Introduction
Ganoderma lucidum, also called ‘Ling-Zhi’ in
China and ‘Rei-Shi’ in Japan, is a basidiomycete white rot fungus
mostly distributed in tropical regions and usually growing on cut
or rotten trees (1). Among the many
traditional medicines, Ganoderma lucidum has been used for
more than 2,000 years for the prevention or treatment of various
human diseases (2). Even today,
this fungus is still predominantly used by Asian populations to
improve well-being and general health (3), as well as to inhibit cancer while used
alone or alongside chemotherapy treatment (4–6).
Ganoderma lucidum contains various bioactive
molecules, such as triterpenoids, polysaccharides, nucleotides,
fatty acids, glycoproteins, sterols, steroids, proteins and
peptides (7,8). Among these, Ganoderma lucidum
polysaccharides (GLPs), isolated from the spores, mycelia and
fruiting bodies of Ganoderma lucidum, have been suggested to
possess anticancer effects by a large number of basic studies
(9–12). According to the latest laboratory
and preclinical studies both in vitro and in vivo,
the promising anticancer activity of GLPs can be attributed to a
variety of different mechanisms (13). However, clinical studies of GLPs on
patients with cancer are limited, and the results are inadequately
reported concerning various aspects (14). Therefore, further investigation and
evaluation of the mechanism by which GLPs inhibit or kill cancer
cells are necessary to potentiate the use of GLPs as a first-line
treatment for cancer.
A recent survey of the performance of GLPs on 14
human tumor cells with various p53 status revealed that GLP-induced
inhibition of cancer cell growth depends on the existence of
functional p53 (15). However, the
actual role of p53-mediated tumor-suppressing pathways in
facilitating the anticancer effect of GLPs, as well as the
interaction between GLPs and mutant p53 that exist in more than
half of known cancers remain unclear. In the present study, we
chose colorectal cancer cells with different p53 status to analyze
the anticancer effect of GLPs administrated alone or together with
chemotherapy treatment, as well as the involvement of p53-mediated
cell cycle arrest and apoptosis.
Materials and methods
Antibodies
Anti-p53 (sc-126), anti-Bax (sc-493), anti-p21
(sc-397), anti-SDHB (sc-25851) and anti-cytochrome c
(sc-13156) antibodies were purchased from Santa Cruz Biotechnology,
Inc. (Santa Cruz, CA, USA). Anti-GAPDH and anti-H2B rabbit
polyclonal antibodies were raised against bacterially expressed
proteins (Jilin University).
Preparation of GLPs
GLPs were isolated and purified from the fruiting
body of Ganoderma lucidum (Beijing Tong Ren Tang Group Co.,
Ltd.) using a procedure as previously described (16). Crude polysaccharide powder was
obtained through derosination, hot water extraction,
deproteinization, alcohol precipitation and lyophilization. Crude
GLP solution (10 mg/ml in water) was ultra-filtered through 10-kDa
molecular weight cut-off (MWCO) membranes (Millipore, Billerica,
MA, USA). The filtrate (molecular weight of polysaccharides >10
kDa) was freeze-dried and stored at −30°C for subsequent
analysis.
Cell culture and transient
transfection
Human colon cancer cell lines HCT-116
(P53+/+ and P53−/−) were provided by
Professor Yinghua Jin of Jilin University. Human colon cancer cell
line SW480 was obtained from the Department of Gastrointestinal
Surgery, The First Bethune Hospital of Jilin University. Human
colon cancer cell line HT29 (ATCC® HTB-38™) was
purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA). Cells were cultured in McCoys 5A medium
modified (for HCT-116 and HT29) or Leibovitz's L-15 medium (for
SW480) (both from Gibco, Grand Island, NY, USA) with 10% fetal
bovine serum, 100 U/ml penicillin and 100 mg/ml streptomycin at
37°C in a humidified atmosphere of 5% CO2. For transient
transfection, the HCT-116 P53−/− cells were cultured and
transfected with mutant p53 cDNAs in pcDNA3.1 plasmid using
polyethylenimine (PEI). After 24 h of transfection, 5-fluorouracil
(5-FU) and GLPs were added into the medium and incubated for
another 24 h. The cells were then analyzed by MTT assay or
harvested and lysed for western blotting.
Cell viability and growth assay
For the viability assay, cells (5×103
cells/well) were seeded into 96-well plates and cultured overnight.
Then, the cells were treated with 5-FU and GLPs. After 24 h, cell
proliferation and viability were determined by the MTT assay. Upon
termination of treatment, MTT was applied to each well (10 µl) at a
final concentration of 0.5 mg/ml. After incubation for 4 h at 37°C,
the supernatant was removed and 100 µl dimethyl sulfoxide (DMSO)
was applied, and the MTT-formazan products were extracted. The
absorbance was read at 570 nm using a 96-well microplate reader
(BioTek, Winooski, VT, USA).
For the growth assay, cells (1×105
cells/well) were seeded in 6-well plates and cultured overnight.
Then, the cells were treated with 5-FU and GLPs for up to 96 h.
Cell growth curves were plotted by trypan blue staining and cell
counting (living cell number/well).
Isolation of mitochondrial
fraction
Cells (1×108) were suspended in 2 ml
mitochondria isolation buffer [0.225 M mannitol, 0.075 M sucrose, 1
mM EGTA (pH adjusted to 7.4 with 0.5 M Tris)] in a 10-ml Wheaton
homogenizer tube and carefully homogenized for 30 strokes on ice.
The cell debris was removed by centrifugation at 2,500 rpm (~600 ×
g) twice for 5 min. The supernatant was filtered through a nylon
screen cloth (Small Parts, Inc., Miami Lakes, FL, USA), and then
centrifuged at 10,000 rpm (~9,000 × g) for 10 min. The supernatant
was maintained as the cytosolic fraction and the pellet was washed
by adding 0.5 ml of mitochondria isolation buffer and centrifuged
at 10,000 rpm for 5 min. This washing step was repeated twice. The
mitochondrial pellet was resuspended in 50–100 µl of mitochondria
isolation buffer containing protease inhibitor cocktail (Research
Products International Corp. Mount Prospect, IL, USA). The purity
of the mitochondrial and cytosolic fractions was further examined
by western blot analysis.
Western blotting
Whole cell lysate or the mitochondrial or cytosolic
fractions from cultured cells was mixed with 4X SDS loading buffer
(0.25 M Tris-HCl pH 6.8, 8% SDS, 30% glycerol, 0.02% bromophenol
blue containing 10% BME), and boiled for 5 min at 95°C. Denatured
proteins were then separated by 12 or 18% SDS-PAGE, and specific
proteins were detected using the indicated antibodies.
Apoptosis assays
Cells were collected and stained with the Annexin
V-FLUOS staining kit (Roche) following the instructions for users.
Sample cells were then analyzed using a FACS flow cytometer
(Becton-Dickinson, Franklin Lakes, NJ, USA). We recorded Annexin
V-positive and propidium iodide (PI)-negative cells as early stage
apoptotic cells.
Cell cycle analysis
Cells (1.0×106) were harvested and rinsed
with phosphate-buffered saline (PBS). The cell pellets were fixed
in 70% ethanol at 4°C for 30 min. After washing twice with PBS, the
cells were stained with 1.0 ml of PI solution (Dingguo Changsheng
Biotechnology Co., Ltd., Beijing, China) containing 50 mg/l of PI
and 10 mg/l of RNase, followed by incubation on ice in a dark
condition for 30 min. The samples were then analyzed by FACS
(Becton-Dickinson).
In vitro cell-free apoptosis
assays
Preparation of cytosol extract: 1×108
HCT-116 P53−/− cells were disrupted in 0.5 ml of
cytoplasmic extraction buffer (CEB) buffer (50 mM PIPES, pH 7.4, 50
mM KCI, 5 mM EGTA, 2 mM MgCl2, 1 mM DTT, protease
inhibitor cocktail) by freezing-thawing cycles with liquid
nitrogen. The cell debris, organelles and nuclei were removed by
centrifugation at 50,000 rpm (~300,000 × g) for 1 h. The
supernatant was collected as the cytosol extract.
Preparation of nuclei extract: 2×107
HCT-116 P53−/− cells were harvested and resuspended in
0.5 ml of NEB buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM
MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 0.1%
Triton X-100, protease inhibitor cocktail) and incubated on ice for
15 min. Lysate was centrifuged at 3,500 rpm (~1,200 × g) for 5 min
and the pellet (nuclei) was washed and resuspended in DB buffer (10
mM HEPES, pH 7.0, 5 mM EGTA, 50 mM NaCl, 2 mM MgCl2, 1
mM DTT, protease inhibitor cocktail).
Basic reaction mix (50 µl) was prepared with DB
buffer containing the cytosol extract (7.5 µg/ml), mitochondrial
extract (40 µg/ml, isolated as described above), dATP (1 mM),
phosphocreatine (5 mM), creatine kinase (25 µg/ml) (all from
Sigma), and nuclear extract (1×106 nuclei/ml).
Cytochrome c (10 µM) (Sigma; served as positive control for
the assay), prokaryotically expressed p53 (5 µg/ml) and GLPs (100
µg/ml) were added to the basic reaction mix with a designed
combination. The final mix was incubated at 37°C for 4 h. After
reaction, the nuclei were collected and fixed with 4%
paraformaldehyde solution (in PBS), and were then stained with
4,6-diamidino-2-phenylindole (DAPI) (H-1200; Vector Laboratories,
Inc.). Fluorescence images of the nuclei were observed with an
Olympus Bx40F microscope (Olympus Corporation, Tokyo, Japan). At
least 500 nuclei were randomly counted and determined as being
apoptotic or having a normal morphological phenotype.
Statistical analysis
Statistical analysis was achieved using GraphPad
Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA). Data are
reported as the mean ± SEM (n=3). Statistically significant
differences were determined by one-way or two-way ANOVA tests.
Values of p<0.05 were considered to indicate a statistically
significant result.
Results
Wild-type p53 is essential for
5-FU-induced inhibition of colorectal cancer cells
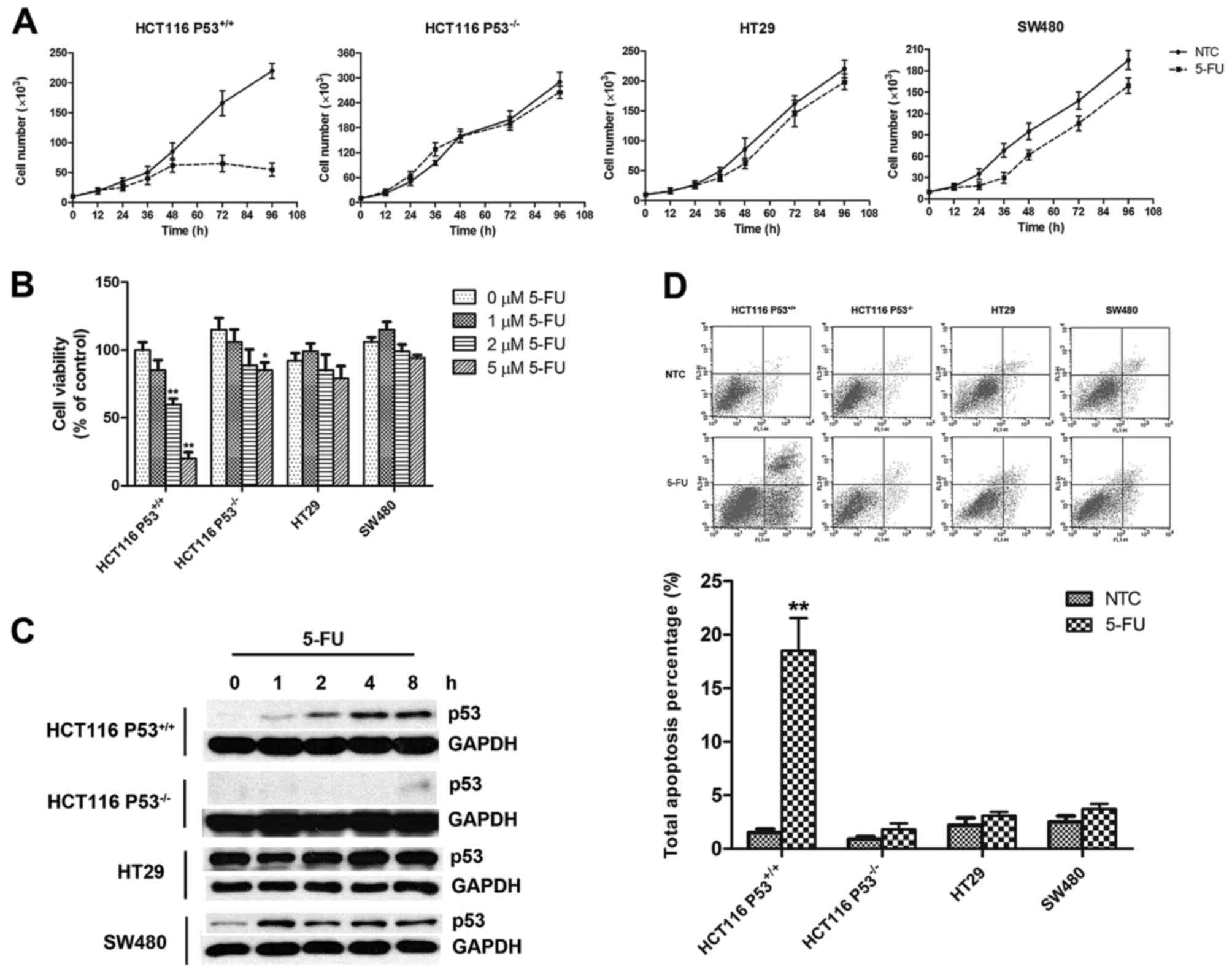
Initially, a series of studies of cell growth were
performed to detect the response of the colorectal cancer cells
bearing a different p53 status to 5-FU, a chemotherapy drug widely
used in clinical practice for patients with colorectal cancer. An
obvious growth inhibition was observed in the HCT-116
P53+/+ cells, a cell line expressing wild-type p53
(17), after treatment with 5 µM
5-FU. In contrast, the HCT-116 P53−/− cells, a
homogenous P53 gene-knockout cell line, exhibited resistance to
5-FU under the same treatment condition (Fig. 1A). Similar drug tolerance was also
observed in the HT29 and SW480 cells (Fig. 1A); these two cell lines both express
mutant p53 while HT29 carries a single mutation (R273H) and SW480
carries a double mutation (R273H and P309S) (18). To further confirm the role of p53 in
5-FU-mediated cell growth inhibition, we carried out an MTT assay
to measure the viability of the cells treated with 0–5 µM of 5-FU.
As expected, only the cells with wild-type p53 (HCT-116
P53+/+) sensitively responded to treatment with
different concentrations of 5-FU in the MTT assay (Fig. 1B). Indeed, 5-FU treatment stabilized
wild-type p53 in the HCT-116 cells (Fig. 1C, row 1) and thus probably triggered
associated downstream cellular events such as apoptosis (Fig. 1D, column 1 and 2). Moreover, even
though the mutant p53 presented a different response pattern to
5-FU treatment (Fig. 1C, rows 5 and
7), there was no apoptosis signal detected in these two cell lines
(Fig. 1D, column 5–8). It was also
not surprising to find negative results for both the p53 protein
level and the apoptosis signal in the 5-FU-treated HCT-116
P53−/− cells (Fig. 1C,
row 3 and D, column 3 and 4). All these results indicate an
important role of functional p53 and its tumor-suppressing effects
in 5-FU treatment.
GLPs administered in combination with
5-FU synergistically suppress mutant p53-bearing colorectal cancer
cells
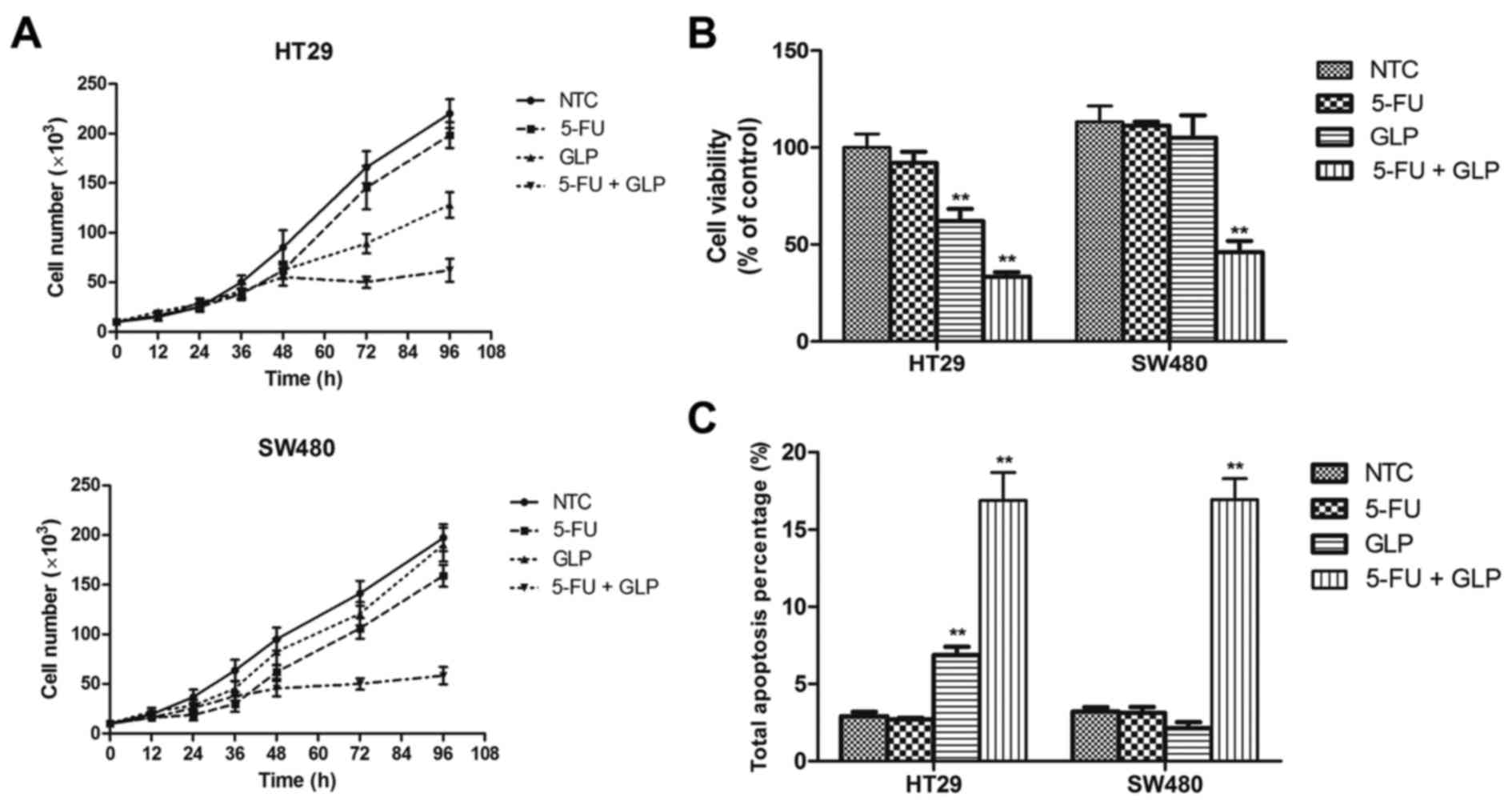
In order to determine the relationship between the
anticancer effects of GLPs and p53 status, we introduced GLPs to
the treatment of mutant p53-bearing colorectal cancer cells with or
without 5-FU co-treatment, since GLPs are widely used as an
anticancer drug alone or combined with chemotherapeutics in many
Asian countries. As shown in Fig. 2A
and B, combined treatment with 5-FU and GLPs significantly
suppressed the growth and viability of both mutant p53-bearing
cancer cell lines compared to 5-FU treatment alone. These results
suggest that GLPs may have a type of reactivating effect to mutant
p53 that facilitates the p53-dependent tumor-suppressing activity
of 5-FU. In line with that of the cell growth assay, the results of
the apoptosis assay also showed a synergistic increase while
introducing GLPs to 5-FU treatment (Fig. 2C). However, the effect of GLP
treatment alone exhibited different results in the two cell lines
tested. GLP treatment alone inhibited the growth and viability and
triggered apoptosis of HT29 cells, while in contrast showed trivial
effect on the SW480 cells (Fig.
2).
GLPs reactivate several types of
exogenous mutant p53 in the HCT116 cells
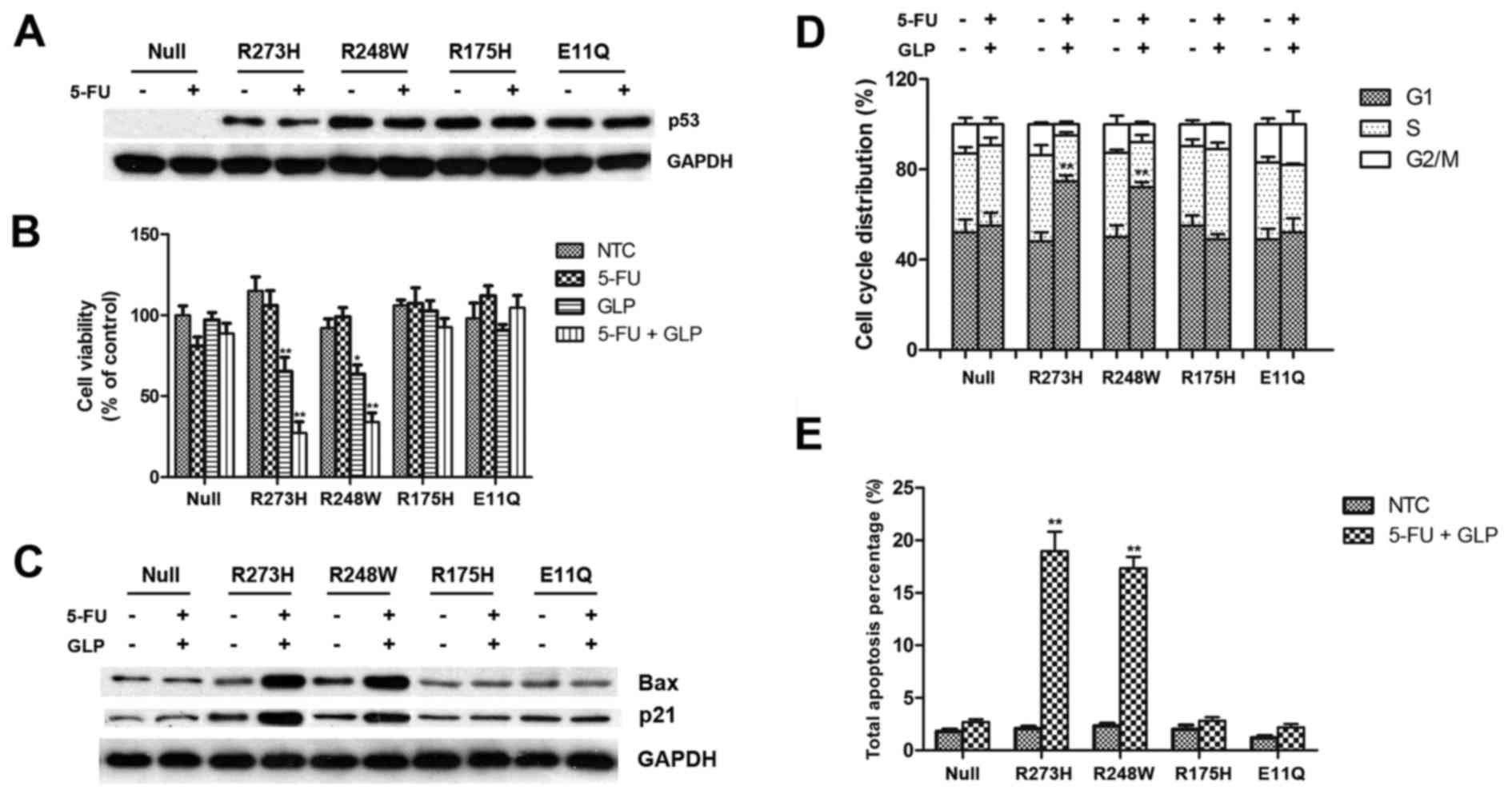
It has been well documented that various types of
p53 mutations that perturb p53 function exist in more than half of
our known cancer cases (19). Thus,
to evaluate the potential reactivating ability of GLPs on mutant
p53, we constructed plasmids expressing exogenous p53 with
different types of hot mutations and transfected HCT-116
P53−/− cells with them, followed by treatment with 5-FU
and GLPs in various combinations. All types of exogenous mutant p53
have a high level of basal expression so that 5-FU treatment did
not induce further accumulation of whole cellular p53 in this
situation (Fig. 3A). As revealed by
the MTT assay, cell viability of the cells transfected with
mutation R273H and R248W were significantly inhibited by GLPs
either alone or combined with 5-FU (Fig. 3B). To confirm the reactivation of
p53-mediated tumor-suppressing pathways, we performed western blot
assay to measure the expression level of downstream genes that are
regulated by p53. As shown in Fig.
3C, the expression of Bax and p21 were upregulated in the
P53R273H and P53R248W transfected cells after
treatment with 5-FU and GLPs, suggesting a restoration of
transactivation function to mutant p53 by GLPs. As two major
downstream outputs of tumor-suppressor pathways, significant G1
arrest and apoptosis were also detected in the cells transfected
with mutation P53R273H and P53R248W (Fig. 3D and E), indicating full
reactivation of p53 function.
GLPs facilitate the interaction
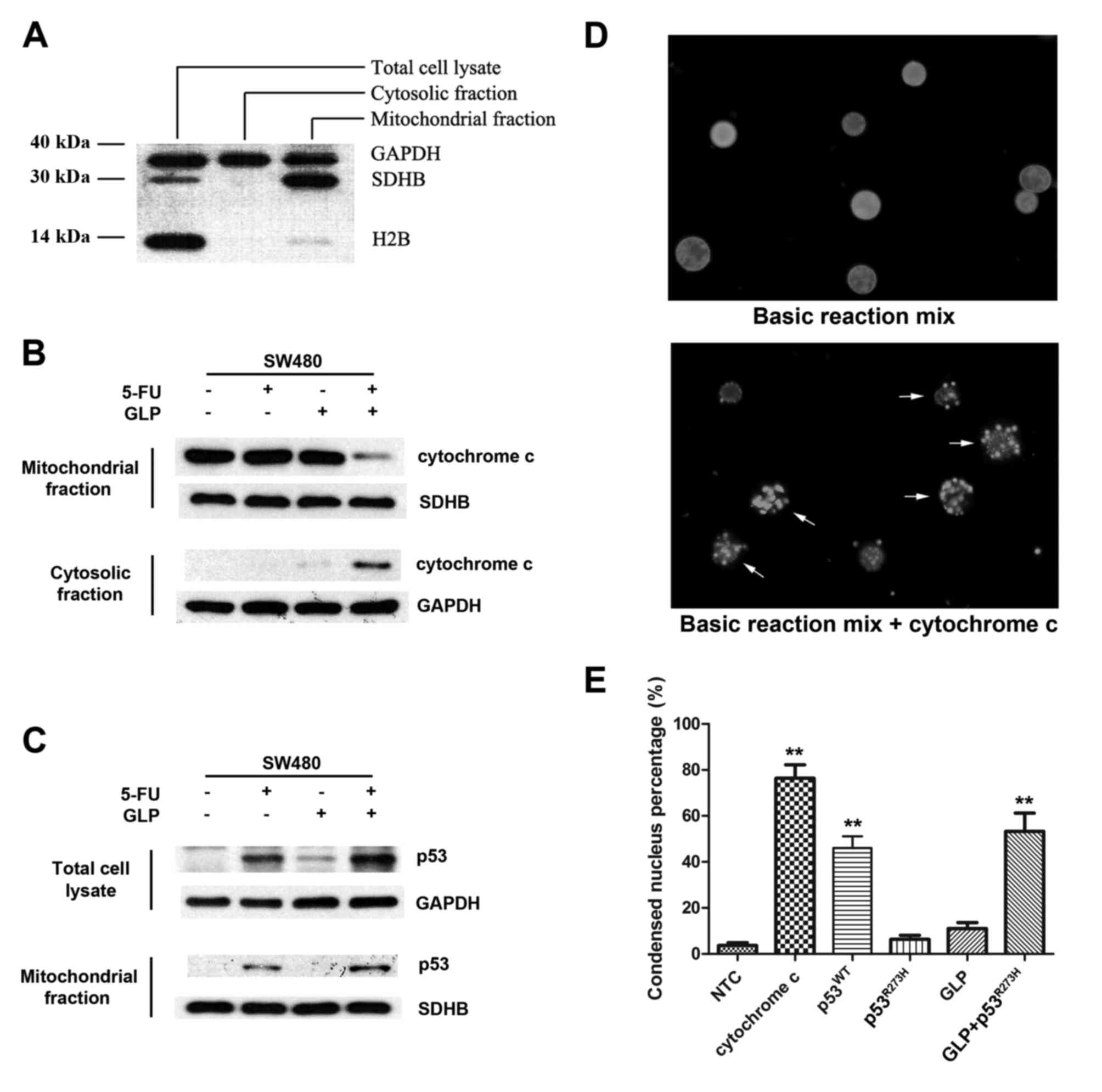
between mutant p53 and mitochondria
In addition to the transactivation of downstream
genes encoding proapoptotic proteins, p53 is also able to
physically interact with mitochondria and induce apoptosis through
a so called transcriptional-independent pathway (20–22).
To investigate whether or not the transcriptional-independent
apoptotic pathway is involved in GLP-mediated mutant p53
reactivation, we designed a series of assays that bypass the
nuclear p53 function. Cellular components from the cytosol and
mitochondria were respectively separated with differential
centrifugation (Fig. 4A) and the
levels of the proteins of interest were detected with specific
antibodies. As shown in Fig. 4C, in
the SW480 cells, 5-FU treatment induced whole cellular accumulation
of mutant p53, and in the meantime a fraction of mutant p53
translocated to thevmitochondria. This distribution pattern of
mutant p53 was further strengthened by adding GLPs to the
treatment, indicating that GLPs facilitated 5-FU-induced p53
mitochondrial translocation. It is believed that functional p53
translocating to mitochondria can interact with Bax, a Bcl-2 family
protein, and thus induce mitochondrial membrane permeabilization
(MMP) and cytochrome c release (23,24).
In SW480 cells, mutant p53 accumulation in the mitochondria by 5-FU
treatment was not sufficient to induce cytochrome c release
(Fig. 4B, lane 2). However,
co-treatment with GLPs resulted in obvious cytochrome c
release which was clearly detected in both mitochondrial and
cytosolic fraction (Fig. 4B, lane
4), suggesting the restoration of the p53 ability to regulate Bax
and MMP by GLPs. To further exclude the influence of the
transactivation effect of p53, we introduced an in vitro
cell-free system to determine the regulatory role of GLPs in the
interaction between p53 and mitochondria. Isolated nuclei, cytosol
and mitochondria from HCT-116 P53−/− cells were mixed
together and incubated with prokaryotically expressed p53 and GLPs.
After interaction between p53 and mitochondria, cytochrome c
released from mitochondria activated the downstream caspase cascade
and eventually induced chromatin condensing and bubbling of
isolated nuclei (Fig. 4D). Based on
this in vitro assay system, our results showed that
wild-type p53 induced MMP and cytochrome c release, while in
contrast mutant p53R273H lacked this activity but was
reactivated by GLPs (Fig. 4E).
Discussion
Polysaccharides, along with triterpenes, are
regarded as major biologically active compounds in Ganoderma
lucidum (25). The most
frequent classes of polysaccharides isolated from Ganoderma
lucidum are glucans, glycoproteins, glycopeptides and
water-soluble heteropolysaccharides (26). Although GLPs have been proven to
possess strong anticancer effects, the anticancer effects of GLPs
are generally believed to depend on modulation of immune cell
response, which further targets cancer cells (27). Specifically, GLPs affect immune
cells and immune-related cells such as T and B lymphocytes,
macrophages, dendritic and natural killer cells (9). However, whether GLPs have an
anticancer effect that is independent of its immunostimulatory
effects remain unclarified. Several pharmacological studies have
revealed that GLPs are able to directly suppress the proliferation
of various types of cancer cell lines (10–12,28),
but the mechanisms underlying the cytotoxic and cytostatic
activities are yet to be determined.
p53 is a potent tumor suppressor which can prevent
the propagation of cells carrying oncogenic lesions via a multitude
of pathways such as growth arrest, senescence or apoptosis,
modulation of tumor stroma, angiogenesis and metabolism, as well as
the blockage of invasion and metastasis (29). This explains why loss of p53
function is opted for during tumor development, resulting in p53
inactivation in the majority of human tumors (19,30).
Therefore, restoration of mutant p53 function in tumor cells could
be an attractive and specific strategy for treating cancer. Indeed,
it has already been reported that various natural or artificially
synthesized compounds are able to reactivate mutant p53 in tumor
cells (31–34).
In the present study, we demonstrated a direct
reactivation effect of GLPs on mutant p53 in colorectal cancer
cells. This reactivation effect influenced both the transcriptional
and non-transcriptional activities of p53. It is notable that in
the absence of 5-FU, GLPs alone were not sufficient to induce
growth inhibition and apoptosis in the SW480 cells (Figs. 2 and 4C), while in contrast, it was effective in
the HT29 cells (Fig. 2), exogenous
mutant p53-transfected HCT-116 P53−/− cells (Fig. 3B), as well as in vitro
cell-free system (Fig. 4E). In
addition, an obvious difference between SW480 cells and other
experimental models is that there is no or little p53 protein
accumulated without 5-FU treatment (Figs. 1C and 3A). Thus, it is reasonable to speculate
that GLPs can reactivate mutant p53 that must already exist
abundantly at the right location in the cell, but may lose target
whether the cells lack appropriate p53 accumulation or
localization, which may be triggered by other stimuli such as 5-FU
through post-translational modification of p53 or other pathways.
Another point worth noting is the target site of GLPs on mutant
p53. In the present study, using 4 different types of hot mutations
of p53, only R273H and R248W were sensitive to GLP treatment, which
are both mutations in the DNA binding domain (Fig. 3). This result indicates strong
target site specificity of GLP-mediated mutant p53 reactivation and
suggests further structural studies to verify the interaction model
between GLPs and major hot mutations of p53.
In conclusion, the present study offers a novel
insight into the mechanism of GLP-induced cytotoxicity and
apoptosis in human colorectal cancer cells, suggesting promising
usage of GLPs along with other chemotherapeutics to reactivate
mutant p53 in the treatment of cancer. Our results also provide
potential perspectives for further research on the pharmacology of
GLPs as a possible candidate for cancer prevention or treatment of
cancer with mutant p53.
Acknowledgements
The present study was supported by Jilin Province
Science and Technology Development Program (20140520005JH), the
National Natural Science Foundation of China (31100999), and the
China Postdoctoral Science Foundation Grant (2014M561288).
Glossary
Abbreviations
Abbreviations:
|
GLPs
|
Ganoderma lucidum
polysaccharides
|
|
5-FU
|
5-fluorouracil
|
|
MMP
|
mitochondrial membrane
permeabilization
|
References
|
1
|
Wang XC, Xi RJ, Li Y, Wang DM and Yao YJ:
The species identity of the widely cultivated Ganoderma, ‘G.
lucidum’ (Ling-zhi), in China. PLoS One. 7:e408572012. View Article : Google Scholar
|
|
2
|
Li J, Zhang J, Chen H, Chen X, Lan J and
Liu C: Complete mitochondrial genome of the medicinal mushroom
Ganoderma lucidum. PLoS One. 8:e720382013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shiao MS: Natural products of the
medicinal fungus Ganoderma lucidum: Occurrence, biological
activities, and pharmacological functions. Chem Rec. 3:172–180.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gordan JD, Chay WY, Kelley RK, Ko AH, Choo
SP and Venook AP: And what other medications are you taking? J Clin
Oncol. 29:e288–e291. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Papadopoulos I, Guo F, Lees S and Ridge M:
An exploration of the meanings and experiences of cancer of Chinese
people living and working in London. Eur J Cancer Care. 16:424–432.
2007. View Article : Google Scholar
|
|
6
|
Sliva D: Ganoderma lucidum (Reishi) in
cancer treatment. Integr Cancer Ther. 2:358–364. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Batra P, Sharma AK and Khajuria R: Probing
Lingzhi or Reishi medicinal mushroom Ganoderma lucidum (higher
Basidiomycetes): A bitter mushroom with amazing health benefits.
Int J Med Mushrooms. 15:127–143. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wachtel-Galor S, Yuen J, Buswell JA and
Benzie IFF: Ganoderma lucidum (Lingzhi or Reishi): A Medicinal
MushroomHerbal Medicine: Biomolecular and Clinical Aspects. Benzie
IFF and Wachtel-Galor S: 92nd. CRC Press/Taylor & Francis; Boca
Raton, FL: 2011, http://dx.doi.org/10.1201/b10787 View Article : Google Scholar
|
|
9
|
Xu Z, Chen X, Zhong Z, Chen L and Wang Y:
Ganoderma lucidum polysaccharides: Immunomodulation and potential
anti-tumor activities. Am J Chin Med. 39:15–27. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shang D, Li Y, Wang C, Wang X, Yu Z and Fu
X: A novel polysaccharide from Se-enriched Ganoderma lucidum
induces apoptosis of human breast cancer cells. Oncol Rep.
25:267–272. 2011.PubMed/NCBI
|
|
11
|
Huang CY, Chen JY, Wu JE, Pu YS, Liu GY,
Pan MH, Huang YT, Huang AM, Hwang CC, Chung SJ, et al: Ling-Zhi
polysaccharides potentiate cytotoxic effects of anticancer drugs
against drug-resistant urothelial carcinoma cells. J Agric Food
Chem. 58:8798–8805. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang J, Zhang L, Yu Y and Cheung PC:
Enhancement of antitumor activities in sulfated and
carboxymethylated polysaccharides of Ganoderma lucidum. J Agric
Food Chem. 57:10565–10572. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Boh B: Ganoderma lucidum: A potential for
biotechnological production of anti-cancer and immunomodulatory
drugs. Recent Patents Anticancer Drug Discov. 8:255–287. 2013.
View Article : Google Scholar
|
|
14
|
Jin X, Beguerie J Ruiz, Sze DM and Chan
GC: Ganoderma lucidum (Reishi mushroom) for cancer treatment.
Cochrane Database Syst Rev. 6:CD0077312012.
|
|
15
|
Zhang J, Chen JM, Wang XX, Xia YM, Cui SW,
Li J and Ding ZY: Inhibitor or promoter? The performance of
polysaccharides from Ganoderma lucidum on human tumor cells with
different p53 statuses. Food Funct. 7:1872–1875. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liang Z, Guo YT, Yi YJ, Wang RC, Hu QL and
Xiong XY: Ganoderma lucidum polysaccharides target a Fas/caspase
dependent pathway to induce apoptosis in human colon cancer cells.
Asian Pac J Cancer Prev. 15:3981–3986. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Y and Bodmer WF: Analysis of P53
mutations and their expression in 56 colorectal cancer cell lines.
Proc Natl Acad Sci USA. 103:976–981. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nigro JM, Baker SJ, Preisinger AC, Jessup
JM, Hostetter R, Cleary K, Bigner SH, Davidson N, Baylin S, Devilee
P, et al: Mutations in the p53 gene occur in diverse human tumour
types. Nature. 342:705–708. 1989. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hainaut P and Hollstein M: p53 and human
cancer: The first ten thousand mutations. Adv Cancer Res.
77:81–137. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vousden KH and Lane DP: p53 in health and
disease. Nat Rev Mol Cell Biol. 8:275–283. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mihara M, Erster S, Zaika A, Petrenko O,
Chittenden T, Pancoska P and Moll UM: p53 has a direct apoptogenic
role at the mitochondria. Mol Cell. 11:577–590. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Moll UM, Wolff S, Speidel D and Deppert W:
Transcription-independent pro-apoptotic functions of p53. Curr Opin
Cell Biol. 17:631–636. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chipuk JE, Kuwana T, Bouchier-Hayes L,
Droin NM, Newmeyer DD, Schuler M and Green DR: Direct activation of
Bax by p53 mediates mitochondrial membrane permeabilization and
apoptosis. Science. 303:1010–1014. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chipuk JE and Green DR: Cytoplasmic p53:
Bax and forward. Cell Cycle. 3:429–431. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Boh B, Berovic M, Zhang J and Zhi-Bin L:
Ganoderma lucidum and its pharmaceutically active compounds.
Biotechnol Annu Rev. 13:265–301. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen Y, Xie MY, Wang YX, Nie SP and Li C:
Analysis of the monosaccharide composition of purified
polysaccharides in Ganoderma atrum by capillary gas chromatography.
Phytochem Anal. 20:503–510. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin ZB: Cellular and molecular mechanisms
of immuno-modulation by Ganoderma lucidum. J Pharmacol Sci.
99:144–153. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li N, Hu YL, He CX, Hu CJ, Zhou J, Tang GP
and Gao JQ: Preparation, characterisation and anti-tumour activity
of Ganoderma lucidum polysaccharide nanoparticles. J Pharm
Pharmacol. 62:139–144. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vousden KH and Prives C: Blinded by the
light: The growing complexity of p53. Cell. 137:413–431. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lawrence MS, Stojanov P, Polak P, Kryukov
GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH,
Roberts SA, et al: Mutational heterogeneity in cancer and the
search for new cancer-associated genes. Nature. 499:214–218. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bykov VJ, Issaeva N, Shilov A, Hultcrantz
M, Pugacheva E, Chumakov P, Bergman J, Wiman KG and Selivanova G:
Restoration of the tumor suppressor function to mutant p53 by a
low-molecular-weight compound. Nat Med. 8:282–288. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang F, Liu J, Robbins D, Morris K, Sit A,
Liu YY and Zhao Y: Mutant p53 exhibits trivial effects on
mitochondrial functions which can be reactivated by ellipticine in
lymphoma cells. Apoptosis. 16:301–310. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu X, Wilcken R, Joerger AC, Chuckowree
IS, Amin J, Spencer J and Fersht AR: Small molecule induced
reactivation of mutant p53 in cancer cells. Nucleic Acids Res.
41:6034–6044. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Selivanova G and Wiman KG: Reactivation of
mutant p53: Molecular mechanisms and therapeutic potential.
Oncogene. 26:2243–2254. 2007. View Article : Google Scholar : PubMed/NCBI
|