Introduction
Esophageal cancer (EC) is the eighth most common
cancer in the world; more than 450,000 people worldwide are
affected. It has an extremely aggressive nature and poor survival
rate with a 5-year overall survival rate of approximately 10–25%.
The two main types are squamous cell carcinoma (SCC) and
adenocarcinoma (AC). The incidence of esophageal adenocarcinoma
(EAC) has risen during the last 20 years, mainly in the Western
population. The single most important risk factor is Barrett's
metaplasia. Esophageal SCC has the highest rates in areas of the
East and Middle-East. The most important risk factors for SCC
include smoking and alcohol intake. Most of the tumors are
diagnosed in advanced stages and a cure can only be anticipated in
patients with superficial cancers (1–4). Only
about 30–40% of patients respond to chemotherapy. Several
biomarkers with possible predictive power have been described.
However, a pre-selection of patients with possible benefit of
neoadjuvant treatment is not in clinical practice to date (5).
TP53 mutations are the most frequent mutations in
ECs detected so far in several studies. Thus, between 40 and 50% of
EC cases show a mutation of TP53. Hereby the prevalence of TP53
mutations in esophageal SCC might be slightly higher than that in
EAC (6). However, although the high
mutation rate of TP53 clearly demonstrates the crucial role of the
tumor-suppressor TP53 in EC development, it has not yet led to
individualised treatment or has not influenced prognosis (7–11).
The human homologue of the murine double minute-2
gene (MDM2) is a negative regulator of TP53. MDM2 is an E3
ubiquitinprotein ligase and its overexpression leads to TP53
ubiquitination and subsequent degradation linked to the loss of
TP53 tumor-suppressor activity (12). Overexpression of MDM2 may be
due to gene amplification or to increased promoter activity of
patients carrying the MDM promoter T309G single nucleotide
polymorphism (SNP) (13). Since
many tumors have shown overexpression of MDM2, novel therapeutical
approaches target MDM2 in order to restore TP53 tumor-suppressor
activity (12–14).
In the present study, we investigated MDM2
gene amplification by quantitiative PCR and fluorescence in
situ hybridisation (FISH) in 127 ECs. Importantly,
approximately 18% demonstrated moderately elevated MDM2 copy
numbers by qPCR, which may not affect overall MDM2
overexpression in the tumor. However, 6.3% of all EC showed a
strong enhancement of MDM2 gene copies, that was linked to a
cluster-like amplification pattern by FISH and to strong MDM2
immunostaining.
Materials and methods
Clinical and histological tumor
evaluation
Esophagectomies were performed for patients with EC
at the Department of Surgery, University of Cologne, Germany. Among
146 patients with locally advanced EC (cT3-4, cNx and
M0), tissue samples collected at the Institute for
Pathology, University Hospital of Cologne were applied in our
restrospective study. All specimens were used in accordance with
the local policies of the Institutional Review Board of the
University Hospital of Cologne. TNM staging was performed according
to the criteria of the International Union Against Cancer (UICC)
(17). Clinical staging consisted
of endoscopy, endoscopic ultrasound, barium swallow, CT scanning of
the abdomen and thorax and positron-emission tomography.
Morphologic assessment of tumor regression was performed by an
objective histopathologic examination as described previously
(16). The resected specimens were
fixed in formalin (10%). The whole tumor region including a safety
margin was excised by a pathologist in a topographic order.
Histological sections were prepared according to standard
procedures of pathology.
FISH analysis
FISH was performed on interphase nuclei of
4-µm sections of formalin-fixed and paraffin-embedded (FFPE)
tissue. Deparaffinised slides were subjected to several
pretreatment steps. They were incubated in 0.2 M HCl for 20 min,
followed by washing, and 80°C heat pretreatment in
pretreatment-solution (Abbott, Wiesbaden, Germany) for 30 min and
further washing. Next, the tissue was digested for 1.5 h with
protease, slides were washed again, and fixed in 4% buffered
formalin. After washing, dehydration and air drying, an appropriate
amount of fluorochrome-labeled probe mixture was applied to the
tissue section. Sample DNA and probe mixture were co-denatured in a
thermobrite hybridiser (Abbott) at 85°C for 10 min, cooled down to
37°C, and hybridised overnight in a humidified hybridisation
chamber. After stringent washing and dehydration and air drying,
the sections were counterstained using fluorescence mounting media
containing (4′,6-diamidino-2-phenylindole) (DAPI). FISH slides were
evaluated with a fluorescence microscope (DM 5500; Leica
Microsystems, Wetzlar, Germany) using appropriate filter sets.
Orange (centromeric region) and green (target gene) signals were
counted in 60 non-overlapping tumor cell nuclei from three tumor
areas. Dual color FISH probes were derived from ZytoVision
(Bremerhaven, Germany).
Macrodissection and DNA extraction from
the FFPE tissues
Six 10-µm sections of 127 FFPE esophageal
tumor biopsies were used for macrodissection and subsequent DNA
extraction as previously described (18). Briefly, for genomic DNA extraction,
the lesional areas were marked on a haematoxylin and eosin
(H&E) slide by a senior pathologist. After deparaffinisation,
tumor tissue was macrodissected from the unstained slides and the
tissue was lysed with proteinase K overnight. Subsequently, DNA
purification was carried out with the BioRobot M48 robotic
workstation and the corresponding MagAttract DNA Mini M48 kit
(Qiagen, Hilden, Germany) following the manufacturer's
protocol.
TP53 mutation analysis
The TP53 mutation status was evaluated by data
analyses available from a comprehensive next generation sequencing
approach which was performed on 68 ECs from a total of 127 tumors.
Library construction on a hot spot cancer gene panel (Life
Technologies, Darmstadt, Germany) and ultra-deep sequencing on the
MiSeq platform (Illumina, San Diego, CA, USA) were performed
according to Grünewald et al (19).
Copy number evaluation by real-time
PCR
Evaluation of the MDM2 gene copy number variation
(CNV) of the esophageal tumors was performed by quantitative PCR
(qPCR). DNA from human embryonic kidney epithelial cells (HEK293)
and DNA from 14 macrodissected, non-tumorous colon FPPE tissues
were taken as non-MDM2-amplified reference DNA. Primer and
probes for CNV assays of MDM2, TERT and RNase
P were supplied by Applied Biosystems Life Technologies
(Darmstadt, Germany).
Real-time PCR was carried out in triplicate,
following the manufacturer's protocol. Copy numbers were then
interpreted by the ΔΔCt method using the non-tumorous reference
panel as calibrator and RNAse P as endogenous control. Evaluation
of the RNAse P gene as the best endogenous control gene,
demonstrating stable copy numbers of two alleles per cell in
different reference panels (data not shown), was determined by
standard curves and analysis of different non-amplified reference
DNA.
Results
Efficient MDM2 copy number screening by
qPCR
FISH analysis of gene amplification is laborious,
costly and time-consuming. Therefore, we macrodissected the tumor
areas from 146 EC specimens and extracted DNA, in order to study
MDM2 CNVs by qPCR. DNA qualtity of 127 ECs (Table I) was sufficient to perform qPCR.
Normalization of MDM2 gene quantification was established,
using RNAse P gene as a calibrator. Determination of RNAase P
values in normal tissues ascertained normalisation to two copies
per cell (data not shown).
 | Table ISample cohort used in the present
study (n=127). |
Table I
Sample cohort used in the present
study (n=127).
| AC
| SCC
|
|---|
| Without neoadjuvant
therapy | With neoadjuvant
therapy | Without neoadjuvant
therapy | With neoadjuvant
therapy |
|---|
| Female (n) | 7 | 6 | 6 | 4 |
| Male (n) | 38 | 36 | 11 | 19 |
| Total (n) | 45 | 42 | 17 | 23 |
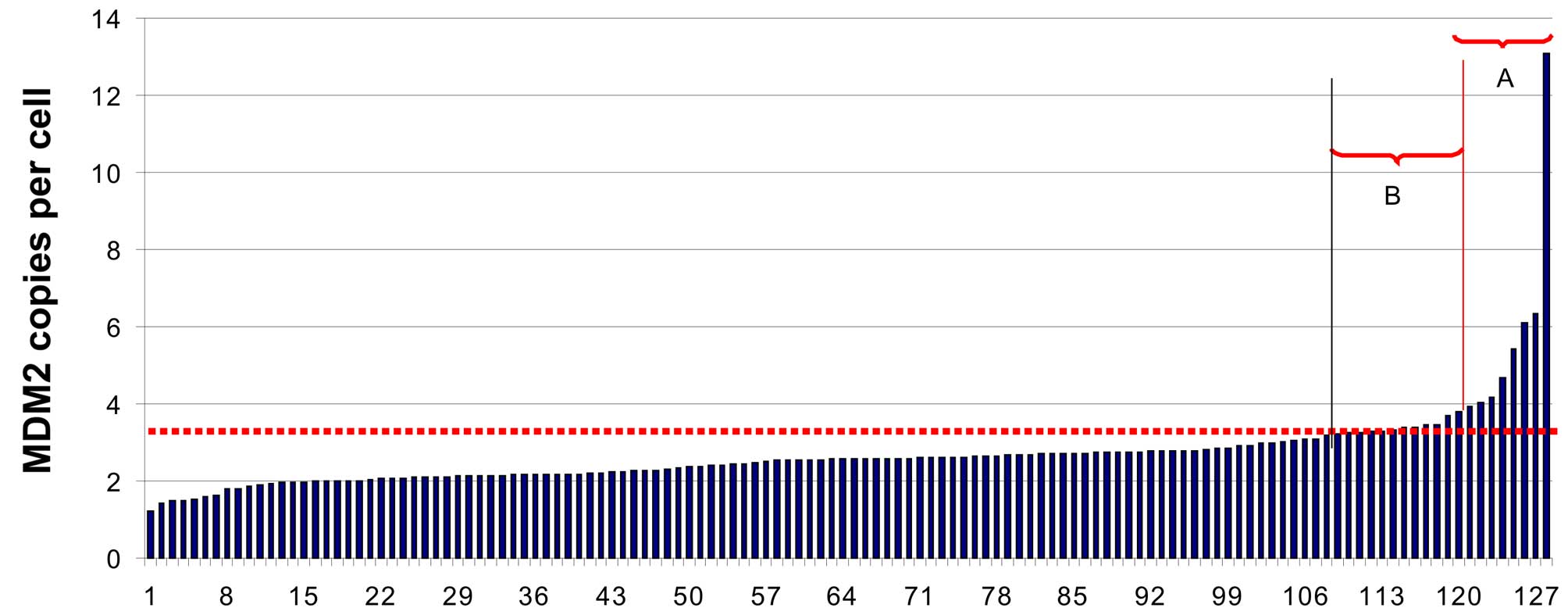
Then, we screened the 127 ECs (Table I) for MDM2 gene
amplification. Fig. 1 demonstrates
that eight EC (6.3%) samples showed an MDM2 copy number of 4
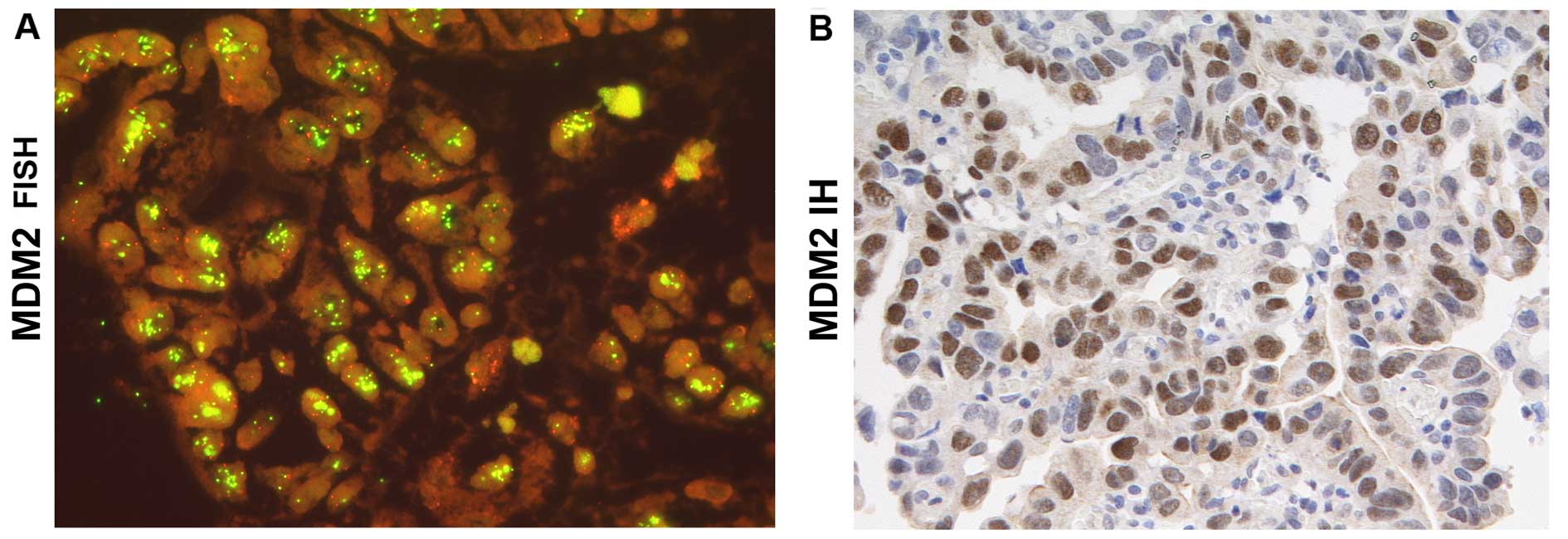
and higher. These tumors were then studied by FISH showing tumor
areas with high MDM2 gene clusters (Fig. 2) and increased MDM2 protein
expression in the nuclei of EC tumor cells.
Importantly, in some samples (n=15, 11.8%) we found
mean values of more than three MDM2 gene copies per cell by
qPCR, indicating that these ECs might be also positve for
MDM2 gene amplification (shown in area B of Fig. 1). Therefore, we studied the tumor
histology and the MDM2 protein expression of these ECs.
Furthermore, gene amplification was analysed by FISH. In most
cases, no or only very low gene amplification was shown by FISH and
importantly, we did not observe the typical cluster-like
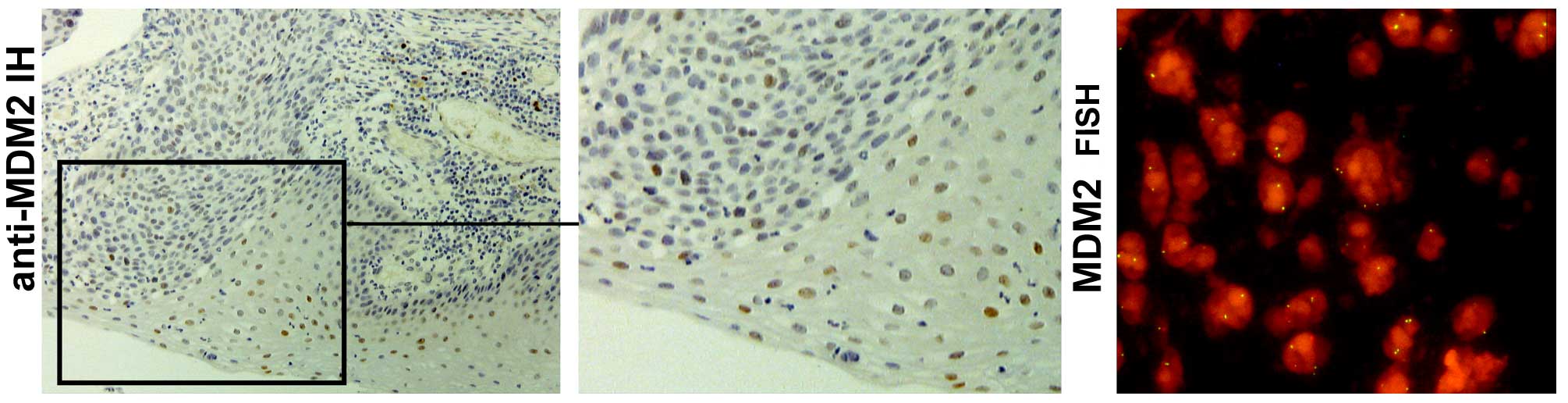
amplification pattern of MDM2-positive tumor cells. In addition,
ECs with slightly elevated MDM2 copy numbers showed
moderately enhanced MDM2 protein expression by immunohistology
(Fig. 3).
However, a slightly increased MDM2 copy
number by PCR (shown in area B of Fig.
1) might also be due to low tumor cell content in the
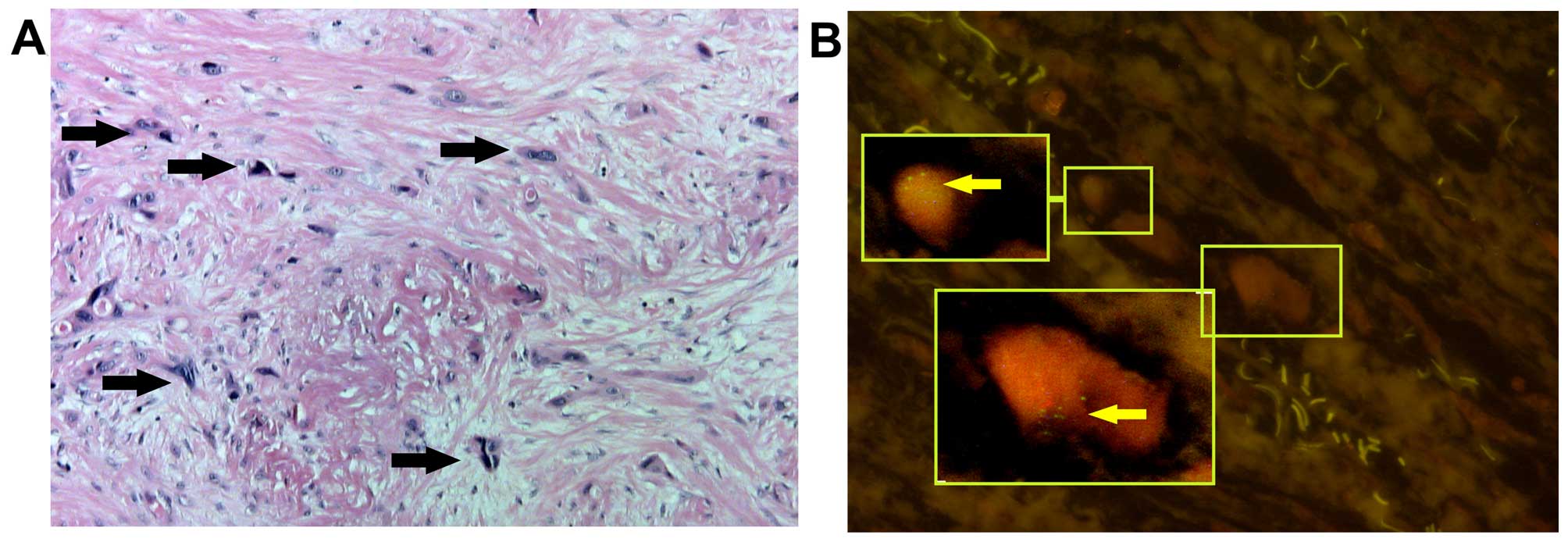
macrodissected EC area. Indeed, in some case with an MDM copy
number >3 (Fig. 2B) only a few
tumor cells were observed, that were scattered over the tumorous
area, embedded by a high proportion of non-tumorous stroma cells.
These cells clearly showed MDM2 gene amplification by a
cluster-like pattern of the MDM2 FISH signals (Fig. 4).
MDM2 gene amplification in esophageal SCC
and AC
From a total of 127 ECs, we identified by qPCR 8
tumors (6.3%) that carry highly elevated numbers of the MDM2
gene (Table II and Fig. 2). There was no prevalence of
MDM2 gene amplification in AC or SCC (Table II).
 | Table IITP53 mutation status of the
esophageal carcinoma samples with MDM2 gene
amplification. |
Table II
TP53 mutation status of the
esophageal carcinoma samples with MDM2 gene
amplification.
| Sample no. | Esophageal
carcinoma | TP53 mutation
status |
|---|
| 1 | SCC | p.C176W |
| 2 | SCC | p.R213a |
| 3 | AC | wt |
| 4 | AC | p.E286a |
| 5 | AC | p.R248Q |
| 6 | SCC | wt |
| 7 | AC | wt |
| 8 | AC | wt |
Treatment with MDM2 inhibitors is a novel important
therapeutical option, but depends on the TP53 wild-type form.
Therefore, we searched the NGS data of the EC cohort and analysed
whether tumors with MDM2 amplification and overexpression
harbor the mutated or wild-type TP53. From a total of 68 ECs, NGS
data were available and revealed that 63% of the ECs had a
pathogenic mutation in TP53. From the 8 samples, which were
positive for MDM2 amplification, 4 samples carried the
wild-type and 4 were shown to carry the TP53 mutants.
Discussion
MDM2 is an E3 ubiquitin ligase, which functions as a
crucial oncogene by its interaction with TP53 leading to TP53
degradation. Due to its high impact on TP53 regulation, MDM2 is an
important target of novel therapeutic strategies (12,14).
MDM2 overexpression was previously shown in
many tumor types such as soft tissue sarcoma (20) and in non-small cell lung cancer
(NSCLC) (21,22). In the present study, we investigated
MDM2 gene amplification in 127 EC specimens. Using qPCR
assay, we identified 6.3% of the EC cases with a pronounced
MDM2 gene amplification, which were validated by a
cluster-like signal pattern by FISH. In addition, the qPCR
screening approach detected 15 borderline ECs with a moderate
MDM2 gene amplification. Although these ECs showed also no
or only low MDM2 gene alterations by FISH, in the ECs in
which only a low number of tumor cells were recorded, high
MDM2 gene amplification was confirmed by the FISH assay.
Since FISH is very laborious, costly and time consuming, qPCR is a
highly efficient method by which to first screen tumors for
MDM2 gene amplification. The selected putative positives can
then be further studied by MDM2 immunostaining and FISH. This
economic approach is suggested to be of great importance for future
EC treatment, in particular, since presently several small MDM2
inhibitors are in phase I of clinical trials, as summarised by Zhao
et al (23).
There was no prevalence of MDM2 gene
amplification in the SCC or in AC cases. Taniere et al
reported that MDM2 amplifications were noted in SCC and AC
in a small Asian cohort (n=23) with a frequency of 4% (24,25).
Interestingly, the authors also described that the prevalence of
MDM2 amplifications was much higher in Barrett's mucosa and
adenocarcinoma of the cardia (25)
reaching 19% of the cases.
In our study, the ECs with higher MDM2 copy
numbers showed also a pronounced staining for the MDM2 protein.
Primary findings of Soslov et al demonstrated that >50%
of AC cases showed MDM2 overexpression. However, beside gene
amplification, MDM2 expression depends also on the T309G SNP
which is associated with increased MDM2 promoter activity
(13,26,27,29).
Furthermore, MDM2 gene amplification might be due to an
increased activity of the eukaryotic translation initiation factor
4E (eIF4E), shown to elevate MDM2 expression in EC (28).
From a total of 127 patients, we studied the TP53
mutation status in 68 tumors demonstrating an overall TP53 mutation
frequency of 63%. There was no significant difference in the TP53
mutation frequency in the MDM2-positive and -negative ECs.
Thus, in summary, our findings on MDM2 gene amplification
and TP53 mutations revealed that therapeutic approaches with MDM2
inhibitors may be an important option for patients with EC
independently in the case of the development of AC or SCC.
Acknowledgments
We thank Hannah Eischeid for the excellent technical
support in macrodissection and DNA extraction. Furthermore, we are
very grateful for the great technical assistance of Elke Binot in
the FISH experiments. The present study was supported by the
Fortune Program of the Medical Faculty to P.G.
Abbreviations:
|
AC
|
adenocarcinoma
|
|
CNV
|
copy number variation
|
|
EC
|
esophageal cancer
|
|
FFPE
|
formalin-fixed and
paraffin-embedded
|
|
FISH
|
fluorescence in situ hybridisation
|
|
MDM2
|
murine double minute-2
|
|
qPCR
|
quantitative PCR
|
|
SCC
|
squamous cell carcinoma
|
|
SNP
|
single nucleotide polymorphism
|
|
TP53
|
tumor protein 53
|
References
|
1
|
Bosman FT, Carneiro F, Hruban RH and
Theise ND: WHO Classification of Tumours of the Digestive System.
3. 4th edition. WHO Press; Lyon; 2010
|
|
2
|
Zhang Y: Epidemiology of esophageal
cancer. World J Gastroenterol. 19:5598–5606. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pennathur A and Luketich JD: Resection for
esophageal cancer: Strategies for optimal management. Ann Thorac
Surg. 85:S751–S756. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vallböhmer D, Brabender J, Metzger R and
Hölscher AH: Genetics in the pathogenesis of esophageal cancer:
Possible predictive and prognostic factors. J Gastrointest Surg.
14(Suppl 1): S75–S80. 2010. View Article : Google Scholar
|
|
6
|
Agrawal N, Jiao Y, Bettegowda C, Hutfless
SM, Wang Y, David S, Cheng Y, Twaddell WS, Latt NL, Shin EJ, et al:
Comparative genomic analysis of esophageal adenocarcinoma and
squamous cell carcinoma. Cancer Discov. 2:899–905. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gibson MK, Abraham SC, Wu TT, Burtness B,
Heitmiller RF, Heath E and Forastiere A: Epidermal growth factor
receptor, p53 mutation, and pathological response predict survival
in patients with locally advanced esophageal cancer treated with
preoperative chemoradiotherapy. Clin Cancer Res. 9:6461–6468.
2003.PubMed/NCBI
|
|
8
|
Makino T, Yamasaki M, Miyata H, Yoshioka
S, Takiguchi S, Fujiwara Y, Nakajima K, Nishida T, Mori M and Doki
Y: p53 mutation status predicts pathological response to
chemoradiotherapy in locally advanced esophageal cancer. Ann Surg
Oncol. 17:804–811. 2010. View Article : Google Scholar
|
|
9
|
Egashira A, Morita M, Yoshida R, Saeki H,
Oki E, Sadanaga N, Kakeji Y, Tsujitani S and Maehara Y: Loss of p53
in esophageal squamous cell carcinoma and the correlation with
survival: Analyses of gene mutations, protein expression, and loss
of heterozygosity in Japanese patients. J Surg Oncol. 104:169–175.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Silveira AP, Da Silva Manoel-Caetano F,
Aoki S, Yamasaki LH, Rahal P and Silva AE: Gene mutations and
polymorphisms of TP53 and FHIT in chronic esophagitis and
esophageal carcinoma. Anticancer Res. 31:1685–1690. 2011.PubMed/NCBI
|
|
11
|
Sengpiel C, König IR, Rades D, Noack F,
Duchrow M, Schild SE, Ludwig D and Homann N: p53 Mutations in
carcinoma of the esophagus and gastroesophageal junction. Cancer
Invest. 27:96–104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Forslund A, Zeng Z, Qin LX, Rosenberg S,
Ndubuisi M, Pincas H, Gerald W, Notterman DA, Barany F and Paty PB:
MDM2 gene amplification is correlated to tumor progression but not
to the presence of SNP309 or TP53 mutational status in primary
colorectal cancers. Mol Cancer Res. 6:205–211. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen B, Cao L, Hu KW, Zhang JW, Meng XL
and Xiong MM: MDM2 SNP309 is an ethnicity-dependent risk factor for
digestive tract cancers. Tumour Biol. 35:3431–3438. 2014.
View Article : Google Scholar
|
|
14
|
Nag S, Zhang X, Srivenugopal KS, Wang MH,
Wang W and Zhang R: Targeting MDM2-p53 interaction for cancer
therapy: Are we there yet? Curr Med Chem. 21:553–574. 2014.
View Article : Google Scholar
|
|
15
|
Bollschweiler E, Hölscher AH and Metzger
R: Histologic tumor type and the rate of complete response after
neoadjuvant therapy for esophageal cancer. Future Oncol. 6:25–35.
2010. View Article : Google Scholar
|
|
16
|
Schneider PM, Baldus SE, Metzger R, Kocher
M, Bongartz R, Bollschweiler E, Schaefer H, Thiele J, Dienes HP,
Mueller RP, et al: Histomorphologic tumor regression and lymph node
metastases determine prognosis following neoadjuvant
radiochemotherapy for esophageal cancer: Implications for response
classification. Ann Surg. 242:684–692. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sobin L, Gospodarowicz M and Wittekind C:
TNM Classification of Malignant Tumours. 7th edition.
Wiley-Blackwell; 2009
|
|
18
|
Merkelbach-Bruse S, Dietmaier W, Füzesi L,
Gaumann A, Haller F, Kitz J, Krohn A, Mechtersheimer G, Penzel R,
Schildhaus HU, et al: Pitfalls in mutational testing and reporting
of common KIT and PDGFRA mutations in gastrointestinal stromal
tumors. BMC Med Genet. 11:1062010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Grünewald I, Vollbrecht C, Meinrath J,
Meyer MF, Heukamp LC, Drebber U, Quaas A, Beutner D, Hüttenbrink
KB, Wardelmann E, et al: Targeted next generation sequencing of
parotid gland cancer uncovers genetic heterogeneity. Oncotarget.
6:18224–18237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Leach FS, Tokino T, Meltzer P, Burrell M,
Oliner JD, Smith S, Hill DE, Sidransky D, Kinzler KW and Vogelstein
B: p53 mutation and MDM2 amplification in human soft tissue
sarcomas. Cancer Res. 53:2231–2234. 1993.PubMed/NCBI
|
|
21
|
Higashiyama M, Doi O, Kodama K, Yokouchi
H, Kasugai T, Ishiguro S, Takami K, Nakayama T and Nishisho I: MDM2
gene amplification and expression in non-small cell lung cancer:
Immunohistochemical expression of its protein is a favourable
prognostic marker in patients without p53 protein accumulation. Br
J Cancer. 75:1302–1308. 1997. View Article : Google Scholar
|
|
22
|
Marchetti A, Buttitta F, Pellegrini S,
Merlo G, Chella A, Angeletti CA and Bevilacqua G: MDM2 gene
amplification and overexpression in non-small cell lung carcinomas
with accumulation of the p53 protein in the absence of p53 gene
mutations. Diagn Mol Pathol. 4:93–97. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao Y, Aguilar A, Bernard D and Wang S:
Small-molecule inhibitors of the MDM2-p53 protein-protein
interaction (MDM2 inhibitors) in clinical trials for cancer
treatment. J Med Chem. 58:1038–1052. 2015. View Article : Google Scholar :
|
|
24
|
Tanière P, Martel-Planche G, Puttawibul P,
Casson A, Montesano R, Chanvitan A and Hainaut P: TP53 mutations
and MDM2 gene amplification in squamous-cell carcinomas of the
esophagus in south Thailand. Int J Cancer. 88:223–227. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tanière P, Martel-Planche G, Maurici D,
Lombard-Bohas C, Scoazec JY, Montesano R, Berger F and Hainaut P:
Molecular and clinical differences between adenocarcinomas of the
esophagus and of the gastric cardia. Am J Pathol. 158:33–40. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen B, Xiong MM and Meng XL: Current
evidence on the relationship between murine double minute 2 T309G
polymorphism and esophageal cancer susceptibility. Dis Esophagus.
28:593–601. 2015. View Article : Google Scholar
|
|
27
|
Renouf DJ, Zhai R, Sun B, Xu W, Cheung WY,
Heist RS, Kulke MH, Cescon D, Asomaning K, Marshall AL, et al:
Association of MDM2 T309G and p53 Arg72Pro polymorphisms and
gastroesophageal reflux disease with survival in esophageal
adenocarcinoma. J Gastroenterol Hepatol. 28:1482–1488. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hsu HS, Chen HW, Kao CL, Wu ML, Li AF and
Cheng TH: MDM2 is overexpressed and regulated by the eukaryotic
translation initiation factor 4E (eIF4E) in human squamous cell
carcinoma of esophagus. Ann Surg Oncol. 18:1469–1477. 2011.
View Article : Google Scholar
|
|
29
|
Soslow RA, Altorki NK, Yang G, Xie D and
Yang CS: mdm-2 expression correlates with wild-type p53 status in
esophageal adenocarcinoma. Mod Pathol. 12:580–586. 1999.PubMed/NCBI
|