Introduction
Breast cancer is a major health problem and the
leading cause of cancer-related mortality in women. Clinically,
breast cancer is a highly heterogeneous adenocarcinoma with various
molecular and genetic alterations. For example, Perou et al
(1), were the first to carry out a
gene expression profile analysis of human breast tissue specimens
using gene microarray technology and identified a number of
differentially expressed genes associated with breast cancer
development. Subsequent studies using cDNA microarray were able to
classify breast cancer into four different subtypes: luminal-like,
normal breast-like Her-2 overexpression, and basal-like subtype
(2,3). Each subtype has unique gene expression
characteristics, biological behavior, and clinical prognosis
(4). For example, luminal-like
breast cancer is characterized by a high level of ER expression and
better prognosis, whereas the other three subtypes have negative ER
expression. The basal-like breast cancer subtype has a unique
phenotype, poorer prognosis, and insensitivity to endocrine and
targeted therapy. Thus, research attention has focused on the
basal-like subtype of breast cancer.
Basal-like breast cancer accounts for 8−20% of all
invasive breast cancers or ~25% of advanced and difficult-to-treat
breast cancer (5). Clinically,
basal-like breast cancer occurs at a relatively young age and often
expresses basal-cell cytokeratin subtypes (CK5/6, CK14, and CK17),
but does not express hormone receptors (ER or PR) or Her-2.
Eighty-five percent of tumor tissues have mutated p53 protein and
60% of cases express epidermal growth factor receptor (EGFR)
(6,7), indicating that this subtype of breast
cancer has a highly proliferative and invasive clinical behavior
with poor survival rate. At the gene level, multiple gene signaling
pathways are involved in growth and metastasis of breast cancer,
such as the mitogen-activated protein kinase (MEK)-extracellular
regulated protein kinase (ERK) pathway and phosphatidylinositol
3-kinase (PI3K)-serine/threonine protein kinase (AKT) pathway
(8). The MEK-ERK pathway is closely
related to tumor cell proliferation and resistance to apoptosis
and, thus, contributes to tumor development and progression, while
the PI3K/AKT pathway is one of the main anti-apoptotic mechanisms
utilized by tumor cells to create a survival advantage (9,10).
The SIAH family of proteins are homologues of the
Drosophila SIAH protein, and humans have two highly
conserved SIAHs: SIAH1 and SIAH2 (11). SIAH2, localized at chromosome
3q25, contains two exons and a 9.8-kb intron and encodes a
324-amino-acid protein (12) that
is only expressed in proliferating cells. Schmidt et al
(13) showed that the SIAH2 protein
is expressed in pancreatic, breast, lung, and cervical cancers and
the level of SIAH2 protein is associated with increased tumor
malignancy. Thus, SIAH2 is considered to be an oncogene. Recent
studies revealed that SIAH2-regulated cell proliferation and
apoptosis occur through activation of the ERK pathway (13). Inhibition of SIAH2 activity led to
upregulation of Spry2 expression, one of its substrates, and
subsequently a decrease in ERK activity (14,15)
and inhibition of tumor growth and metastasis. For example, Ahmed
et al (15) found that
inhibition of SIAH2 expression using SIAH2 shRNA downregulated the
level of p-ERK and, thus, inhibited tumor growth and proliferation
and promoted apoptosis. Schmidt et al (13) found that inhibition of SIAH2
expression using RNAi inhibited Ras-mediated malignant
transformation of fibroblasts and xenograft growth of pancreatic
tumor cells in nude mice, and that knockdown of SIAH2 expression in
K-Ras-transformed pancreatic tumor cells led to downregulation of
the p-ERK level. Thus, in this study, it was hypothesized that
SIAH2 or its substrates may be part of the feedback loop of the
Ras/Raf/MEK/ERK pathway in breast cancer cells. Therefore,
immunohistochemistry, western blotting, and gene knockdown studies
were performed to detect SIAH2 expression in basal cell-like and
non-basal cell-like breast cancer tissue samples. The effects of
SIAH2 knockdown on the regulation of the biological behavior of
breast cancer cells and the underlying mechanism were then
investigated.
Materials and methods
Patients and specimens
In this study, a total of 200 cases of breast
tissues were retrospectively collected between 2000 and 2009 from
the Central Hospital, Shenyang Medical College (Shenyang, China).
The study was in compliance with the published Helsinki Declaration
and was approved by the Medical Ethics Committee of the Chinese
Medical University. Informed consent was obtained prior to surgery
from all enrolled patients. The formalin-fixed and
paraffin-embedded surgical tissue blocks were retrieved, sectioned,
and stained routinely with hematoxylin and eosin (H&E) and then
reviewed by two senior pathologists to carry out a histological
diagnosis according to the World Health Organization breast
carcinoma histological classification criteria (2012). In addition,
30 cases out of these 155 invasive breast cancer tissues had paired
tumor and normal tissues (at least 5 cm away from the primary tumor
lesion) and were flash-frozen in liquid nitrogen and stored in a
−80°C freezer until protein extraction.
Immunohistochemistry
Four-micron-thick sections were prepared for
immunostaining with a streptavidin-peroxidase (S-P) method
(Ultrasensitive; Maixin, Fuzhou, China) according to the
manufacturer's instructions. The primary antibody used was a
monoclonal mouse anti-SIAH2 antibody (1:200; Santa Cruz
Biotechnology, Santa Cruz, CA, USA). The colorimetric reaction was
performed using 3,3-diaminobenzidine tetrahydrochloride (DAB;
Maixin). For the negative control, the primary antibody was
replaced with a non-immunized normal goat serum. The stained tissue
sections were reviewed and scored by two pathologists
independently; brown particles appearing in the nucleus were
regarded as positive staining of the cell. The intensity of
immunostaining was scored as 1, weak; 2, moderate; and 3, intense
and the percentage of positive cells as 0%, negative; 1−50%, 1;
51−75%, 2; ≥76%, 3. Scoring was determined from at least five high
power fields (x400 magnification). The scores of each sample were
multiplied to reach a final score of 0, 1, 2, 3, 4, 6, or 9, and
the tissues were determined as negative if the final score was
<4 and positive if the score was ≥4.
Cell lines and culture
Human breast cancer MDA-MB-231 and MCF-7 cell lines
and normal human mammary epithelial cell line MCF-10A were obtained
from the American Type Culture Collection (ATCC; Manassas, VA,
USA). MDA-MB-231 cells were maintained in Leibovitz L-15 medium
with 10% fetal bovine serum (FBS), and MCF-7 cells in Dulbecco's
modified Eagle's medium (DMEM) with 10% FBS, while MCF-10A cells
were maintained in DMEM with 5% FBS, 10 µg/ml insulin, and
20 ng/ml EGF in a humidified incubator with 5% CO2 at
37°C.
According to previous studies of gene expression
profiling and gene copy number (1,16−20), MDA-MB-231 cells belong
to the basal-like subtype of breast cancer cells, while MCF-7 cells
represent luminal breast cancer cells.
Construction of vectors, transient gene
transfection, and treatment with ERK and PI3K inhibitors
The SIAH2 expression vector pcDNA3.1-flag-SIAH2 and
pcDNA3.1-flag empty vector were kindly provided by Dr Ze'ev Ronai
of The Burnham Institute (La Jolla, CA, USA). SIAH2 shRNA and the
negative control shRNA were purchased from Santa Cruz
Biotechnology. For transient gene transfections, cells were grown
and transfected with pcDNA3.1-flag-SIAH2, pcDNA3.1-flag, SIAH2
shRNA, or control shRNA using Attractene or HiperFect (both from
Qiagen) following the manufacturer's instructions. Afterwards, the
cells were subjected to western blotting and the transfected cells
were also subjected to treatment with ERK (PD98059) or PI3K
(LY294002) (both from Sigma, St. Louis, MO, USA) inhibitor at 10
µmol/l for 24 h.
Protein extraction and western
blotting
Total cellular protein was extracted using lysis
buffer and the supernatants were collected by centrifugation of the
cell lysis at 15,000 rpm for 30 min at 4°C. The protein lysis was
boiled for 5 min and 60 µg from each protein sample each was
separated on a 12% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) gel and transferred onto PVDF membranes.
After blocking with 5% skim milk solution in PBS, the membranes
were incubated with a primary antibody against SIAH2 (at a dilution
of 1:200), p-ERK (1:400), p-AKT (1:400), or β-actin (1:200)
overnight at 4°C. Afterwards, membranes were incubated with each
corresponding secondary antibody at room temperature for 2 h. The
protein bands were visualized using an enhanced chemiluminescence
(ECL) kit and then quantified using EC3 imaging system (UVP, LLC).
The gray scale values of each protein band were normalized to the
values of the corresponding β-actin band. The experiments were
repeated once independently.
Cell viability MTT assay
The transfected cells were seeded in 96-well plates
at 1×104 cells/well and the cell viability was evaluated
using the MTT assay. The absorbance, which was directly
proportional to the number of living cells in culture, was measured
at 490 nm using a microplate reader. A blank with dimethyl
sulfoxide alone was measured and subtracted from all values. The
experiments were performed in triplicate and repeated at least
three times independently.
Tumor cell Matrigel invasion assay
Cell invasive ability was assessed using a 24-well
Transwell with 8-µm pore polycarbonate membrane inserts.
Matrigel (100 µg/ml at dilution of 1:7 with cell culture
medium) was applied to the upper surface of the membranes. After
transfection for 24 h, cells were seeded in the upper chamber at a
density of 5×104 cells/well and the bottom of the
chamber was filled with cell culture medium with 20% FBS. Next the
plates were incubated for 24 h. At the end of the experiments,
cells that had invaded into the surface of the membrane were fixed
with methanol and stained with hematoxylin. Five high-magnification
microscope fields were randomly selected per filter to count the
number of invaded cells.
Flow cytometric apoptosis assay
Annexin V/FITC double staining was carried out as
described by the manufacturer (Invitrogen, Carlsbad, CA, USA).
Cells were immediately analyzed by flow cytometry and quantified
using CellQuest software (both from BD Biosciences) for the
percentage of apoptotic cells in each experimental group.
Statistical analysis
All statistical calculations were performed by SPSS
version 13.0 for Windows software (SPSS, Chicago, IL, USA). The
χ2 test and one-way analysis of variance (ANOVA) were
performed to assess statistically significant differences between
groups of tissues or cell treatments. A P<0.05 was considered
statistically significant.
Results
Characteristics of the patients
The 200 breast cancer cases included 10 normal
breast tissues (NBTs), 35 ductal carcinoma in situ (DCIS)
tissues, and 155 invasive breast carcinoma (IBC) tissues. Among the
155 IBCs, 74% (115/155) were invasive ductal carcinomas, 12%
(18/155) were invasive lobular carcinomas, 10% (15/155) were
medullary carcinomas, and 4% (7/155) were of other histological
types. Clinicopathological data and expression of p53 and Ki67
proteins were also collected. In brief, the median age of the
patients was 50 years old (ranging between 28 and 84 years) and the
median tumor size was 25 mm (ranging between 6 and 90 mm) (Table I). Moreover, 28 patients were
identified as basal-like breast cancers by immunohistochemistry on
the basis of Nielsen's (21)
method. Specifically, the basal-like breast cancers often express
high molecular weight basal cytokeratin (CK5/6, CK14, and CK17)
and/or EGFR, while they do not express hormone receptors [estrogen
receptor (ER) and progesterone receptor (PR)] or Her-2/neu.
However, according to Nielsen's criteria, the 155 breast cancer
cases were categorized into five intrinsic subgroups: the
basal-like group (ER−, HER-2−,
CK5/6+ and/or EGFR+), the luminal A group
(ER+ and HER-2−), the luminal B group
(ER+ and Her-2+), the Her-2 group
(ER− and Her-2+) and the negative (null)
group (ER−, Her-2−, CK5/6− and
EGFR−).
 | Table IAssociation of SIAH2 expression with
clinicopathological factors in 155 invasive breast carcinomas. |
Table I
Association of SIAH2 expression with
clinicopathological factors in 155 invasive breast carcinomas.
| Clinicopathological
factors | N | SIAH2 expression
| P-value |
|---|
| Positive | Negative |
|---|
| Age (years) | | | | 0.883 |
| <50 | 56 | 27 | 29 | |
| ≥50 | 99 | 51 | 48 | |
| Tumor size
(mm) | | | | 0.693 |
| <25 | 87 | 45 | 42 | |
| ≥25 | 68 | 33 | 35 | |
| Nuclear grade | | | | 0.000 |
| I | 17 | 3 | 14 | |
| II | 60 | 21 | 39 | |
| III | 78 | 54 | 24 | |
| Tumor stage | | | | 0.572 |
| I | 40 | 18 | 22 | |
| II | 81 | 44 | 37 | |
| III | 34 | 16 | 18 | |
| Lymph node
metastasis | | | | 0.284 |
| No | 90 | 42 | 48 | |
| Yes | 65 | 36 | 29 | |
| Genotyping | | | | 0.000 |
| Basal-like | 28 | 25 | 3 | |
| Luminal A | 49 | 9 | 40 | |
| Luminal B | 10 | 5 | 5 | |
| Her-2 | 22 | 15 | 7 | |
| Null | 46 | 24 | 22 | |
Differential expression of SIAH2 protein
in breast cancer tissues and cell lines
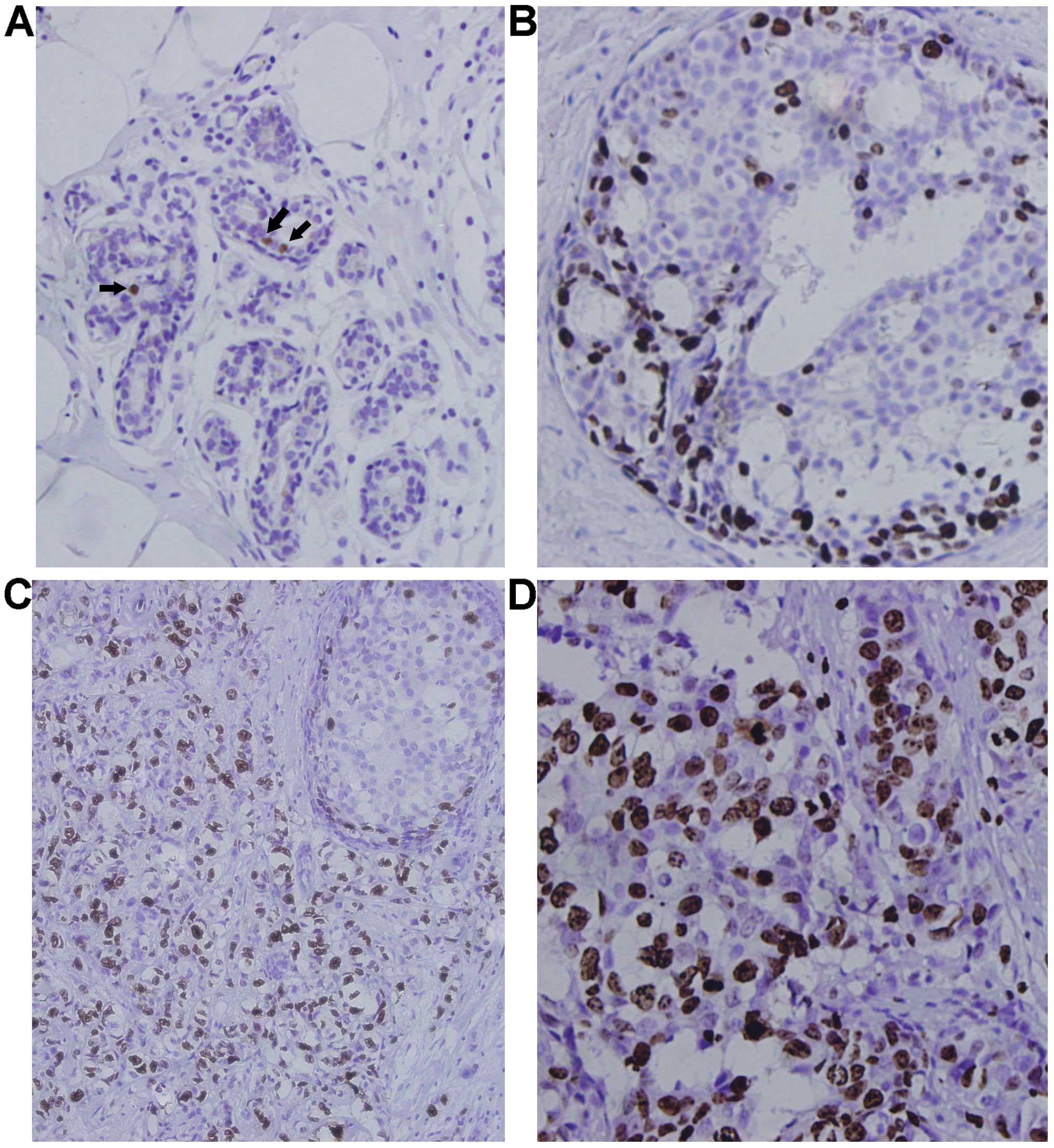
SIAH2 protein was expressed in the nuclei of
occasional cells within the luminal layer of ducts and acini in
normal mammary glands in 4 out of the 10 cases (40%). The level of
SIAH2 expression was usually of mild to moderate intensity and, if
used as a cut-off point for the tumor tissues, SIAH2 expression was
considered negative in normal tissues (Fig. 1A). SIAH2 expression was positive and
of moderate to strong intensity with a homogeneous distribution in
the nuclei of 5 out of 35 (14.29%) DCIS cases (Fig. 1B and C). In contrast, the SIAH2
protein was expressed in 50.32% (78/155) of invasive breast
carcinoma tissues with the majority of a strong intensity (Fig. 1C and D). SIAH2 expression was
significantly upregulated in IBCs compared to normal or DCIS
tissues (P=0.000; Table II and
Fig. 1E), whereas there was no
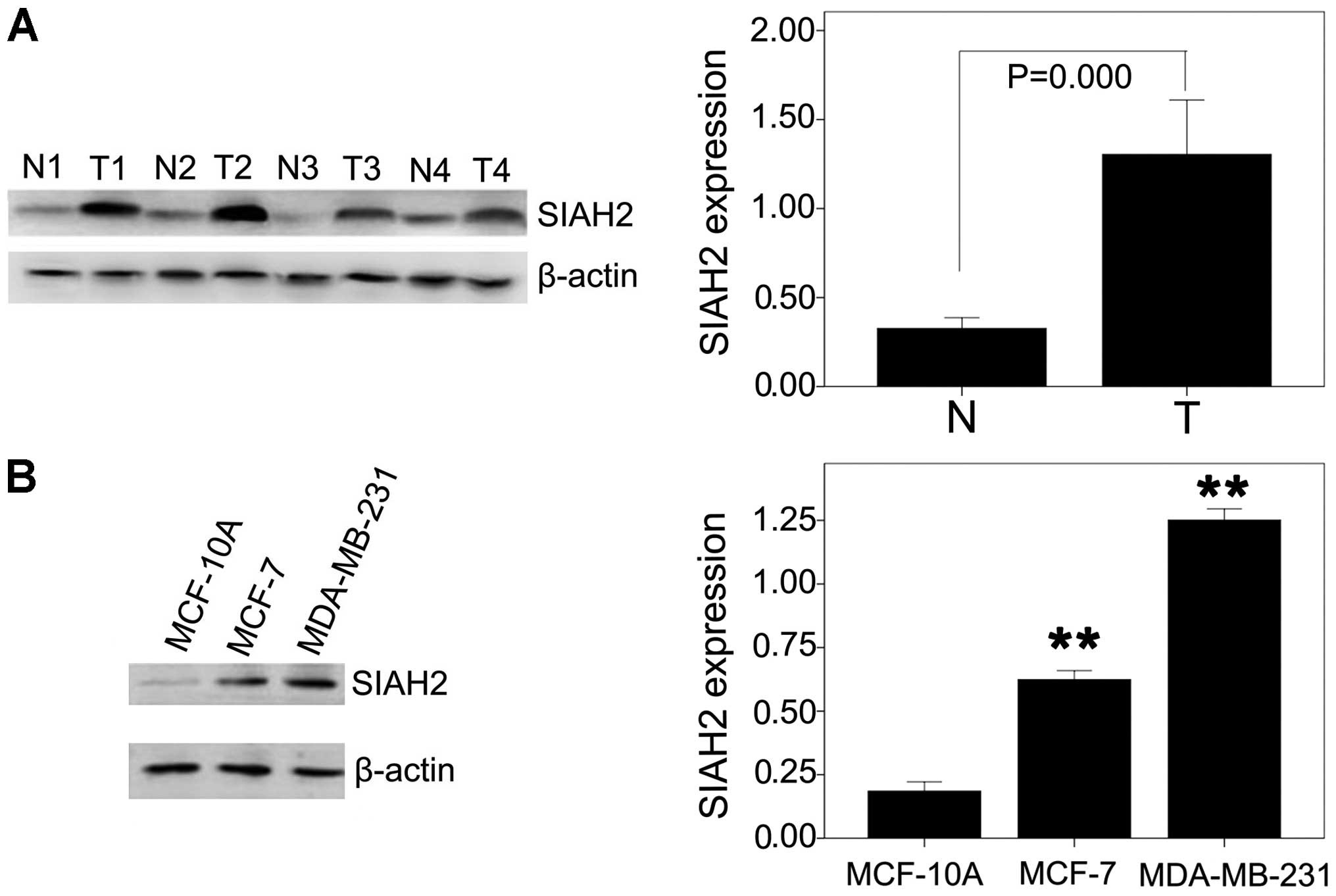
significance between normal and DCIS tissues (P=0.102). Our western
blot data also confirmed upregulation of SIAH2 expression in IBCs
compared to the paired distant normal tissues (t=6.558, P=0.000;
Fig. 2A).
 | Table IIDifferential expression of the SIAH2
protein in breast tissues (n=200). |
Table II
Differential expression of the SIAH2
protein in breast tissues (n=200).
| Diagnosis | N | Mean of IS staining
score | P-value | SIAH2 expression
| P-value |
|---|
| Low (IS<4)
n/total (%) | High (IS≥4) n/total
(%) |
|---|
| NBT | 10 | 1.10±1.45 | 0.000 | 10/10 (100) | 0/10 (0) | 0.000 |
| DCIS | 35 | 2.03±1.52 | | 30/35 (85.71) | 5/35 (14.29) | |
| IBC | 155 | 4.01±1.60 | | 77/155 (49.68) | 78/155 (50.32) | |
Association of SIAH2 expression with
clinicopathological data and p53 and Ki67 expression
Next, the association of SIAH2 expression with
clinicopathological data and p53 and Ki67 expression was
investigated in the 155 IBC cases. The data showed that SIAH2
expression was associated with a higher tumor nuclear grade and
molecular classification of breast cancer subtypes. Moreover, SIAH2
expression was associated with p53 and Ki67 expression and
inversely associated with lymph node metastasis in basal-like IBC.
In the luminal A and null subtype of IBC, SIAH2 expression was
associated with Ki67 expression (Tables
I, III and V). However, there was no association found
between SIAH2 expression and other clinicopathological data.
 | Table IIIAssociation of SIAH2 expression with
p53 level in each subtype of breast cancer. |
Table III
Association of SIAH2 expression with
p53 level in each subtype of breast cancer.
| SIAH2 positive | SIAH2 negative | Spearman's
correlation (rs) | P-value |
|---|
| Basal-like |
|
p53+ | 20 | 0 | 0.548 | |
|
p53− | 5 | 3 | | 0.017 |
| Luminal A |
|
p53+ | 2 | 5 | 0.108 | |
|
p53− | 7 | 35 | | 0.598 |
| Luminal B |
|
p53+ | 2 | 1 | 0.218 | |
|
p53− | 3 | 4 | | 1.00 |
| Her-2 |
|
p53+ | 6 | 2 | 0.111 | |
|
p53− | 9 | 5 | | 0.671 |
| Null |
|
p53+ | 10 | 9 | 0.008 | |
|
p53− | 14 | 13 | | 1.00 |
 | Table VAssociation of SIAH2 expression with
tumor lymph node metastasis in each subtype of breast cancer. |
Table V
Association of SIAH2 expression with
tumor lymph node metastasis in each subtype of breast cancer.
| SIAH2 positive | SIAH2 negative | Spearman's
correlation (rs) | P-value |
|---|
| Basal-like |
| Lymph node
metastasis positive | 6 | 3 | −0.503 | |
| Lymph node
metastasis negative | 19 | 0 | | 0.026 |
| Luminal A |
| Lymph node
metastasis positive | 3 | 14 | −0.014 | |
| Lymph node
metastasis negative | 6 | 26 | | 1.00 |
| Luminal B |
| Lymph node
metastasis positive | 3 | 2 | 0.200 | |
| Lymph node
metastasis negative | 2 | 3 | | 1.00 |
| Her-2 |
| Lymph node
metastasis positive | 12 | 4 | 0.239 | |
| Lymph node
metastasis negative | 3 | 3 | | 0.334 |
| Null |
| Lymph node
metastasis positive | 12 | 6 | 0.233 | |
| Lymph node
metastasis negative | 12 | 16 | | 0.14 |
Change in breast cancer cell malignant
behaviors after knockdown of SIAH2 expression
According to the genotyping and gene expression
analysis by Neve et al (18), MCF-7 breast cancer cells were chosen
as a luminal breast cancer cell line and MDA-MB-231 cells as a
basal-like type of breast cancer in the current study. A MCF-10A
normal mammary epithelial cell line was used as a control. It was
found that the SIAH2 protein was highly expressed in the MDA-MB-231
and MCF-7 cells, whereas the MCF-10A cell line had a very low
expression level of SIAH2 protein (P<0.05; Fig. 2B). MDA-MB-231 cells expressed a much
higher level of SIAH2 protein than MCF-7 cells (P<0.05, Fig. 2B).
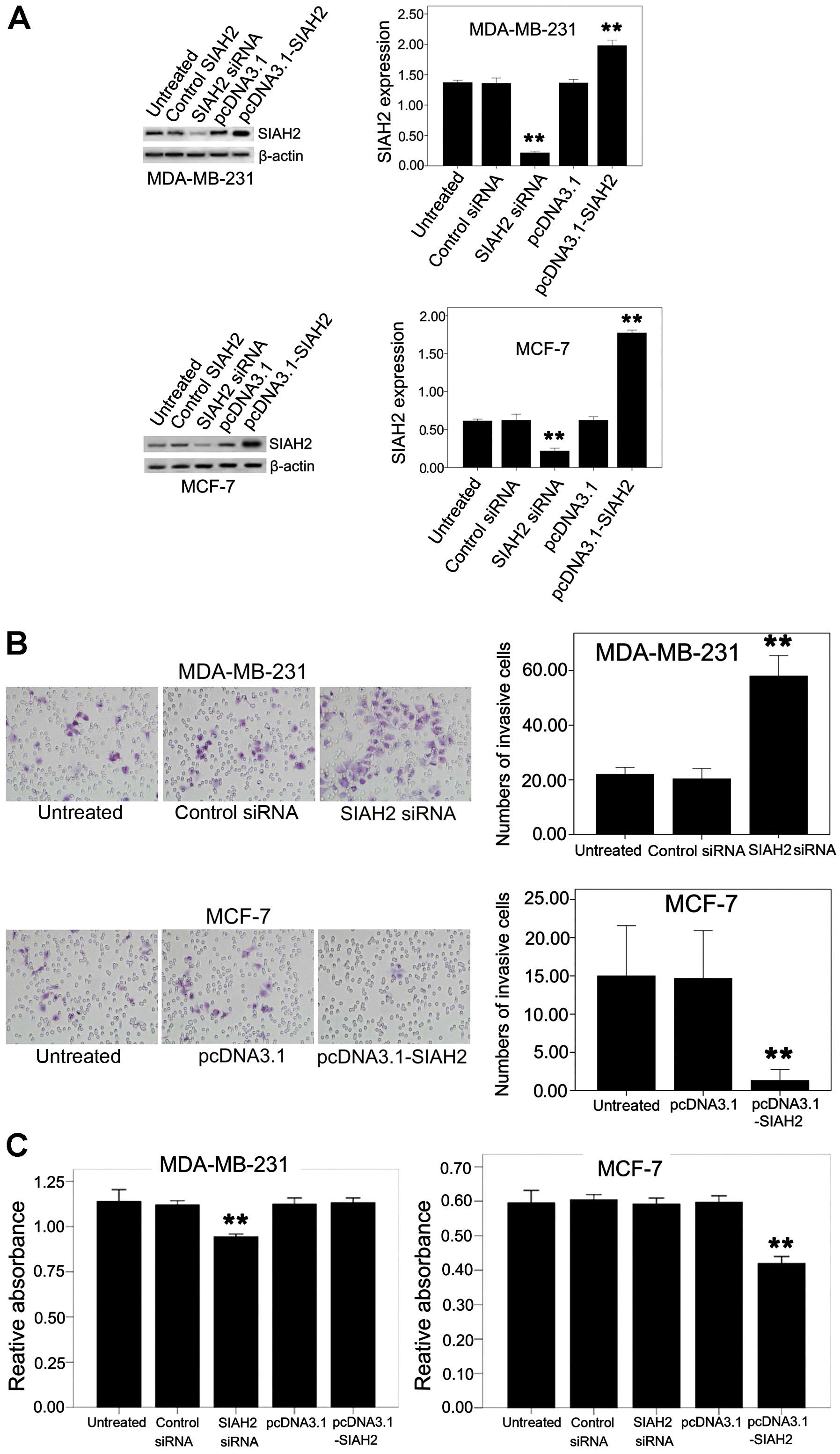
Next, the effects of SIAH2 expression and knockdown
on the regulation of breast cancer cell proliferation and invasion
capacity was investigated by transient transfection of
pcDNA3.1-flag-SIAH2 and SIAH2 siRNA, respectively, into breast
cancer cell lines. Western blotting data showed SIAH2 expression or
knockdown in breast cancer cell lines (Fig. 3A). A tumor cell invasion assay
showed that knockdown of SIAH2 expression in the MDA-MB-231 cells
with high SIAH2 expression resulted in induction of tumor cell
invasion capacity, whereas restoration of SIAH2 expression in the
MCF-7 cells reduced tumor cell invasion (Fig. 3B).
Furthermore, knockdown of SIAH2 expression reduced
the viability of breast cancer MDA-MB-231 cells, whereas
overexpression of SIAH2 protein also reduced cell viability in the
MCF-7 luminal-like breast cancer cell line (Fig. 3C).
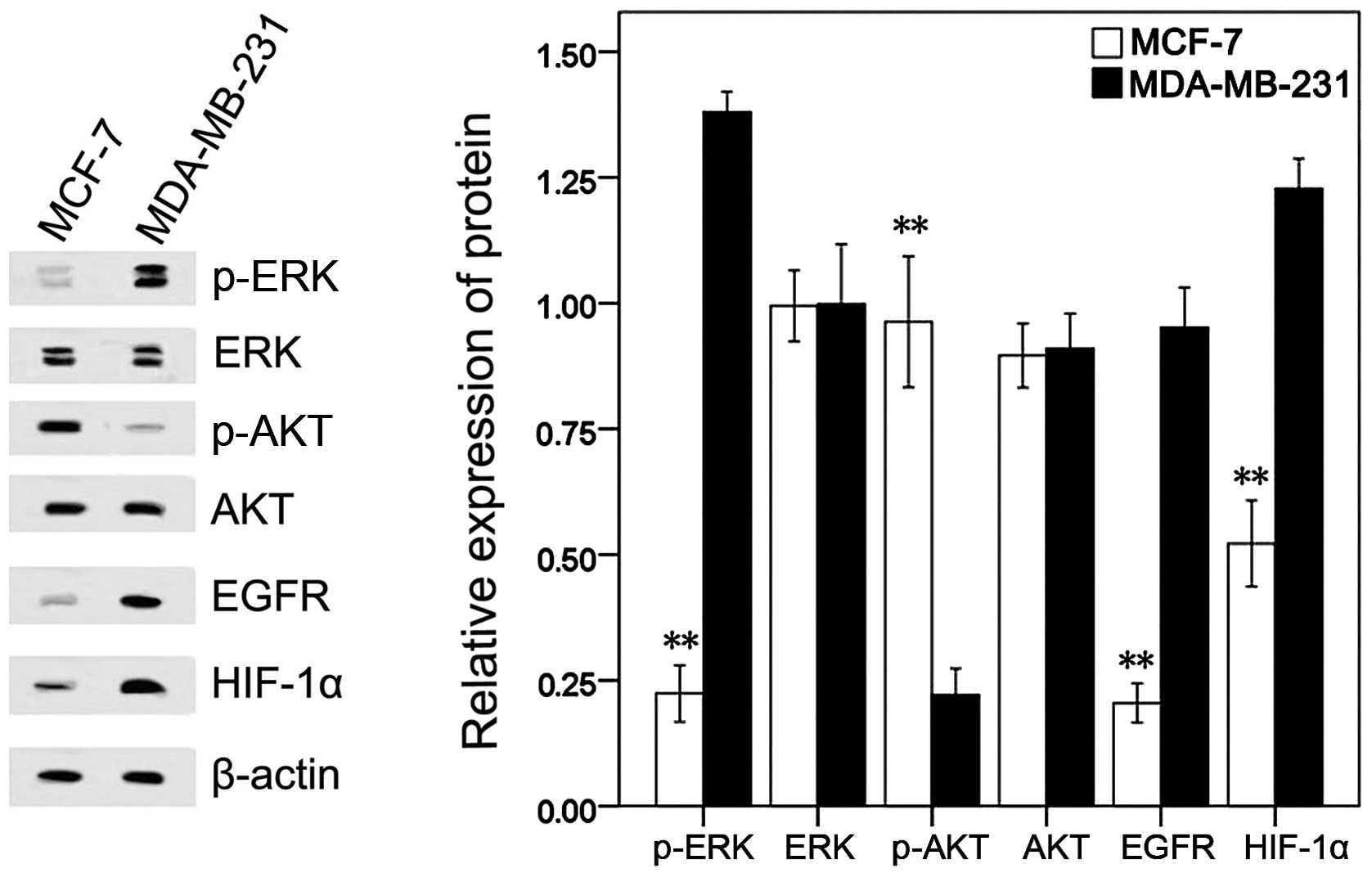
Additionally, the underlying mechanism for the
different roles of the SIAH2 protein in the various breast cancer
cells was explored. Our data showed that the MDA-MB-231 cells had a
higher level of p-ERK, EGFR and HIF-1α expression, but a lower
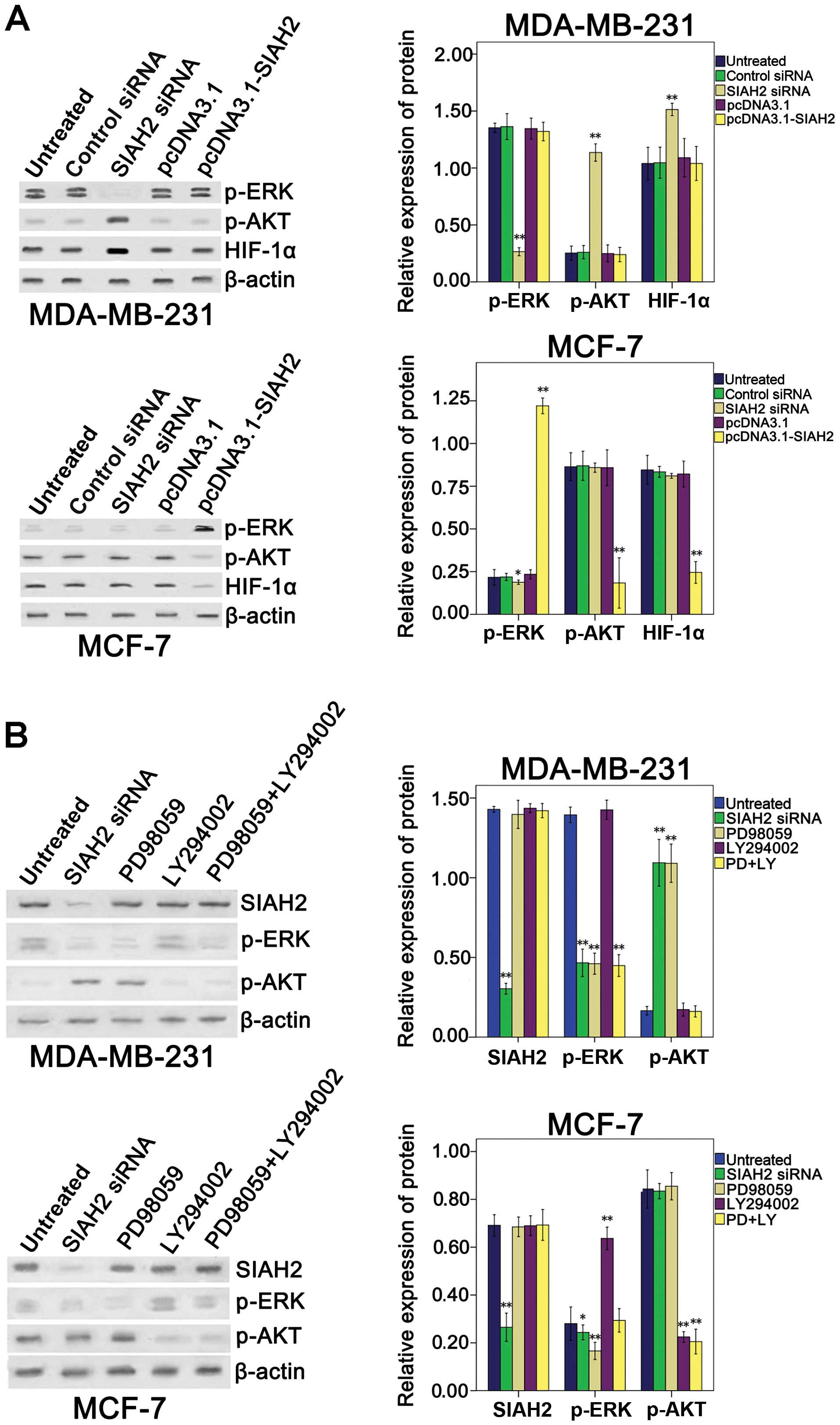
level of p-AKT than the MCF-7 cells (Fig. 4).
'Cross-talk' between the ERK and PI3K
pathways in breast cancer cell lines
To investigate the effect of SIAH2 on regulation of
p-ERK, p-AKT and HIF-1α expression, SIAH2 shRNA or
pcDNA3.1-flag-SIAH2 was transfected into the breast cancer cell
lines. Overexpression of SIAH2 in the MDA-MB-231 cell line had no
effect on p-ERK, p-AKT or HIF-1α expression, whereas knockdown of
SIAH2 expression was able to reduce the level of p-ERK, but induced
the levels of p-AKT and HIF-1α. In contrast, overexpression of
SIAH2 protein in the MCF-7 cell line increased the p-ERK level, but
decreased the p-AKT and HIF-1α levels, whereas knockdown of SIAH2
expression slightly decreased p-ERK expression, but had no effects
on the p-AKT level (Fig. 5A).
To determine the relationship between the ERK and
PI3K pathways in the breast cancer cells, the cells were
transfected with SIAH2 shRNA and then treated with p-ERK and/or
PI3K inhibitor (PD98059 and/or LY294002, respectively). The results
showed that, in the MDA-MB-231 basal-like breast cancer cell line,
inhibition of SIAH2 expression or blocking the activity of the ERK
pathway caused negative feedback activation of the PI3K pathway
(Fig. 5B). Similarly, in the MCF-7
luminal breast cancer cell line, blockage of the PI3K pathway
caused negative feedback activation of the ERK pathway (Fig. 5B).
Effects of SIAH2 on regulation of breast
cancer cell invasion capacity through feedback
activation/inhibition of the PI3K pathway
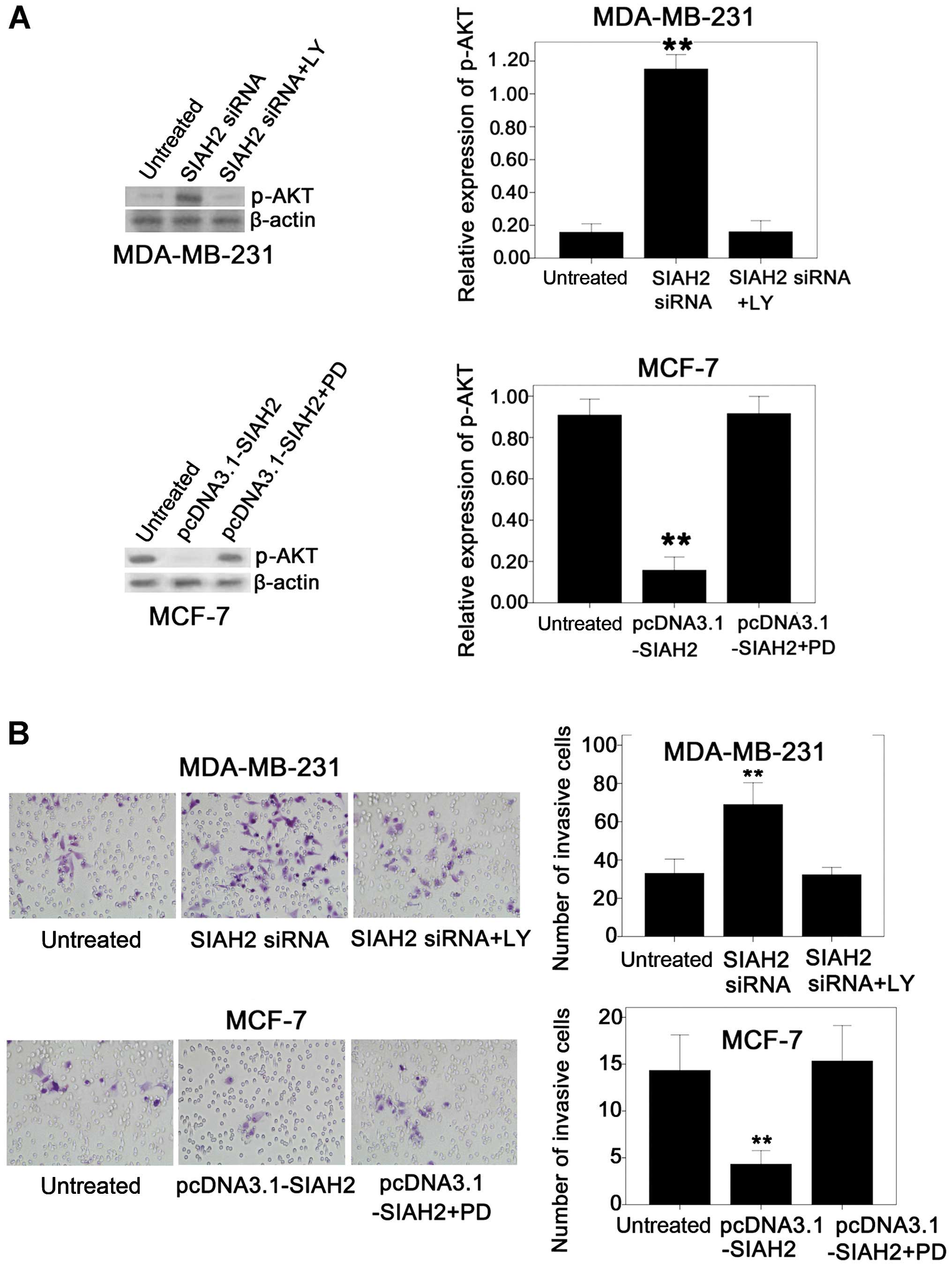
Next, the effects of SIAH2 on regulation of breast
cancer cell invasion capacity and the underlying molecular events
were assessed. An invasion assay showed that the invasion capacity
of the MDA-MB-231 cells was increased after knockdown of SIAH2
expression, whereas the invasion capacity of the MCF-7 cells was
decreased after transfection with SIAH2 cDNA. We speculate that
these results can be partially explained by the feedback activation
of the PI3K pathway and increased expression of HIF-1α after SIAH2
silencing, or blockage of the PI3K pathway and a decrease in HIF-1α
expression due to overexpression of SIAH2.
To prove whether the increase in tumor cell invasion
ability after knockdown of SIAH2 expression was caused by feedback
activation of the PI3K pathway in the MDA-MB-231 cell line, the
cells were treated with SIAH2 siRNA and PI3K inhibitor (LY294002)
simultaneously. The data showed that the induced tumor cell
invasion capacity after SIAH2 knock down was reversed by treatment
with the PI3K inhibitor in the MDA-MB-231 cells (Fig. 6A). In contrast, treatment with
LY294002 alone had no effect on MDA-MB-231 cell invasion capacity
(Fig. 6B).
Similarly, in the MCF-7 cells, SIAH2 overexpression
and treatment with the ERK inhibitor (PD98059) simultaneously
recovered p-AKT expression and activity of the PI3K pathway
activity (Fig. 6A); thus, tumor
cell invasion capacity was also recovered (Fig. 6B). These data indicate that the
effects of SIAH2 on the regulation of breast cancer cell invasion
capacity occurred through feedback activation/inhibition of the
PI3K pathway.
Effect of SIAH2 shRNA and ERK and PI3K
inhibitors on regulation of cell viability
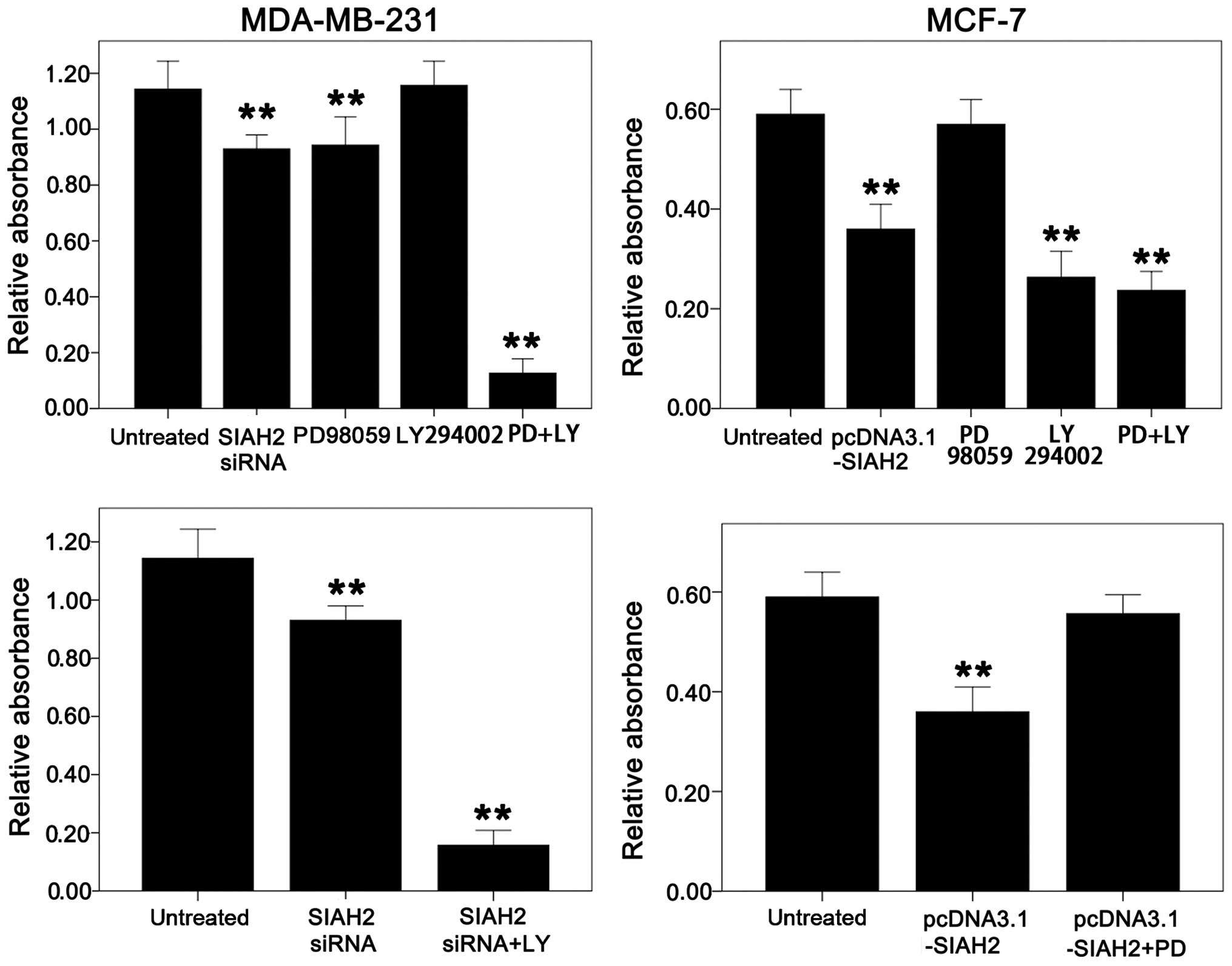
Next, the effect of SIAH2 on the regulation of cell
viability in the MDA-MB-231 and MCF-7 cells was assessed. MTT data
showed that knockdown of SIAH2 expression suppressed MDA-MB-231
cell viability, whereas overexpression of SIAH2 protein did not
affect cell viability. In contrast, knockdown of SIAH2 expression
did not significantly affect MCF-7 cell viability, but
overexpression of SIAH2 protein decreased MCF-7 cell viability.
Furthermore, MDA-MB-231 cell viability was decreased after
treatment with the p-ERK inhibitor (PD98059) and addition of SIAH2
siRNA, while addition of the PI3K inhibitor (LY294002) had no
effect on MDA-MB-231 cell viability. Upon combined treatment with
these two inhibitors (PD98059 and LY294002), MDA-MB-231 cell
viability was significantly suppressed (Fig. 7). This result could be due to
feedback activation of the PI3K pathway after knockdown of SIAH2
expression or inhibition of the ERK pathway by PD98059 treatment.
The PI3K pathway could mediate the biological effects on inhibition
of apoptosis, which partly counteracted the effects on inhibition
of cell proliferation caused by SIAH2 knockdown or PD98059
treatment. Addition of these two inhibitors (PD98059 and LY294002)
together terminated the decreased apoptosis by activation of the
PI3K pathway. Therefore, MDA-MB-231 cell viability was slightly
reduced after addition of SIAH2 siRNA (0.93±0.017) or PD98059
(1.10±0.018 in LY294002-treated cells vs. PD98059-only-treated
cells (0.9487±0.025) and control cells (1.133±0.021). Cell
viability was significantly reduced after addition of these two
inhibitors (PD98059 and LY294002) together (1.10±0.018 in
LY294002-treated cells or 0.14±0.014 in LY294002- and
PD98059-treated cells vs. 1.133±0.025 in control cells). However,
in the MCF-7 cells, addition of PD98059 had no significant effect
on cell viability, but addition of LY294002 significantly decreased
cell viability. The combination of these two inhibitors (PD98059
and LY294002) caused the same degree of cell proliferation
reduction as LY294002 alone (Fig.
7), indicating that there was no effect on cell viability after
activation of the ERK pathway plus treatment with LY294002 in the
MCF-7 cells. All in all, these results suggest that MDA-MB-231
cells are sensitive to the ERK inhibitor, whereas MCF-7 cells are
sensitive to the PI3K inhibitor.
To prove whether the slight decrease in cell
viability after knockdown of SIAH2 expression was caused by
feedback activation of the PI3K pathway in the MDA-MB-231 cells,
the cells were transfected with SIAH2 siRNA and then treated with
the PI3K inhibitor (LY294002). It was found that cell viability was
reduced significantly (Fig. 7).
Similarly, the MCF-7 cells were transfected with SIAH2 cDNA and
then treated with the ERK inhibitor (PD98059); it was found that
with the recovery of p-AKT expression, cell viability was also
recovered (Fig. 7).
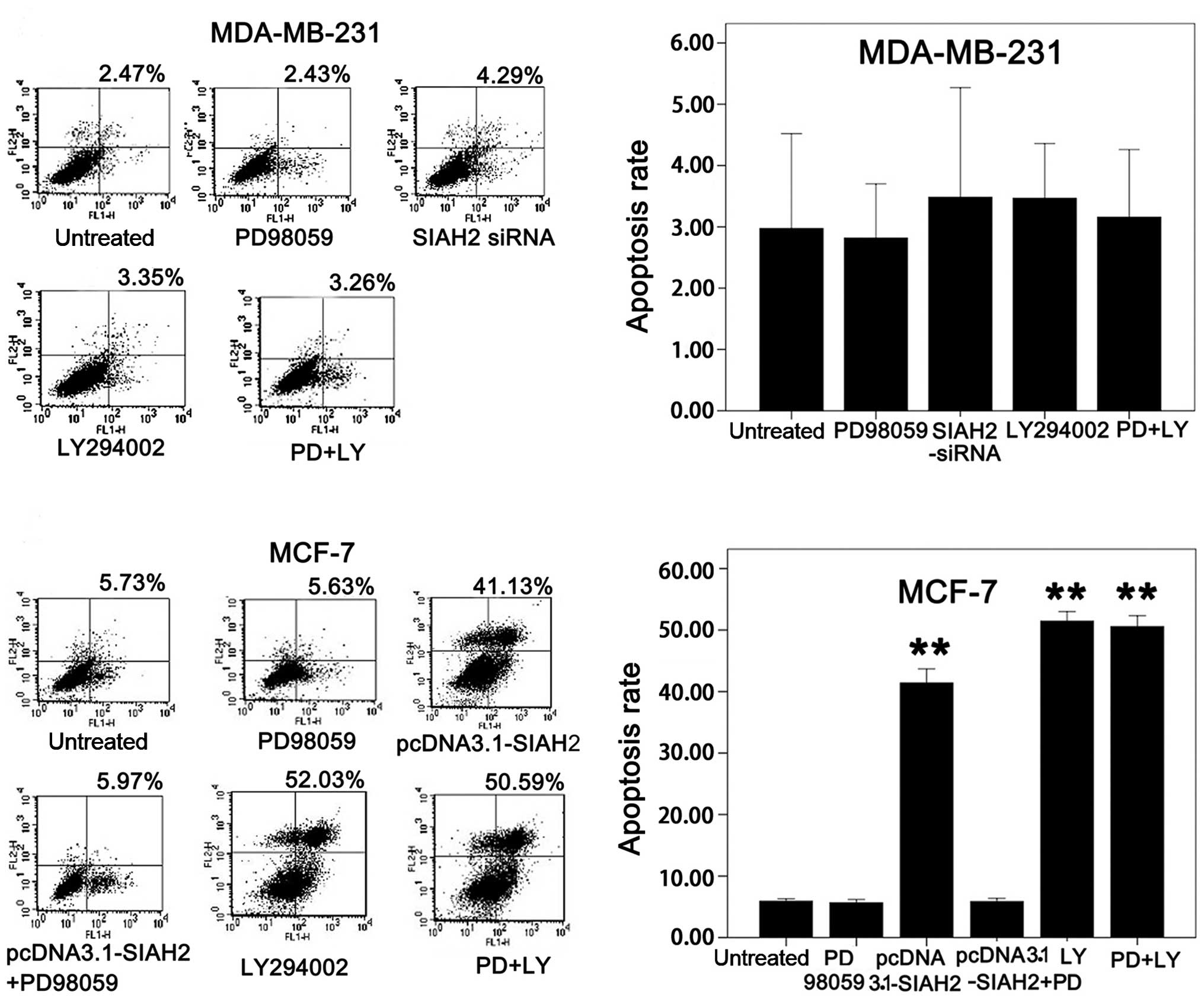
Similar data were obtained in SIAH2 knockdown as
well as treatment with the ERK and PI3K inhibitors on regulation of
tumor cell apoptosis between MDA-MB-231 and MCF-7 cells (Fig. 8).
Discussion
In the present study, the expression of the SIAH2
protein in normal, premalignant, and malignant breast tissues was
analyzed for association with clinicopathological data from IBC
patients. The effects of SIAH2 expression or knockdown on the
regulation of breast cancer cell biology behavior and the
underlying molecular events were then explored. It was found that
expression of the SIAH2 protein was upregulated in IBCs compared to
normal or DCIS tissues. SIAH2 expression was associated with a
higher tumor nuclear grade and molecular classification of breast
cancer subtypes. The in vitro data showed that manipulation
of SIAH2 expression (overexpression or knockdown) led to
'cross-talk' of the ERK and PI3K pathways in different subtypes of
breast cancer cell lines (luminal- vs. basal-like), which could be
one of the mechanisms by which SIAH2 regulates viability,
apoptosis, and invasion capacity in breast cancer cells.
SIAH2 protein is a ring finger E3 ubiquitin ligase
involved in hypoxia response and regulation of the Ras/Raf and
p38/JNK/NF-κB signaling pathway. SIAH2 substrates include DCC
(22), OBF-1 (23,24),
TRAF2 (25), PHD1/3 (26,27),
Vav1 (28), and Spry2 (29). SIAH2 substrates can interact with
the SIAH2 protein in an AXVXP motif-dependent or independent way to
affect multiple aspects of cell functions, such as cell survival,
mitochondrial biogenesis, and hypoxia and stress response. Thus,
overexpression of the SIAH2 protein may lead to breast cancer
development, which is consistent with our current data.
Furthermore, our current study also demonstrated that SIAH2
expression was associated with tumor nuclear grade and molecular
classification of breast cancer subtypes. Higher expression of
SIAH2 protein led to higher IBC nuclear grade and contributed to
basal-like breast cancer. A previous study also showed that SIAH2
was expressed in pancreatic, breast, lung, and cervical cancers and
different tumor cell lines and that the level of SIAH2 expression
increased with tumor cell malignant behavior (13).
Furthermore, our current data also showed that SIAH2
expression or knockdown led to 'cross-talk' of the ERK and PI3K
pathways, which may be one of the mechanisms by which SIAH2
regulates viability, apoptosis, and invasion capacity in breast
cancer cells. Indeed, alteration of multiple signaling pathways has
been shown to occur in breast cancer, including the commonly
observed MAPK and PI3K pathways (30). Previous studies revealed that
SIAH2-regulated cell proliferation and inhibition of apoptosis
occurred through activation of the ERK pathway (13,16,17).
SIAH2 was also closely related to HIF-1α; an increase in SIAH2
activity under a hypoxic condition is able to upregulate HIF-1α
level and, in turn, promote expression of its downstream target
genes, including VEGF, c-Met, CXCR4 and lysine oxidase, for tumor
growth and metastasis (31).
Moreover, our current study showed differential
effects of SIAH2 expression or knockdown on the regulation of cell
viability, apoptosis, and invasion capacity of luminal-like and
basal-like breast cancer cells (MDA-MB-231 and MCF-7 cells,
respectively). Furthermore, it was also found that overexpression
of SIAH2 in the MDA-MB-231 cell line had no effect on p-ERK, p-AKT
or HIF-1α expression, whereas knockdown of SIAH2 expression was
able to reduce the level of p-ERK, but induce p-AKT and HIF-1α
levels. In contrast, overexpression of the SIAH2 protein in the
MCF-7 cell line increased the p-ERK level, but decreased the p-AKT
and HIF-1α levels, whereas knockdown of SIAH2 expression slightly
decreased p-ERK expression, but had no effects on the p-AKT level.
These data suggest that the role of SIAH2 in different breast
cancer cell lines may be based on differential gene expression
profiles in the given cell lines. In addition, our ex vivo
data showed that SIAH2 may function differently in different
molecular subtypes (e.g., luminal- vs. basal-like type of breast
cancer) of breast cancer.
In addition, our current study assessed that the
effect of SIAH2 on breast cancer cell proliferation occurring
through activation of the ERK and PI3K pathways by transfecting
breast cancer cells with SIAH2 shRNA and then treating them with
the ERK and/or PI3K inhibitors (PD98059 and/or LY294002,
respectively). Our results showed that SIAH2 regulated the MCF-7
luminal-like breast cancer cell line growth mainly through
apoptosis in a PI3K-dependent manner, whereas suppression of the
MDA-MB-231 basal-like tumor cell line growth was mainly regulated
by the ERK pathway with little or no involvement of apoptosis.
Indeed, ERK is a member of the MAPK family and plays a crucial role
in the regulation of cell growth and division. In the ERK pathway,
Ras acts as an upstream activation protein to form the
Ras-Raf-MEK-ERK pathway. Spry2, an inhibitor of the Ras/Raf
signaling pathway, is a SIAH2 substrate and plays an important role
in cancer development. A previous study showed that changes in
Spry2 expression are associated with downregulation of Ras
signaling and that knockdown of SIAH2 expression upregulated Spry2
and weakened Ras signaling (14).
Furthermore, PI3K/AKT is an important pathway in mediating the
survival signal in cells and inhibition of AKT may trigger feedback
that activates other signaling pathways to maintain cell survival.
It has been confirmed that after inhibition of AKT signaling,
expression of the tyrosine kinase receptor is upregulated and
mTORC2 is activated in a feedback manner to activate MAPK activity,
while inhibition of mTOR downstream of AKT by rapamycin or
temsirolimus can also activate the PI3K/AKT pathway in a feedback
manner. Our current study showed cross-reaction or a competitive
relationship between the ERK and PI3K pathways and SIAH2 regulation
of breast cancer invasion mainly depended on activation of the PI3K
pathway. Thus, an unsatisfactory efficacy of a single MEK or AKT
inhibitor on breast cancer may be associated with feedback
activation, which may be the major obstacle in the development of
anticancer drugs.
However, our current study is a proof-of-principle
and additional research needs to be performed to confirm our
results. Future studies are required to study additional cell lines
to investigate SIAH2 in classifying breast cancer molecularly and
developing the protein as a biomarker for early or differential
diagnosis of breast cancer.
Abbreviations:
|
SIAH
|
seven in absentia homolog
|
|
PKB
|
protein kinase B
|
|
PDK
|
phosphatidylinositol-dependent
kinase
|
|
Ras
|
rat sarcoma
|
|
Raf
|
rapidly accelerated fibrosarcoma
|
|
MAPK
|
mitogen-activated protein kinase
|
|
ERK
|
extracellular signal-regulated
kinases
|
|
MEK
|
MAPK/ERK kinase
|
|
PI3K
|
phosphatidylinositol-3-kinase
|
|
HIF-1α
|
hypoxia-inducible factor 1α
subunit
|
|
VEGF
|
vascular endothelial growth factor
|
|
K-Ras
|
Kirsten rat sarcoma
|
|
HIF
|
hypoxia-inducible factor
|
|
VHL
|
von Hippel-Lindau
|
|
IGF
|
insulin-like growth factor
|
|
EGFR
|
epidermal growth factor receptor
|
|
NF-κB
|
nuclear factor-κB
|
|
Spry2
|
Sprouty2
|
|
mTOR
|
mammalian target of rapamycin
|
|
ER
|
estrogen receptor
|
|
PR
|
progesterone receptor
|
|
Her-2
|
human epidermal growth factor-2
|
|
RNAi
|
ribonucleic acid interference
|
|
siRNA
|
small interference ribonucleic
acid
|
|
shRNA
|
short hairpin ribonucleic acid
|
|
MTT
|
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltet-razolium
|
|
TBS
|
Tris-buffered saline
|
|
H&E
|
hematoxylin and eosin
|
|
PBS
|
phosphate-buffered saline
|
|
S-P
|
streptavidin-peroxidase method
|
|
DAB
|
3,3′-diaminobenzidine
|
|
PAGE
|
polyacrylamide gel electrophoresis
|
|
SDS
|
sodium dodecyl sulfate
|
|
ECL
|
enhanced chemiluminescence
|
|
PI
|
propidium iodide
|
|
BSA
|
bovine serum albumin
|
|
FCM
|
flow cytometry
|
|
SPSS
|
Statistical Package for Social
Science
|
|
NBT
|
normal breast tissues
|
|
DCIS
|
ductal carcinoma in situ
|
|
IBC
|
invasive breast carcinoma
|
Acknowledgments
We thank Dr Ze'ev Ronai of The Burnham Institute (La
Jolla, CA, USA) for the SIAH2 expression vector
(pcDNA3.1-flag-SIAH2).
References
|
1
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triple-negative breast cancer: Clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
West M, Blanchette C, Dressman H, Huang E,
Ishida S, Spang R, Zuzan H, Olson JA Jr, Marks JR and Nevins JR:
Predicting the clinical status of human breast cancer by using gene
expression profiles. Proc Natl Acad Sci USA. 98:11462–11467. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carey LA, Perou CM, Livasy CA, Dressler
LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S,
et al: Race, breast cancer subtypes, and survival in the Carolina
Breast Cancer Study. JAMA. 295:2492–2502. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kreike B, van Kouwenhove M, Horlings H,
Weigelt B, Peterse H, Bartelink H and van de Vijver MJ: Gene
expression profiling and histopathological characterization of
triple-negative/basal-like breast carcinomas. Breast Cancer Res.
9:R652007. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Abd El-Rehim DM, Ball G, Pinder SE, Rakha
E, Paish C, Robertson JF, Macmillan D, Blamey RW and Ellis IO:
High-throughput protein expression analysis using tissue microarray
technology of a large well-characterised series identifies
biologically distinct classes of breast cancer confirming recent
cDNA expression analyses. Int J Cancer. 116:340–350. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Savage K, Leung S, Todd SK, Brown LA,
Jones RL, Robertson D, James M, Parry S, Rodrigues Pinilla SM,
Huntsman D, et al: Distribution and significance of caveolin 2
expression in normal breast and invasive breast cancer: An
immunofluorescence and immunohistochemical analysis. Breast Cancer
Res Treat. 110:245–256. 2008. View Article : Google Scholar
|
|
8
|
Kim JY, Ahn HJ, Ryu JH, Suk K and Park JH:
BH3-only protein Noxa is a mediator of hypoxic cell death induced
by hypoxia-inducible factor 1α. J Exp Med. 199:113–124. 2004.
View Article : Google Scholar
|
|
9
|
Oliver PL, Bitoun E, Clark J, Jones EL and
Davies KE: Mediation of Af4 protein function in the cerebellum by
Siah proteins. Proc Natl Acad Sci USA. 101:14901–14906. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Habelhah H, Laine A, Erdjument-Bromage H,
Tempst P, Gershwin ME, Bowtell DD and Ronai Z: Regulation of
2-oxoglu-tarate (alpha-ketoglutarate) dehydrogenase stability by
the RING finger ubiquitin ligase Siah. J Biol Chem.
279:53782–53788. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Carthew RW and Rubin GM: seven in
absentia, a gene required for specification of R7 cell fate in the
Drosophila eye. Cell. 63:561–577. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hu G, Chung YL, Glover T, Valentine V,
Look AT and Fearon ER: Characterization of human homologs of the
Drosophila seven in absentia (sina) gene. Genomics. 46:103–111.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schmidt RL, Park CH, Ahmed AU, Gundelach
JH, Reed NR, Cheng S, Knudsen BE and Tang AH: Inhibition of
RAS-mediated transformation and tumorigenesis by targeting the
downstream E3 ubiquitin ligase seven in absentia homologue. Cancer
Res. 67:11798–11810. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qi J, Nakayama K, Gaitonde S, Goydos JS,
Krajewski S, Eroshkin A, Bar-Sagi D, Bowtell D and Ronai Z: The
ubiquitin ligase Siah2 regulates tumorigenesis and metastasis by
HIF-dependent and -independent pathways. Proc Natl Acad Sci USA.
105:16713–16718. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ahmed AU, Schmidt RL, Park CH, Reed NR,
Hesse SE, Thomas CF, Molina JR, Deschamps C, Yang P, Aubry MC, et
al: Effect of disrupting seven-in-absentia homolog 2 function on
lung cancer cell growth. J Natl Cancer Inst. 100:1606–1629. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Finn RS, Dering J, Ginther C, Wilson CA,
Glaspy P, Tchekmedyian N and Slamon DJ: Dasatinib, an orally active
small molecule inhibitor of both the src and abl kinases,
selectively inhibits growth of basal-type/“triple-negative” breast
cancer cell lines growing in vitro. Breast Cancer Res Treat.
105:319–326. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kao J, Salari K, Bocanegra M, Choi YL,
Girard L, Gandhi J, Kwei KA, Hernandez-Boussard T, Wang P, Gazdar
AF, et al: Molecular profiling of breast cancer cell lines defines
relevant tumor models and provides a resource for cancer gene
discovery. PLoS One. 4:e61462009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Neve RM, Chin K, Fridlyand J, Yeh J,
Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al: A
collection of breast cancer cell lines for the study of
functionally distinct cancer subtypes. Cancer Cell. 10:515–527.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Payne RE, Yagüe E, Slade MJ,
Apostolopoulos C, Jiao LR, Ward B, Coombes RC and Stebbing J:
Measurements of EGFR expression on circulating tumor cells are
reproducible over time in metastatic breast cancer patients.
Pharmacogenomics. 10:51–57. 2009. View Article : Google Scholar
|
|
20
|
Charafe-Jauffret E, Ginestier C, Monville
F, Finetti P, Adélaïde J, Cervera N, Fekairi S, Xerri L, Jacquemier
J, Birnbaum D, et al: Gene expression profiling of breast cell
lines identifies potential new basal markers. Oncogene.
25:2273–2284. 2006. View Article : Google Scholar
|
|
21
|
Nielsen TO, Hsu FD, Jensen K, Cheang M,
Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler
L, et al: Immunohistochemical and clinical characterization of the
basal-like subtype of invasive breast carcinoma. Clin Cancer Res.
10:5367–5374. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu G, Zhang S, Vidal M, Baer JL, Xu T and
Fearon ER: Mammalian homologs of seven in absentia regulate DCC via
the ubiquitin-proteasome pathway. Genes Dev. 11:2701–2714. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Boehm J, He Y, Greiner A, Staudt L and
Wirth T: Regulation of BOB.1/OBF1 stability by SIAH. EMBO J.
20:4153–4162. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tiedt R, Bartholdy BA, Matthias G, Newell
JW and Matthias P: The RING finger protein Siah-1 regulates the
level of the transcriptional coactivator OBF-1. EMBO J.
20:4143–4152. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Habelhah H, Frew IJ, Laine A, Janes PW,
Relaix F, Sassoon D, Bowtell DD and Ronai Z: Stress-induced
decrease in TRAF2 stability is mediated by Siah2. EMBO J.
21:5756–5765. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nakayama K, Frew IJ, Hagensen M, Skals M,
Habelhah H, Bhoumik A, Kadoya T, Erdjument-Bromage H, Tempst P,
Frappell PB, et al: Siah2 regulates stability of
prolyl-hydroxylases, controls HIF1α abundance, and modulates
physiological responses to hypoxia. Cell. 117:941–952. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nakayama K, Gazdoiu S, Abraham R, Pan ZQ
and Ronai Z: Hypoxia-induced assembly of prolyl hydroxylase PHD3
into complexes: Implications for its activity and susceptibility
for degradation by the E3 ligase Siah2. Biochem J. 401:217–226.
2007. View Article : Google Scholar :
|
|
28
|
Germani A, Romero F, Houlard M, Camonis J,
Gisselbrecht S, Fischer S and Varin-Blank N: hSiah2 is a new Vav
binding protein which inhibits Vav-mediated signaling pathways. Mol
Cell Biol. 19:3798–3807. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nadeau RJ, Toher JL, Yang X, Kovalenko D
and Friesel R: Regulation of Sprouty2 stability by mammalian
Seven-in-Absentia homolog 2. J Cell Biochem. 100:151–160. 2007.
View Article : Google Scholar
|
|
30
|
Hu Z, Fan C, Oh DS, Marron JS, He X,
Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, et al: The
molecular portraits of breast tumors are conserved across
microarray platforms. BMC Genomics. 7:962006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Khurana A, Nakayama K, Williams S, Davis
RJ, Mustelin T and Ronai Z: Regulation of the ring finger E3 ligase
Siah2 by p38 MAPK. J Biol Chem. 281:35316–35326. 2006. View Article : Google Scholar : PubMed/NCBI
|