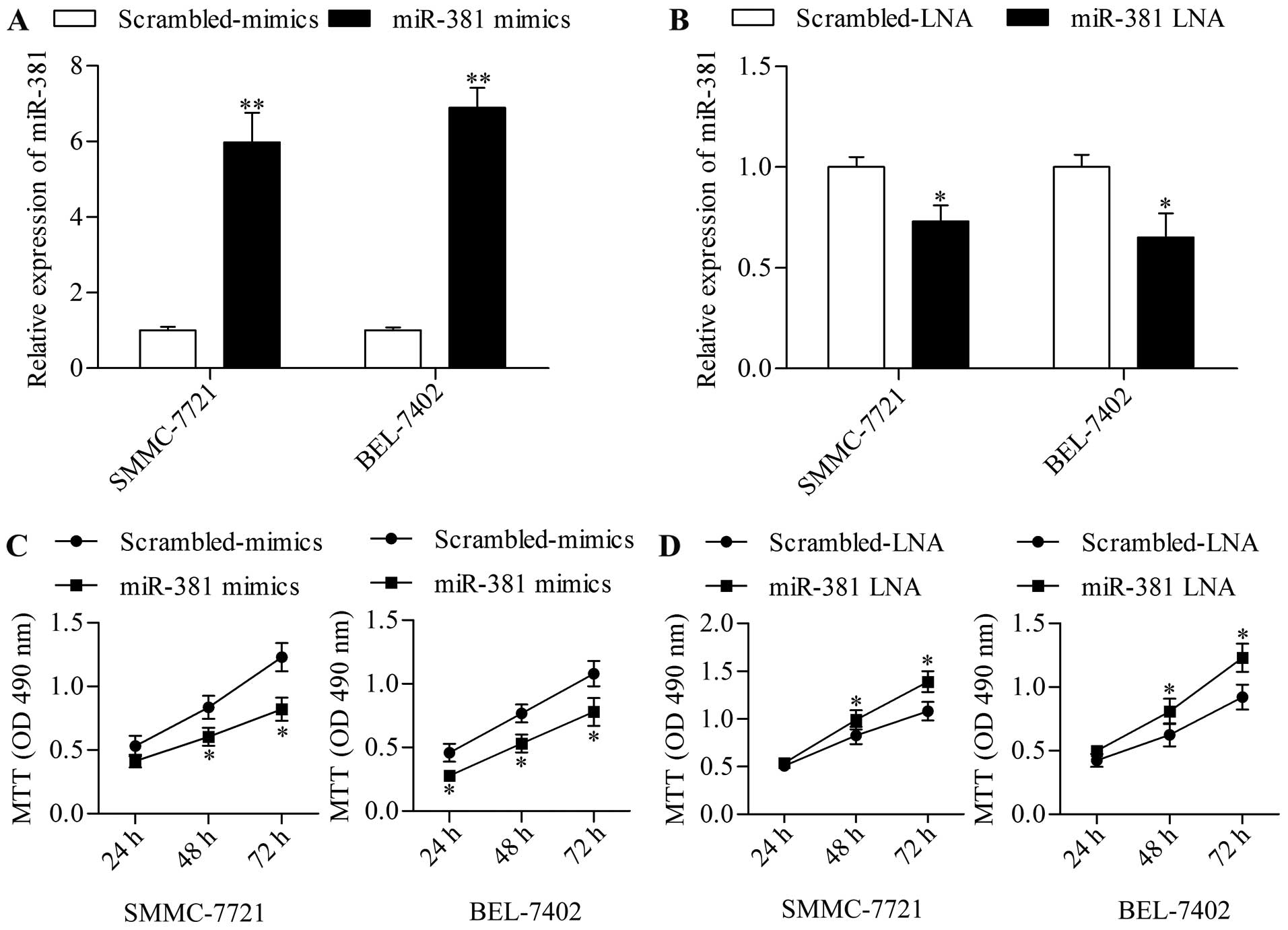
miR-381 inhibits HCC cell growth
To explore the precise function of miR-381 in HCC,
we next assessed the effect of miR-381 on HCC growth by using a
gain-of-function approach with miR-381 mimics. SMMC-7721 and
BEL-7402 cells were transfected with miR-381 mimics followed by
detection using MTT assay. The results showed that miR-381
overexpression (Fig. 2A)
significantly decreased HCC cell growth (Fig. 2C) in SMMC-7721 and BEL-7402 cells.
By contrast, miR-381 suppression (Fig.
2B) significantly promoted HCC cell growth (Fig. 2D). To further verify its effect on
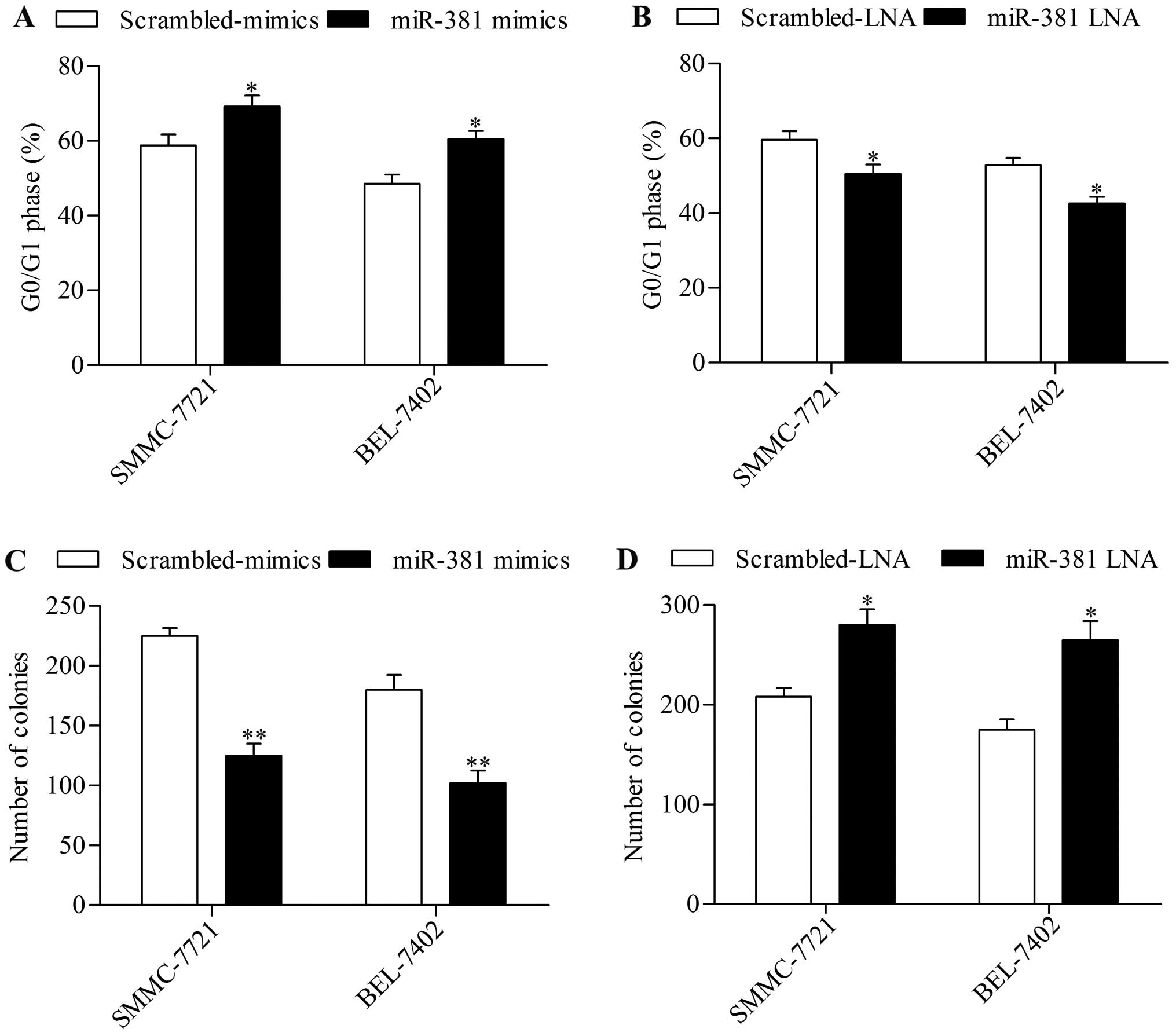
cell growth, we examined the effect of miR-381 overexpression on
cell cycle progression and found that miR-381 overexpression
induced an increase in cell cycle arrest in G0/G1 phase (Fig. 3A). Moreover, the colony-forming
capacity of HCC cells was remarkably suppressed by miR-381
overexpression (Fig. 3C). miR-381
suppression by transfection of miR-381 LNA conversely exhibited the
opposite effect on cell cycle (Fig.
3B) and colony-forming capacity (Fig. 3D). These results implied that
miR-381 functioned as a potential tumor suppressor in HCC.
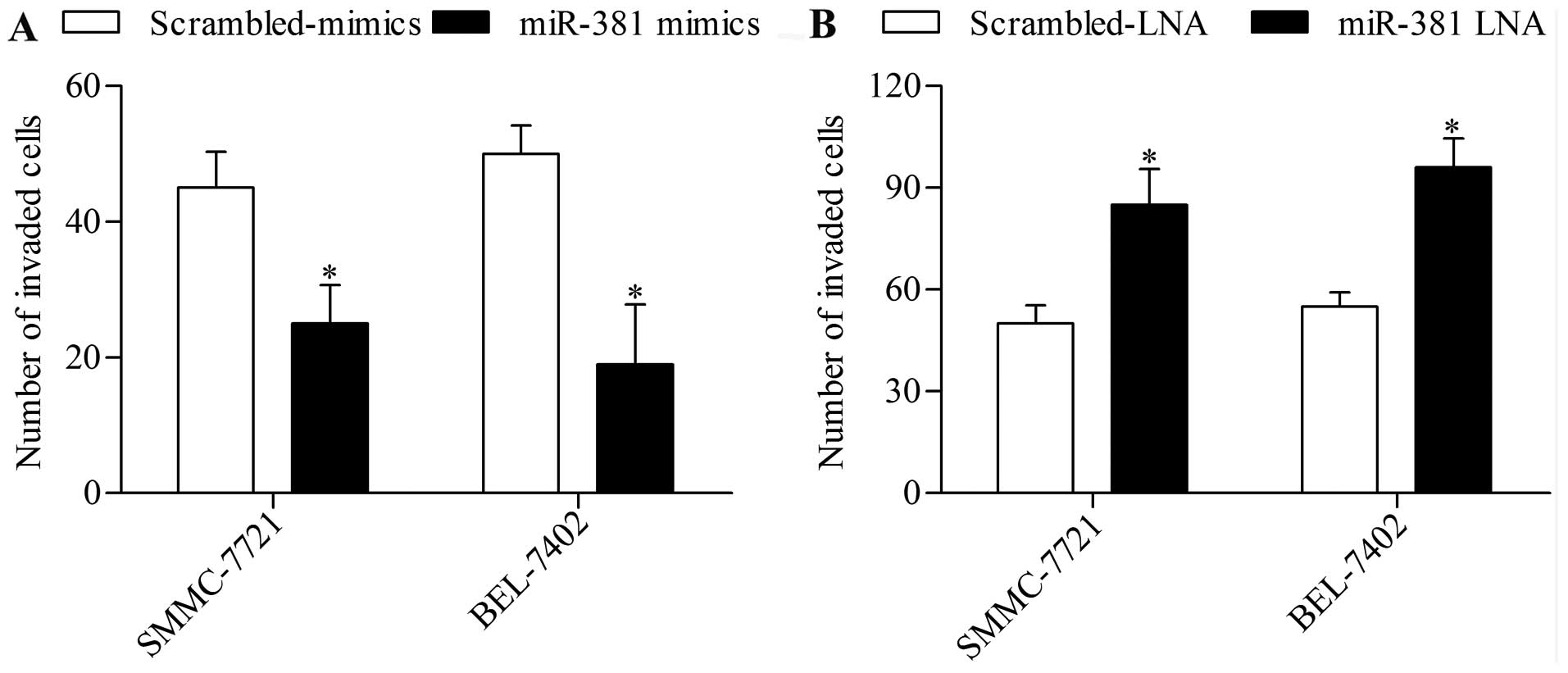
miR-381 suppresses HCC cell invasion in
vitro
To further verify the antitumor effect of miR-381 on
HCC, we next examined the effect of miR-381 overexpression or
suppression on HCC cell invasion by using Transwell invasion assay.
The results showed that miR-381 overexpression significantly
decreased the invasion of HCC cells (Fig. 4A), whereas miR-381 suppression
further promoted this ability (Fig.
4B).
miR-381 directly targets and mediates
LRH-1 expression
To uncover the molecular basis of miR-381 in
regulating HCC, we sought to screen the putative targets of miR-381
by using bioinformatics analysis. Notably, among those targets,
LRH-1 attracted our particular interests because of its important
role in various cancers (16). As
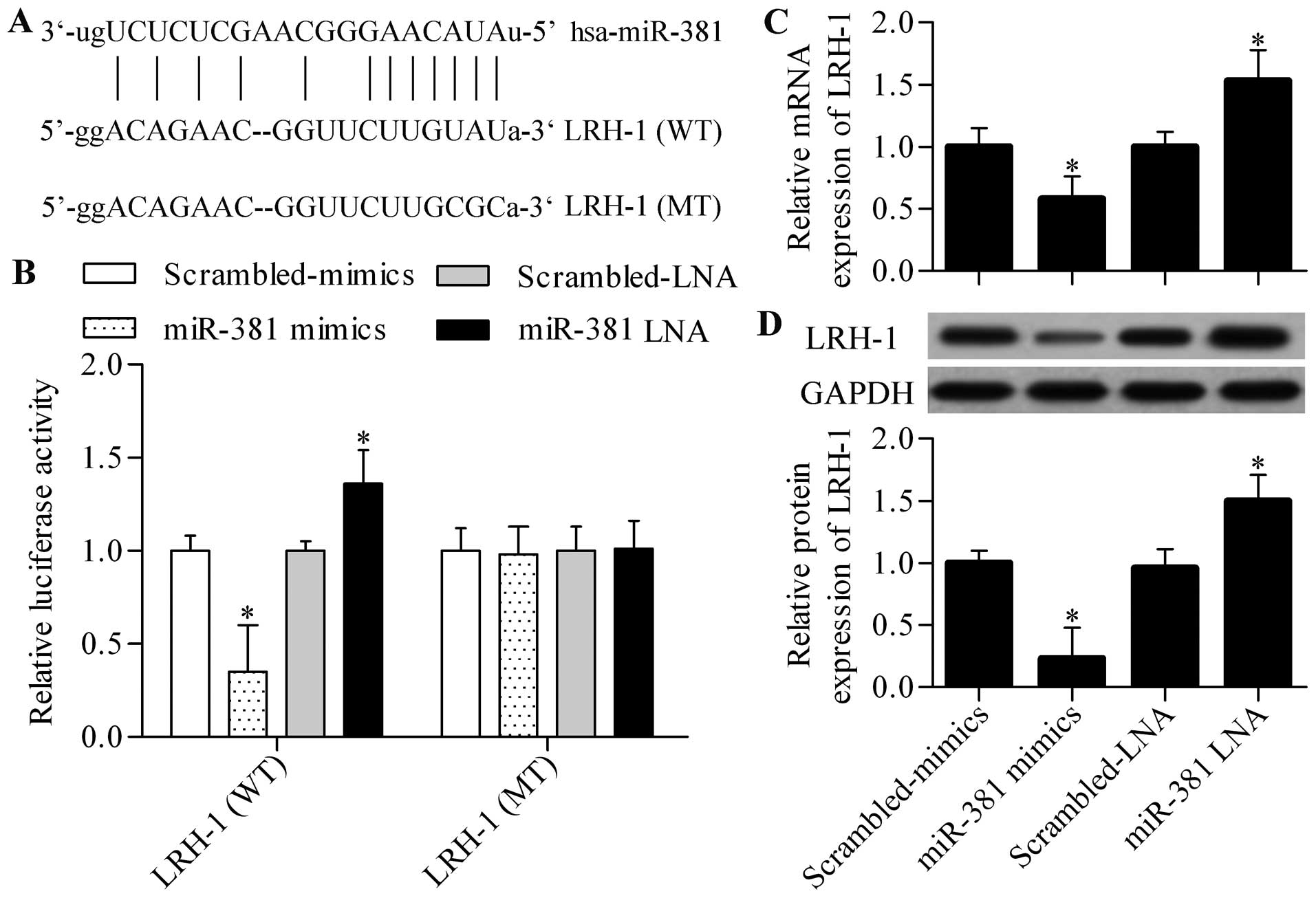
shown in Fig. 5A, the predicted
complementary site of miR-381 was found in the 3′-UTR of LRH-1. We
evaluated the dual-luciferase reporter activity by using pmirGLO
vector harbouring the wild-type (WT) or mutant (MT) 3′-UTR of LRH-1
to verify whether LRH-1 was the direct target gene of miR-381.
Luciferase assay displayed that miR-381 mimics significantly
decreased the luciferase activity of pmirGLO-WT LRH-1, whereas
miR-381 LNA markedly increased this activity (Fig. 5B). However, neither miR-381 mimics
nor miR-381 LNA showed any effects on the luciferase activity of
pmirGLO-MT LRH-1 (Fig. 5B). The
data indicated that miR-381 directly targeted the predicted
complementary site in the 3′-UTR of LRH-1. To further investigate
whether miR-381 could regulate endogenous LRH-1 expression, we
transfected miR-381 mimics or miR-381 LNA into SMMC-7721 cells and
detected LRH-1 expression by using qPCR and western blot analysis.
The results showed that both mRNA and protein expression of LRH-1
was significantly decreased by miR-381 overexpression (Fig. 5C and D). On the contrary, miR-381
suppression further increased LRH-1 expression in SMMC-7221 cells
(Fig. 5C and D). Consistent results
were obtained by using BEL-7402 cells (data not shown).
miR-381 functions as a tumor suppressor
to repress HCC cell growth and invasion through LRH-1
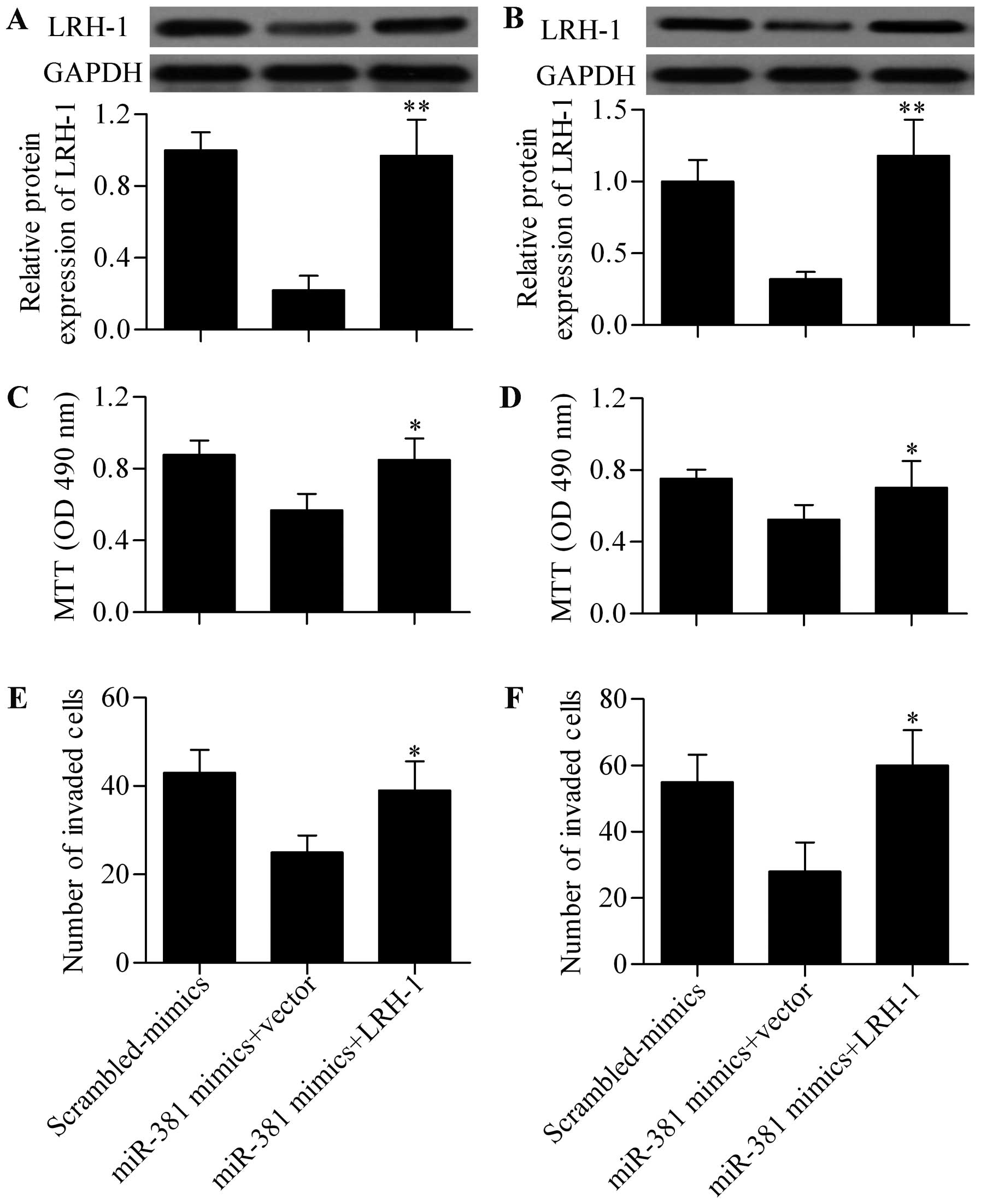
We performed a rescue experiment to validate miR-381
functions through LRH-1. HCC cells were co-transfected with miR-381
mimics and LRH-1 expressing vectors without 3′-UTR. Western blot
analysis showed that LRH-1 protein expression was significantly
restored by LRH-1 expressing vectors transfection even though with
miR-381 mimics transfection in SMMC-7721 (Fig. 6A) and BEL-7402 (Fig. 6B) cells. MTT assay showed that LRH-1
overexpression significantly reversed the inhibitory effect of
miR-381 on HCC cell growth in SMMC-7721 (Fig. 6C) and BEL-7402 (Fig. 6D) cells. The results further showed
that the decreased cell invasion ability induced by miR-381
overexpression was also remarkably reversed by LRH-1 overexpression
(Fig. 6E and F). These results
indicated that miR-381 exerted its antitumor effect by inhibiting
LRH-1.
miR-381 negatively regulates the Wnt
signaling pathway through LRH-1 in HCC cells
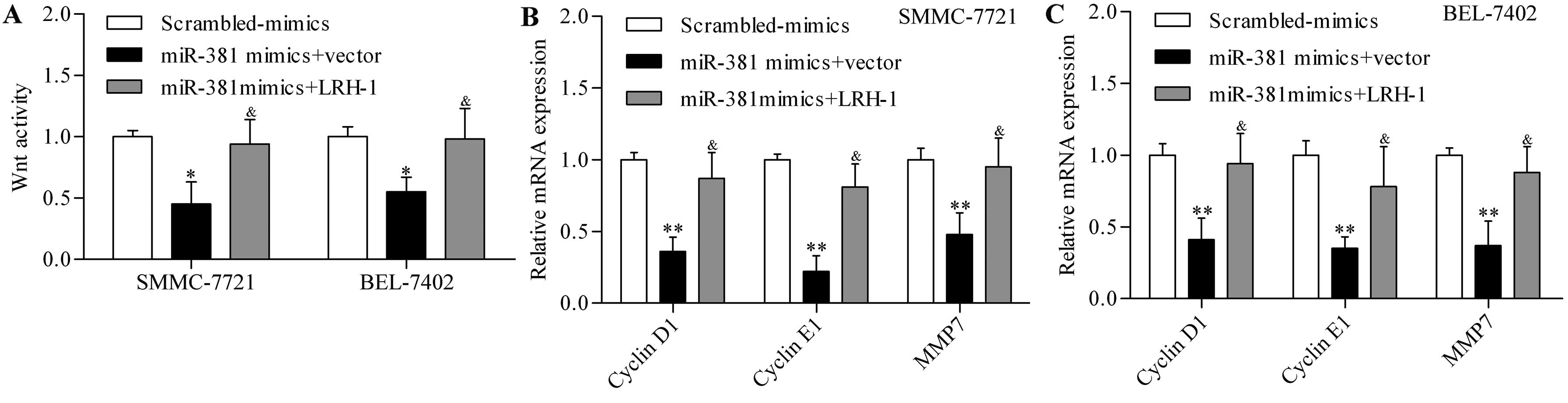
To further elucidate the underlying mechanism of
miR-381 in regulating HCC, we investigated the role of miR-381 in
regulating the downstream signaling pathway of LRH-1. Previous
studies have reported that LRH-1 can promote Wnt signalling
pathways (29), which were
frequently aberrantly activated in cancers including HCC (30). Wnt signaling activity was determined
by using Tcf-dependent TOPFlash reporter activity assay, and the
results showed that miR-381 overexpression significantly
downregulated Wnt signaling activity in SMMC-7721 and BEL-7402
cells (Fig. 7A). To further verify
the regulatory effect of miR-318 on Wnt signaling pathway, we next
detected the transcriptional activity of the Wnt signaling pathway.
The results showed that the expression of Wnt target genes,
including cyclin D1, cyclin E1 and MMP9, were significantly
decreased by miR-381 overexpression in SMMC-7721 (Fig. 7B) and BEL-7402 (Fig. 7C) cells. Moreover, the restoration
of LRH-1 expression significantly reversed the inhibitory effect of
miR-381 overexpression on Wnt signaling activity (Fig. 7). These results indicated that
miR-381 regulated Wnt signaling pathway through LRH-1.
miR-381 expression level inversely
correlates with LRH-1 expression in HCC tissues
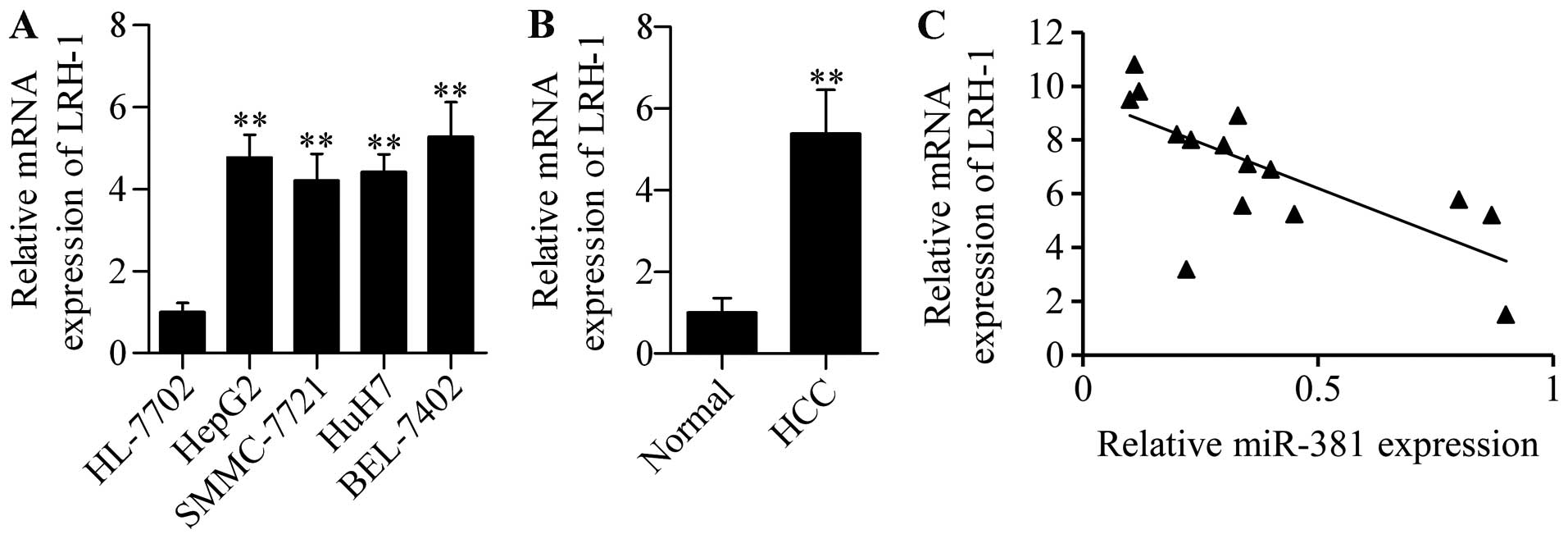
To further explore the functional significance of
miR-381 in HCC, we detected its association with LRH-1 in HCC
tissues. We found that LRH-1 mRNA was highly expressed in HCC cells
(Fig. 8A) and HCC tissues (Fig. 8B). Furthermore, correlation analysis
showed that miR-381 expression was inversely associated with LRH-1
expression in HCC tissues (Fig.
8C). These results indicated that dysregulated miR-381
expression may contribute to increased LRH-1 expression and HCC
development and progression.
Discussion
In the present study, we found that low miR-381
expression and high LRH-1 expression co-occurred in HCC tissues and
cell lines showing an inverse correlation relationship. The study
demonstrated a direct targeted relationship between miR-381 and
LRH-1, in which miR-381 negatively regulated LRH-1 expression. The
decreased miR-381 expression in HCC may contribute to the highly
overexpressed LRH-1 and thus promoted tumor development and
progression. This study provides novel insights into the molecular
mechanism underlying the pathogenesis of HCC malignancy.
The role of LRH-1 has been extensively studied
during the past decades. LRH-1 is required for embryonic
development (33) and for
maintaining the pluripotency of embryonic stem cells (34). LRH-1 is essential for pregnancy, and
a lack of LRH-1 results in infertility (19,35).
Moreover, LRH-1 regulated bile acid biosynthesis and glucose
sensing processes (20,22). However, the deregulated LRH-1
contributed to disease occurrences such as cancer. The most studied
cancer type is breast cancer, wherein LRH-1 was overexpressed,
which was associated with aromatase production (36–38).
Annicotte et al reported that LRH-1 was an estrogen receptor
target gene that exhibited potential oncogenic effects during
tumorigenesis of breast cancer (39). LRH-1 was also capable of regulating
the estrogen receptor expression and estrogen receptor-mediated
target gene expression that promoted breast cancer cell
proliferation (27,40,41).
LRH-1 overexpression promoted breast cancer cell migration and
invasion (42). LRH-1 can directly
target the promoter region of cyclin-dependent kinase inhibitor p21
and inhibit p21 expression leading to breast cancer cell
proliferation (43). Benod et
al found that LRH-1 was overexpressed in human pancreatic
cancer cells, and LRH-1 knockdown significantly repressed
pancreatic cancer cell proliferation (24). Silencing of LRH-1 inhibited colon
cancer cell proliferation and induced G0/G1 cycle arrest (25). LRH-1 was highly expressed in gastric
cancer tissues, and LRH-1 overexpression promoted cell
proliferation of gastric cancer (26). All these studies pointed out the
oncogenic role of LRH-1. The present results of our study also
demonstrated that LRH-1 was overexpressed in HCC tissues and cell
lines, and LRH-1 downregulation by miR-381 impeded the tumorigenic
potential of HCC cells, including cell growth arrest and decreased
invasion ability. These results were consistent with the findings
of Wang et al, who reported that LRH-1 knockdown inhibited
cell growth and induced cell apoptosis of HCC cells (32). Interestingly, it was reported that
LRH-1 could facilitate hepatitis B virus DNA replication and gene
transcription, which promoted the risk for HCC tumorigenesis
(44). Overall, these findings
supported the notion that LRH-1 can be used as a potential and
promising molecular target for HCC prevention and treatment.
Several studies have been performed to develop a
novel antagonist for LRH-1 (45,46).
Corzo et al reported that a small molecule repressor of
LRH-1 and SR1848 exhibited anti-proliferation activity against
LRH-1-expressing cancer cells by inactivating LRH-1 (31). miRs have received particular
attention for cancer detection and treatment in recent years
because of their negative regulator effect on gene expression
(47). A variety of miRs have been
found to be potential molecular target for HCC treatment by
targeting oncogenes or tumor suppressors. For instance, Wang et
al reported that miR-302b inhibited HCC cell invasion and
metastasis by inhibiting AKT2 expression (10). miR-99b accelerated HCC cell
metastasis by suppressing claudin 11 expression (48). Recent studies have demonstrated that
miR-381 plays an important role in various cancer types. For
example, Chen et al revealed that miR-381 inhibited renal
cancer cell proliferation and sensitised renal cancer cells to
chemotherapeutics by inhibiting WEE1 (14,49).
miR-381 overexpression suppressed multidrug resistance of cancer
cells (50). In lung cancers,
miR-381 overexpression inhibits cancer cell migration and invasion
by inhibiting the inhibitor of differentiation 1 (15). miR-381 was downregulated in prostate
cancer, and the lower expression level was correlated with higher
incidence of metastatic events (51). Consistent with these studies, the
present study demonstrated that miR-381 was also significantly
downregulated in HCC, and miR-381 overexpression markedly
suppressed HCC cell growth and invasion. The critical role of
miR-381 in HCC was shown for the first time. miR-381 exerted
antitumor effects by targeting LRH-1, thereby implying that LRH-1
was one of the miR-381 targets. miR-381 exhibited different
functions due to regulating different targets. Tian et al
reported that miR-381 functioned as an oncomiR that promoted glioma
growth by inhibiting leucine-rich repeat C4 expression (52,53).
miR-381 inhibition sensitised glioma cells to temozolomide by
upregulating neurofilament light polypeptide (54). Moreover, Papp et al
elucidated that miR-381 may promote epithelioid sarcoma by
inhibiting SWI/SNF-related, matrix-associated, actin-dependent
regulator of chromatin, subfamily b, member 1 (55). These conflicting findings implicated
that miR-381 may exert different functions by regulating different
targets in different cells or conditions.
LRH-1 is a co-activator of Wnt signaling pathway
that synergistically promoted the transcriptional activity of Wnt
signaling pathway by inducing cyclin E1, cyclin D1 and c-myc
expression (29). Wnt signaling
pathway is frequently aberrantly activated in HCC, which is
associated with increased cancer cell growth and metastasis
(30). LRH-1 overexpression
promoted gastric adenocarcinoma cell proliferation by inducing
cyclin E1 (26). LRH-1 knockdown
inhibited cyclin E1, cyclin D1 and c-myc expression in pancreatic
cancer cells (24). In the present
study, downregulating LRH-1 by miR-381 inactivated Wnt signaling
and repressed the expression of target genes, including cyclin E1,
cyclin D1 and MMP9. Most importantly, restoration of LRH-1
expression reversed the inhibitory effect of miR-381 on the Wnt
signaling pathway, which may explain why miR-381 overexpression
impeded HCC cell growth and invasion.
To date, the specific miRs that target and regulate
LRH-1 have not been well recognized and investigated. A recent
study elucidated that miR-451 directly targets and suppresses LRH-1
expression in osteosarcoma cells, thereby inhibiting cancer cell
proliferation (56). In the present
study, miR-381 targeted and regulated LRH-1 expression in HCC
cells. These findings raised the possibility that inhibiting LRH-1
by specific miR to treat and prevent cancers in which LRH-1
functioned as an oncogene. The present results therefore suggest
that miR-381 may be a novel tumor suppressor that blocks HCC growth
and invasion by targeting LRH-1 and inhibits the LRH-1-mediated Wnt
signaling pathway. The present study highlighted the functional
significance of miR-381-LRH-1 in HCC, providing novel insights into
the development of future therapeutic interventions for HCC.
Abbreviations:
|
miRs
|
microRNAs
|
|
miR-381
|
microRNA-381
|
|
LRH-1
|
liver receptor homolog-1
|
|
3′-UTR
|
3′-untranslated region
|
|
HCC
|
hepatocellular carcinoma
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Okuda K: Early recognition of
hepatocellular carcinoma. Hepatology. 6:729–738. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kuo KL, Stenehjem D, Albright F, Ray S and
Brixner D: Treatment patterns and outcomes in patients with
hepatocellular carcinoma stratified by stage-guided treatment
categories. J Natl Compr Canc Netw. 13:987–994. 2015.PubMed/NCBI
|
|
4
|
Song S, Nam SW, Bae SH, Kim JD, Jang JW,
Song MJ, Lee SW, Kim HY, Lee YJ, Chun HJ, et al: Outcome of
transarterial chemoembolization-based multi-modal treatment in
patients with unresectable hepatocellular carcinoma. World J
Gastroenterol. 21:2395–2404. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Song Y, Wang F, Huang Q, Cao Y, Zhao Y and
Yang C: MicroRNAs contribute to hepatocellular carcinoma. Mini Rev
Med Chem. 15:459–466. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Winter J, Jung S, Keller S, Gregory RI and
Diederichs S: Many roads to maturity: microRNA biogenesis pathways
and their regulation. Nat Cell Biol. 11:228–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mendell JT and Olson EN: MicroRNAs in
stress signaling and human disease. Cell. 148:1172–1187. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ranganathan K and Sivasankar V: MicroRNAs
- Biology and clinical applications. J Oral Maxillofac Pathol.
18:229–234. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang L, Yao J, Zhang X, Guo B, Le X,
Cubberly M, Li Z, Nan K, Song T and Huang C: miRNA-302b suppresses
human hepato-cellular carcinoma by targeting AKT2. Mol Cancer Res.
12:190–202. 2014. View Article : Google Scholar
|
|
11
|
Liu J, Yan J, Zhou C, Ma Q, Jin Q and Yang
Z: miR-1285-3p acts as a potential tumor suppressor miRNA via
downregulating JUN expression in hepatocellular carcinoma. Tumour
Biol. 36:219–225. 2015. View Article : Google Scholar
|
|
12
|
Kan H, Guo W, Huang Y and Liu D:
MicroRNA-520g induces epithelial-mesenchymal transition and
promotes metastasis of hepatocellular carcinoma by targeting SMAD7.
FEBS Lett. 589:102–109. 2015. View Article : Google Scholar
|
|
13
|
Li T, Yin J, Yuan L, Wang S, Yang L, Du X
and Lu J: Downregulation of microRNA-139 is associated with
hepatocellular carcinoma risk and short-term survival. Oncol Rep.
31:1699–1706. 2014.PubMed/NCBI
|
|
14
|
Chen B, Duan L, Yin G, Tan J and Jiang X:
Simultaneously expressed miR-424 and miR-381 synergistically
suppress the proliferation and survival of renal cancer cells -
Cdc2 activity is up-regulated by targeting WEE1. Clinics (Sao
Paulo). 68:825–833. 2013. View Article : Google Scholar
|
|
15
|
Rothschild SI, Tschan MP, Jaggi R, Fey MF,
Gugger M and Gautschi O: MicroRNA-381 represses ID1 and is
deregulated in lung adenocarcinoma. J Thorac Oncol. 7:1069–1077.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nadolny C and Dong X: Liver receptor
homolog-1 (LRH-1): A potential therapeutic target for cancer.
Cancer Biol Ther. 16:997–1004. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Paré JF, Malenfant D, Courtemanche C,
Jacob-Wagner M, Roy S, Allard D and Bélanger L: The fetoprotein
transcription factor (FTF) gene is essential to embryogenesis and
cholesterol homeostasis and is regulated by a DR4 element. J Biol
Chem. 279:21206–21216. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Duggavathi R, Volle DH, Mataki C, Antal
MC, Messaddeq N, Auwerx J, Murphy BD and Schoonjans K: Liver
receptor homolog 1 is essential for ovulation. Genes Dev.
22:1871–1876. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang C, Large MJ, Duggavathi R, DeMayo
FJ, Lydon JP, Schoonjans K, Kovanci E and Murphy BD: Liver receptor
homolog-1 is essential for pregnancy. Nat Med. 19:1061–1066. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu TT, Makishima M, Repa JJ, Schoonjans K,
Kerr TA, Auwerx J and Mangelsdorf DJ: Molecular basis for feedback
regulation of bile acid synthesis by nuclear receptors. Mol Cell.
6:507–515. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mataki C, Magnier BC, Houten SM, Annicotte
JS, Argmann C, Thomas C, Overmars H, Kulik W, Metzger D, Auwerx J,
et al: Compromised intestinal lipid absorption in mice with a
liver-specific deficiency of liver receptor homolog 1. Mol Cell
Biol. 27:8330–8339. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Oosterveer MH, Mataki C, Yamamoto H,
Harach T, Moullan N, van Dijk TH, Ayuso E, Bosch F, Postic C, Groen
AK, et al: LRH-1-dependent glucose sensing determines intermediary
metabolism in liver. J Clin Invest. 122:2817–2826. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schoonjans K, Dubuquoy L, Mebis J, Fayard
E, Wendling O, Haby C, Geboes K and Auwerx J: Liver receptor
homolog 1 contributes to intestinal tumor formation through effects
on cell cycle and inflammation. Proc Natl Acad Sci USA.
102:2058–2062. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Benod C, Vinogradova MV, Jouravel N, Kim
GE, Fletterick RJ and Sablin EP: Nuclear receptor liver receptor
homologue 1 (LRH-1) regulates pancreatic cancer cell growth and
proliferation. Proc Natl Acad Sci USA. 108:16927–16931. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bayrer JR, Mukkamala S, Sablin EP, Webb P
and Fletterick RJ: Silencing LRH-1 in colon cancer cell lines
impairs proliferation and alters gene expression programs. Proc
Natl Acad Sci USA. 112:2467–2472. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang SL, Zheng DZ, Lan FH, Deng XJ, Zeng
J, Li CJ, Wang R and Zhu ZY: Increased expression of hLRH-1 in
human gastric cancer and its implication in tumorigenesis. Mol Cell
Biochem. 308:93–100. 2008. View Article : Google Scholar
|
|
27
|
Chand AL, Wijayakumara DD, Knower KC,
Herridge KA, Howard TL, Lazarus KA and Clyne CD: The orphan nuclear
receptor LRH-1 and ERα activate GREB1 expression to induce breast
cancer cell proliferation. PLoS One. 7:e315932012. View Article : Google Scholar
|
|
28
|
Chand AL, Pathirage N, Lazarus K, Chu S,
Drummond AE, Fuller PJ and Clyne CD: Liver receptor homologue-1
expression in ovarian epithelial and granulosa cell tumours.
Steroids. 78:700–706. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Botrugno OA, Fayard E, Annicotte JS, Haby
C, Brennan T, Wendling O, Tanaka T, Kodama T, Thomas W, Auwerx J,
et al: Synergy between LRH-1 and beta-catenin induces G1
cyclin-mediated cell proliferation. Mol Cell. 15:499–509. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pez F, Lopez A, Kim M, Wands JR, Caron de
Fromentel C and Merle P: Wnt signaling and hepatocarcinogenesis:
Molecular targets for the development of innovative anticancer
drugs. J Hepatol. 59:1107–1117. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Corzo CA, Mari Y, Chang MR, Khan T,
Kuruvilla D, Nuhant P, Kumar N, West GM, Duckett DR, Roush WR, et
al: Antiproliferation activity of a small molecule repressor of
liver receptor homolog 1. Mol Pharmacol. 87:296–304. 2015.
View Article : Google Scholar :
|
|
32
|
Wang S, Lan F, Huang L, Dong L, Zhu Z, Li
Z, Xie Y and Fu J: Suppression of hLRH-1 mediated by a DNA
vector-based RNA interference results in cell cycle arrest and
induction of apoptosis in hepatocellular carcinoma cell BEL-7402.
Biochem Biophys Res Commun. 333:917–924. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gu P, Goodwin B, Chung AC, Xu X, Wheeler
DA, Price RR, Galardi C, Peng L, Latour AM, Koller BH, et al:
Orphan nuclear receptor LRH-1 is required to maintain Oct4
expression at the epiblast stage of embryonic development. Mol Cell
Biol. 25:3492–3505. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kelly VR and Hammer GD: LRH-1 and Nanog
regulate Dax1 transcription in mouse embryonic stem cells. Mol Cell
Endocrinol. 332:116–124. 2011. View Article : Google Scholar :
|
|
35
|
Gerrits H, Paradé MC, Koonen-Reemst AM,
Bakker NE, Timmer-Hellings L, Sollewijn Gelpke MD and Gossen JA:
Reversible infertility in a liver receptor homologue-1
(LRH-1)-knockdown mouse model. Reprod Fertil Dev. 26:293–306. 2014.
View Article : Google Scholar
|
|
36
|
Zhou J, Suzuki T, Kovacic A, Saito R, Miki
Y, Ishida T, Moriya T, Simpson ER, Sasano H and Clyne CD:
Interactions between prostaglandin E(2), liver receptor
homologue-1, and aromatase in breast cancer. Cancer Res.
65:657–663. 2005.PubMed/NCBI
|
|
37
|
Bouchard MF, Taniguchi H and Viger RS:
Protein kinase A-dependent synergism between GATA factors and the
nuclear receptor, liver receptor homolog-1, regulates human
aromatase (CYP19) PII promoter activity in breast cancer cells.
Endocrinology. 146:4905–4916. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lanzino M, Maris P, Sirianni R, Barone I,
Casaburi I, Chimento A, Giordano C, Morelli C, Sisci D, Rizza P, et
al: DAX-1, as an androgen-target gene, inhibits aromatase
expression: A novel mechanism blocking estrogen-dependent breast
cancer cell proliferation. Cell Death Dis. 4:e7242013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Annicotte JS, Chavey C, Servant N,
Teyssier J, Bardin A, Licznar A, Badia E, Pujol P, Vignon F,
Maudelonde T, et al: The nuclear receptor liver receptor homolog-1
is an estrogen receptor target gene. Oncogene. 24:8167–8175.
2005.PubMed/NCBI
|
|
40
|
Thiruchelvam PT, Lai CF, Hua H, Thomas RS,
Hurtado A, Hudson W, Bayly AR, Kyle FJ, Periyasamy M, Photiou A, et
al: The liver receptor homolog-1 regulates estrogen receptor
expression in breast cancer cells. Breast Cancer Res Treat.
127:385–396. 2011. View Article : Google Scholar
|
|
41
|
Lai CF, Flach KD, Alexi X, Fox SP,
Ottaviani S, Thiruchelvam PT, Kyle FJ, Thomas RS, Launchbury R, Hua
H, et al: Co-regulated gene expression by oestrogen receptor α and
liver receptor homolog-1 is a feature of the oestrogen response in
breast cancer cells. Nucleic Acids Res. 41:10228–10240. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chand AL, Herridge KA, Thompson EW and
Clyne CD: The orphan nuclear receptor LRH-1 promotes breast cancer
motility and invasion. Endocr Relat Cancer. 17:965–975. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bianco S, Jangal M, Garneau D and Gévry N:
LRH-1 controls proliferation in breast tumor cells by regulating
CDKN1A gene expression. Oncogene. 34:4509–4518. 2015. View Article : Google Scholar
|
|
44
|
Cai YN, Zhou Q, Kong YY, Li M, Viollet B,
Xie YH and Wang Y: LRH-1/hB1F and HNF1 synergistically up-regulate
hepatitis B virus gene transcription and DNA replication. Cell Res.
13:451–458. 2003. View Article : Google Scholar
|
|
45
|
Rey J, Hu H, Kyle F, Lai CF, Buluwela L,
Coombes RC, Ortlund EA, Ali S, Snyder JP and Barrett AG: Discovery
of a new class of liver receptor homolog-1 (LRH-1) antagonists:
Virtual screening, synthesis and biological evaluation.
ChemMedChem. 7:1909–1914. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Benod C, Carlsson J, Uthayaruban R, Hwang
P, Irwin JJ, Doak AK, Shoichet BK, Sablin EP and Fletterick RJ:
Structure-based discovery of antagonists of nuclear receptor LRH-1.
J Biol Chem. 288:19830–19844. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Anwar SL and Lehmann U: MicroRNAs:
Emerging novel clinical biomarkers for hepatocellular carcinomas. J
Clin Med. 4:1631–1650. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yang J, Liu X, Yuan X and Wang Z: miR-99b
promotes metastasis of hepatocellular carcinoma through inhibition
of claudin 11 expression and may serve as a prognostic marker.
Oncol Rep. 34:1415–1423. 2015.PubMed/NCBI
|
|
49
|
Chen B, Duan L, Yin G, Tan J and Jiang X:
miR-381, a novel intrinsic WEE1 inhibitor, sensitizes renal cancer
cells to 5-FU by up-regulation of Cdc2 activities in 786-O. J
Chemother. 25:229–238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Xu Y, Ohms SJ, Li Z, Wang Q, Gong G, Hu Y,
Mao Z, Shannon MF and Fan JY: Changes in the expression of miR-381
and miR-495 are inversely associated with the expression of the
MDR1 gene and development of multi-drug resistance. PLoS One.
8:e820622013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Formosa A, Markert EK, Lena AM, Italiano
D, Finazzi-Agró E, Levine AJ, Bernardini S, Garabadgiu AV, Melino G
and Candi E: MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c,
miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p,
mapped to the 14q32.31 locus, regulate proliferation, apoptosis,
migration and invasion in metastatic prostate cancer cells.
Oncogene. 33:5173–5182. 2014. View Article : Google Scholar
|
|
52
|
Tang H, Liu X, Wang Z, She X, Zeng X, Deng
M, Liao Q, Guo X, Wang R, Li X, et al: Interaction of hsa-miR-381
and glioma suppressor LRRC4 is involved in glioma growth. Brain
Res. 1390:21–32. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tang H, Wang Z, Liu Q, Liu X, Wu M and Li
G: Disturbing miR-182 and -381 inhibits BRD7 transcription and
glioma growth by directly targeting LRRC4. PLoS One. 9:e841462014.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang Z, Yang J, Xu G, Wang W, Liu C, Yang
H, Yu Z, Lei Q, Xiao L, Xiong J, et al: Targeting miR-381-NEFL axis
sensitizes glioblastoma cells to temozolomide by regulating
stemness factors and multidrug resistance factors. Oncotarget.
6:3147–3164. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Papp G, Krausz T, Stricker TP, Szendrői M
and Sápi Z: SMARCB1 expression in epithelioid sarcoma is regulated
by miR-206, miR-381, and miR-671-5p on both mRNA and protein
levels. Genes Chromosomes Cancer. 53:168–176. 2014. View Article : Google Scholar
|
|
56
|
Li Z, Wu S, Lv S, Wang H, Wang Y and Guo
Q: Suppression of liver receptor homolog-1 by microRNA-451
represses the proliferation of osteosarcoma cells. Biochem Biophys
Res Commun. 461:450–455. 2015. View Article : Google Scholar : PubMed/NCBI
|