Introduction
Myelodysplastic syndromes (MDSs), one of the five
major categories of myeloid neoplasms, are defined as clonal
stem-cell disorders characterized by ineffective hematopoiesis in
one or more of the lineages of the bone marrow, and the progression
is from blood cytopaenias to acute myeloid leukaemia (AML)
(1). AML, which is the most common
type of acute leukemia in adults, is fatal in more than 50% of
patients that do not receive current treatments (2), and the mortality of MDS is primarily
caused by pancytopenia or transformation to AML. Therefore, the
discovery of the molecular mechanisms would be significant for
better prognoses. It has been discovered that MDS are one of the
clonal haemopathies, in that an aberration within a haematopoietic
progenitor cell gives rise to the entire disease (3). Additionally, some abnormal karyotypes
have been found in the patients with MDS, including del (5q),
monosomy 7 or del (7q), trisomy 8 and del (20q), whereas ~50%
patients have a normal karyotype (4–6).
The pathogenesis of MDS has not yet been completely
elucidated. Recently, uniparental disomy (UPD) has garnered
attention, and some studies have indicated that UPD is related to
hematologic malignancies including both MDS and AML (7,8).
Furthermore, sperm-associated antigen 6 (SPAG6) was shown to be
overexpressed in some patients with MDS in a genome wide single
nucleotide polymorphism analysis, which implies that SPAG6 may play
a role in the pathogenesis of MDS (9).
SPAG6, which is the orthologue of
Chlamydomonas PF16, is essential for sperm flagellar
motility and maintenance of the structural integrity of mature
sperm (10). In the last decade,
several research groups have focused on the relationship between
SPAG6 and malignancies. Seven genes, including WT1 and SPAG6, were
found to be vastly overexpressed in patients with AML as compared
to healthy bone marrow via genome-wide expression profiling
(11). Furthermore, SPAG6 is highly
expressed at the break point of t(10;11)(p12;q14) in patients with
CALM/AF10-positive leukemias (12).
In our previous study, when the expression of SPAG6 was silenced by
SPAG6-shRNA lentivirus, the growth of SKM-1 and K652 was markedly
inhibited, and apoptosis was increased (13). The above evidence indicates that
SPAG6 may promote cellular growth by preventing apoptosis. However,
the specific mechanism of anti-apoptosis is still unknown.
Several studies imply that abnormal apoptosis is one
of the underlying mechanisms in MDS, and tumor necrosis factor
(TNF)-α, Fas ligand, TNF-related apoptosis-inducing ligand (TRAIL)
and other pro-apoptotic cytokines may be the major factors
(14). In the last decades, the
role of TRAIL in the pathogenesis of malignancies has been
observed. TRAIL, also known as Apo2 ligand (Apo2L), is a member of
the TNF ligand superfamily, and it is induced in tumor cells, but
seldom in normal cells (15).
Recently, two researchers explored the possible potential phenotype
related to the TRAIL signal pathway using a genome-wide siRNA
screen, and it was found that SPAG6 and this pathway may have a
correlation, which was recorded in the GenomeRNAi-database
(http://www.genomernai.org) (16).
The aim of this study is to clarify the regulatory
mechanism of SPAG6 in cell apoptosis of MDS by TRAIL signal
pathway, which may provide a new perspective in the development of
MDS.
Materials and methods
Cell culture and infection
The human MDS cell line SKM-1, was kindly provided
by Professor Zhou at the Tongji Medical College, Huazhong
University of Science and Technology (Wuhan, China). The
erythroleukemia K562 and the histiocytic lymphoma U937 cell lines
were kept frozen in our laboratory. Cell line SKM-1, U937 and K562
were cultured in RPMI-1640 containing 10% fetal bovine serum (FBS;
Capricorn Scientific GmbH, Ebsdorfergrund, Germany) and no
antibiotic in 5% CO2, 95% air incubator at 37°C. For the
infection protocol, SKM-1 cells at the exponential stage were
plated in 6-well plates (5×104 cells/well), and were
infected with the SPAG6-shRNA lentivirus or NC-shRNA lentivirus at
a multiplicity of infection (MOI) of 20 in the concentration of 5
µg/ml polybrene, respectively. After 10 h, the cells were washed
and then resuspended in complete medium. After 5 days, the
transfection efficiencies were evaluated by flow cytometry.
Cell treatment
SKM-1, K562 and U937 were seeded in 6-well plates
(1×105), and were treated with recombinant human sTRAIL
(310–04; Peprotech, Rocky Hill, NJ, USA), respectively. The treated
concentrations were as follows: 0, 20, 40, 60, 80, 100 and 50
ng/ml. After 24 h, the cells were collected, and the apoptosis
rates were measured by flow cytometry.
Flow cytometry
The apoptosis rates were detected using Annexin V
and 7-ADD double-staining by flow cytometry. Following the cell
treatments, the cells were collected and washed twice with
phosphate-buffered saline (PBS). The early apoptotic cells
contained Annexin V+/PI−, while the late
apoptotic cells contained Annexin V+/PI+.
Both of the two stages of apoptotic cells were counted, and the
results were illustrated as a percentage of the total cell count.
The techniques were supported by the Life Science Department of
Chongqing Medical University (Chongqing, China).
RNA isolation and real-time PCR
The total cellular RNA from each group was extracted
using the TRIzol reagent, and the reverse transcription reaction
was performed using the Prime Script™ RT reagent kit (Takara
Biotechnology Co., Ltd., Dalian, China). The cDNA was amplified in
a 10-µl PCR mix with 5 µl of SYBR-Green super mixture. The PCR
reactions were performed in a CFX-Connect Real-Time PCR system
(Bio-Rad Laboratories Inc., Hercules, CA, USA). The cycling
parameters were: 95°C for 30 sec, then 40 cycles at 95°C for 5 sec
and 60°C for 30 sec. PCR primers were as follows: SPAG6 forward,
5′-CCTTTCAGCTCTCAGTCAGGTTTC-3′ and reverse,
5′-TCTTCACGTTTCATCCTTGTCCTT-3′; death receptor 4 (DR4) forward,
5′-TCGCTGTCCACTTTCGTCT-3′ and reverse, 5′-GGCGTTCCGTCCAGTTTTG-3′;
DR5 forward, 5′-AAGACCCTTGTGCTCGTTGT-3′ and reverse,
5′-GCTGCAACTGTGACTCCTAT-3′; and GAPDH forward,
5′-CTTTGGTATCGTGGAAGGACTC-3′ and reverse,
5′-GTAGAGGCAGGGATGATGTTCT-3′. The PCR reactions were performed in a
CFX-Connect Real-Time PCR system for 40 cycles (Bio-Rad
Laboratories, Inc.). The relative gene expression levels were
calculated by the 2−ΔΔCt method.
Protein isolation and western blot
analysis
The cells were collected and lysed in RIPA lysis
buffer (Beyotime, Beijing, China) containing 1 µM PMSF, and the
concentration was measured using the BCA Protein Assay kit
(Beyotime). The total cell lysate was denatured via boiling and a
total of 50 µg of protein per lane was separated by sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and
transferred to PVDF membranes. The membranes were blocked with 5%
bovine serum albumin for 90 min at room temperature and then
incubated overnight at 4°C with specific antibodies. The primary
antibodies were as follows: GAPDH (AG019, 1:1,000; Beyotime)
cleaved caspase-8 (9496, 1:500; Cell Signaling Technology, Inc.,
Beverly, MA, USA); SPAG6, PARP, cleaved PARP, caspase-8, DR4, DR5
and FADD (ab155653, ab32138, ab32561, ab32397, ab8414, ab181846,
ab108601, 1:1,000; all from Abcam, Cambridge, UK); caspase-3 and
BID (YT0656, YT0488, 1:500; both from Immunoway, Newark, DE, USA);
and BAX and BAK (RLT0456, RLT0449, 1:500; both from Ruiying
Biological, Suzhou, China). After five washes with TBS-Tween-20,
the membranes were incubated with a goat anti-rabbit of goat
anti-mouse peroxidase-conjugated second antibody (A0216, A0208,
1:1,000; Beyotime) for 1 h at 37°C. Then, the excess antibody was
removed from the blots with TBS-Tween-20 three times before
incubation in ECL. The protein expression levels were analysed
using Vilber Fusion software (Fusion FX5 Spectra; Vilber Lourmat,
Marne-La-Vallée, France).
The Cell Counting Kit-8 (CCK-8)
assay
Apoptosis was measured by a CCK-8 assay. Cells in
exponential growth were seeded in 96-well plates at a density of
8×103 cells/well, and then, they were treated with
recombinant human sTRAIL (rTRAIL) containing different
concentrations for 24 h, respectively. Finally, 10 µl of CCK-8
(Dojindo Laboratories, Kumamoto, Japan) solution was added to each
well, and the plates were incubated at 37°C for 100 min. Cell
apoptosis was estimated by measuring the absorbance at 450 nm using
the plate reader. All conditions were tested in five
replicates.
Immunoprecipitation
The cells infected with the lentivirus were
collected after infection and lysed in RIPA lysis buffer. The
solution was centrifuged at 12,000 rpm for 10 min at 4°C and the
supernatants were kept. Protein A agarose and antibody (ab108601,
1:100, ab14738, ab181846, 1:2,000; Abcam) diluted with RIPA buffer
were mixed and incubated overnight at 4°C. Then the total protein
was mixed and incubated overnight at 4°C. After incubation, the
mixture was washed six times with different types of washing
buffer. Finally, a western blot analysis was performed for
immunoblot analysis.
Statistical analysis
All the results were technically repeated three
times, and were expressed as the mean ± standard deviation (SD).
The significant differences between groups were assessed by one-way
analysis of variance (ANOVA) and Students t-test (SPSS version
20.0; SPSS, Inc., Chicago, IL, USA) and a value of P<0.05 was
considered statistically significant.
Results
Lentivirus-mediated SPAG6 silence in
SKM-1 cells
The lentivirus expressing shRNA was used against
SPAG6 for gene silencing in SKM-1 cells, and the SKM-1 cells
infected with NC-shRNA lentivirus were used as a control group. Six
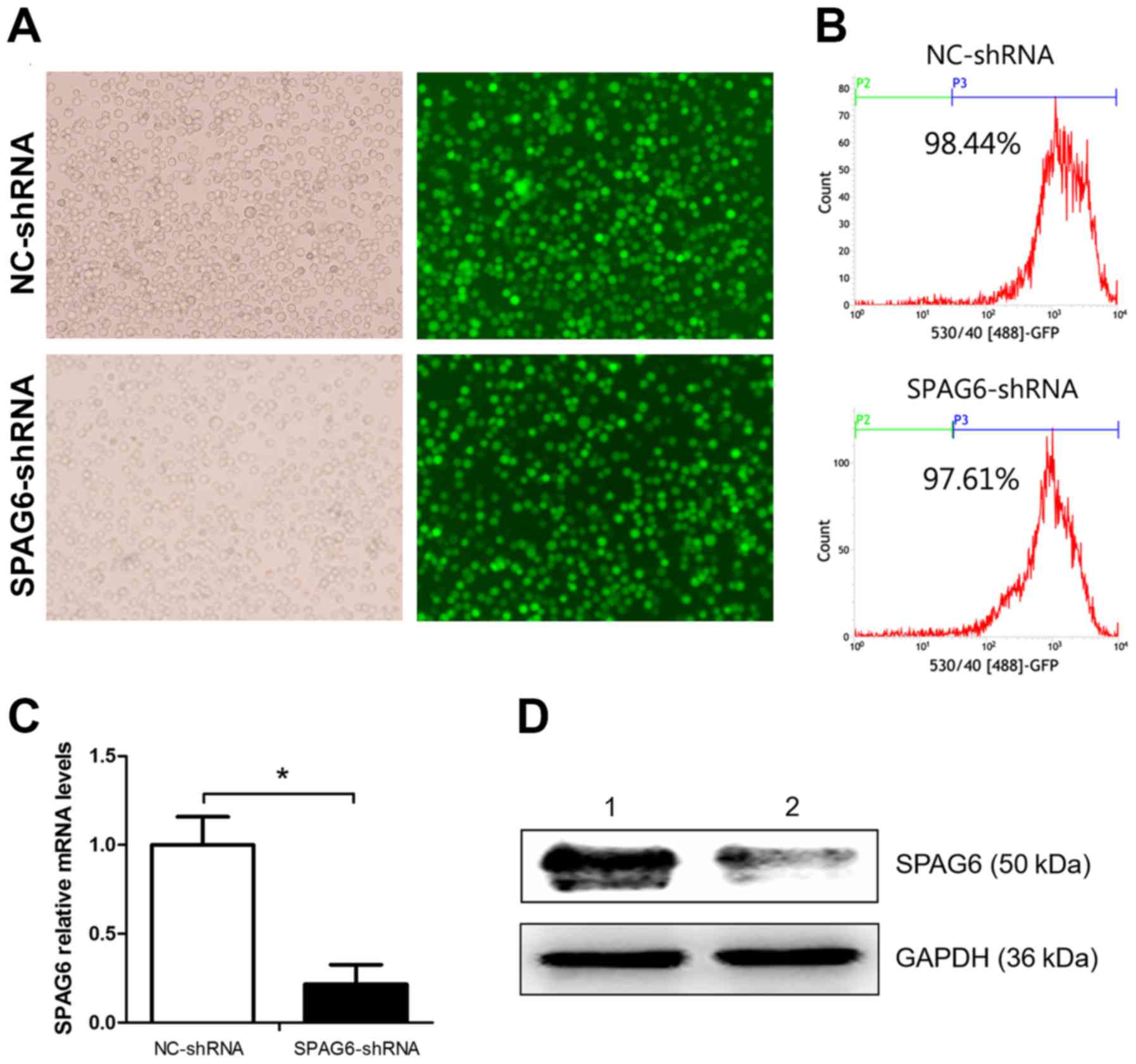
days after infection, abundance of cells with green fluorescence
could be observed under a fluorescence microscope (Fig. 1A). The transfection efficiencies
were detected by FACS, and the results were 97.19±3.21% and
88.31±12.81% (Fig. 1B). This
demonstrated that the SKM-1 cells were successfully infected by the
lentivirus. The inhibitory degree of SPAG6 was measured by qRT-PCR
and western blot analysis. The results showed that both the mRNA
and protein levels were reduced in cells infected with SPAG6
shRNA-lentivirus as compared to the control group (Fig. 1C and D).
Reducing the expression of SPAG6
activates the TRAIL signal pathway
Apoptosis was analysed by Annexin V and 7-AAD
assays, as illustrated in Fig. 2A and
B, and the apoptosis rates in SKM-1 infected with SPAG6-shRNA
were significantly higher than those in the control group
(NC-shRNA, 8.65±2.07% vs. SPAG6-shRNA, 20.20±2.11%; P<0.05). To
explore the activation of the TRAIL signal pathway, the expression
of related proteins was examined. Western blot analysis revealed
that the expression levels of cleaved PARP, caspase-8 and cleaved
caspase-8 were obviously higher in SKM-1 infected with SPAG6-shRNA
lentivirus than in the control group, whereas the expression of
PARP showed no significant difference between the two groups
(Fig. 2C and D). Furthermore, the
expression of apoptotic factors, including BAK, BAX, BID and
caspase-3, also showed statistical differences in the two groups
(Fig. 3E and F). These data
indicate that the TRAIL signal pathway was activated when the
expression of SPAG6 was reduced.
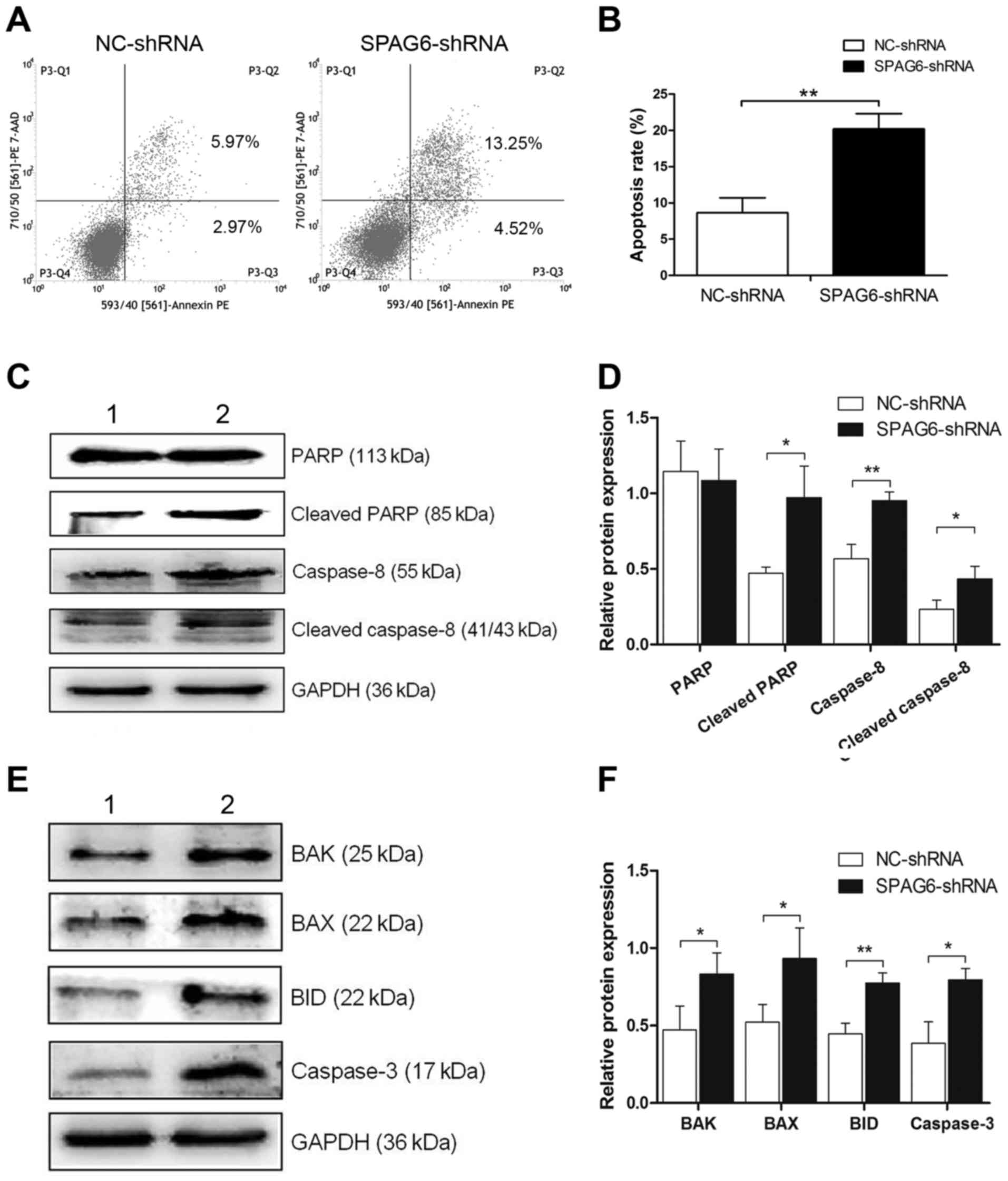 | Figure 2.TRAIL signal pathway is activated when
the expression of SPAG6 is inhibited. (A) After transfection with
lentivirus, the apoptosis rate was evaluated by flow cytometry. (B)
The analysis of the apoptosis rates is shown in (A). The apoptosis
rate of SKM-1 cells infected with SPAG6-shRNA significantly
increased as compared to cells infected with NC-shRNA. (C)
Expression of PARP, cleaved PARP, caspase-8 and cleaved caspase-8
protein was detected by western blot analysis. (D) TRAIL signal
pathway was activated. (E) Expression of BAK, BAX, BID and
caspase-3 protein was detected by western blot analysis. (F) These
proteins showed statistical differences between the groups. The
data are shown as mean ± SD. *P<0.05, **P<0.01. Lane 1,
NC-shRNA group; lane 2, SPAG6-shRNA group. TRAIL, TNF-related
apoptosis-inducing ligand; SPAG6, sperm-associated antigen 6. |
SPAG6 resists apoptosis induced by
TRAIL
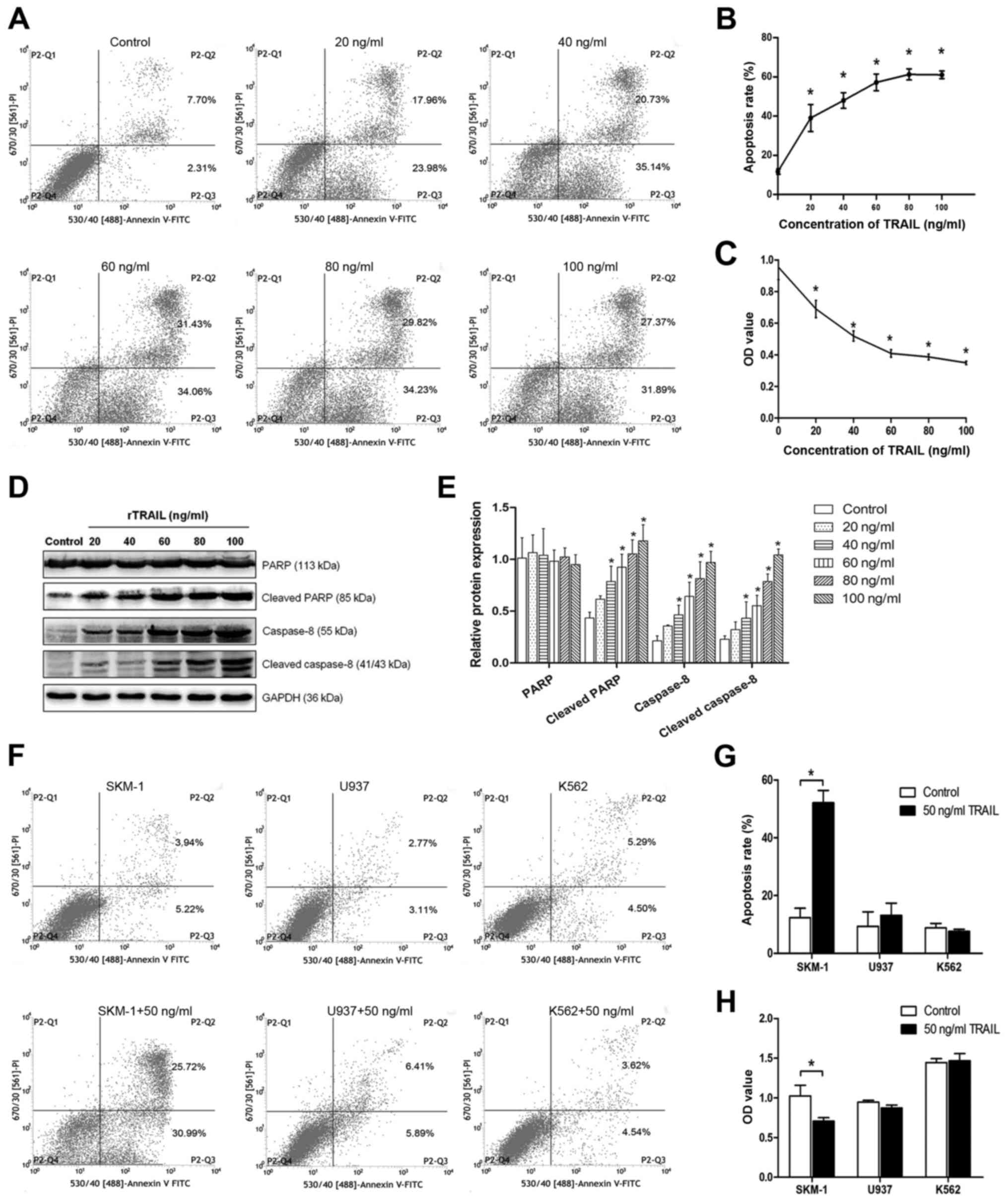
Following rTRAIL treatment, the percentage of
apoptosis cells increased, and the flow cytometry data indicated
that the higher concentration of TRAIL led to higher apoptosis
rates (Fig. 3A and B). To
investigate the growth of SKM-1 cells treated with different
concentrations of TRAIL, CCK-8 assays were performed to analyse
cell proliferation. Cell proliferation was suppressed in the cells
treated with TRAIL as compared to the normal control group
(Fig. 3C). In addition, western
blot analysis showed that the expression of related proteins was
higher after treatment (Fig. 3D and
E). However, as shown in Fig. 3F
and G, in the SKM-1, U937 and K562 cells after treatment with
the same concentration of TRAIL, the apoptosis rates were not
significantly different except for that of SKM-1 (the expression of
SPAG6, SKM-1<U937<K562), and similar results were also
observed in the CCK-8 assays (Fig.
3H). These data illustrate that a high expression of SPAG6 may
resist apoptosis induced by TRAIL.
SPAG6 has no effect on the expression
of TRAIL death receptors, except for FADD
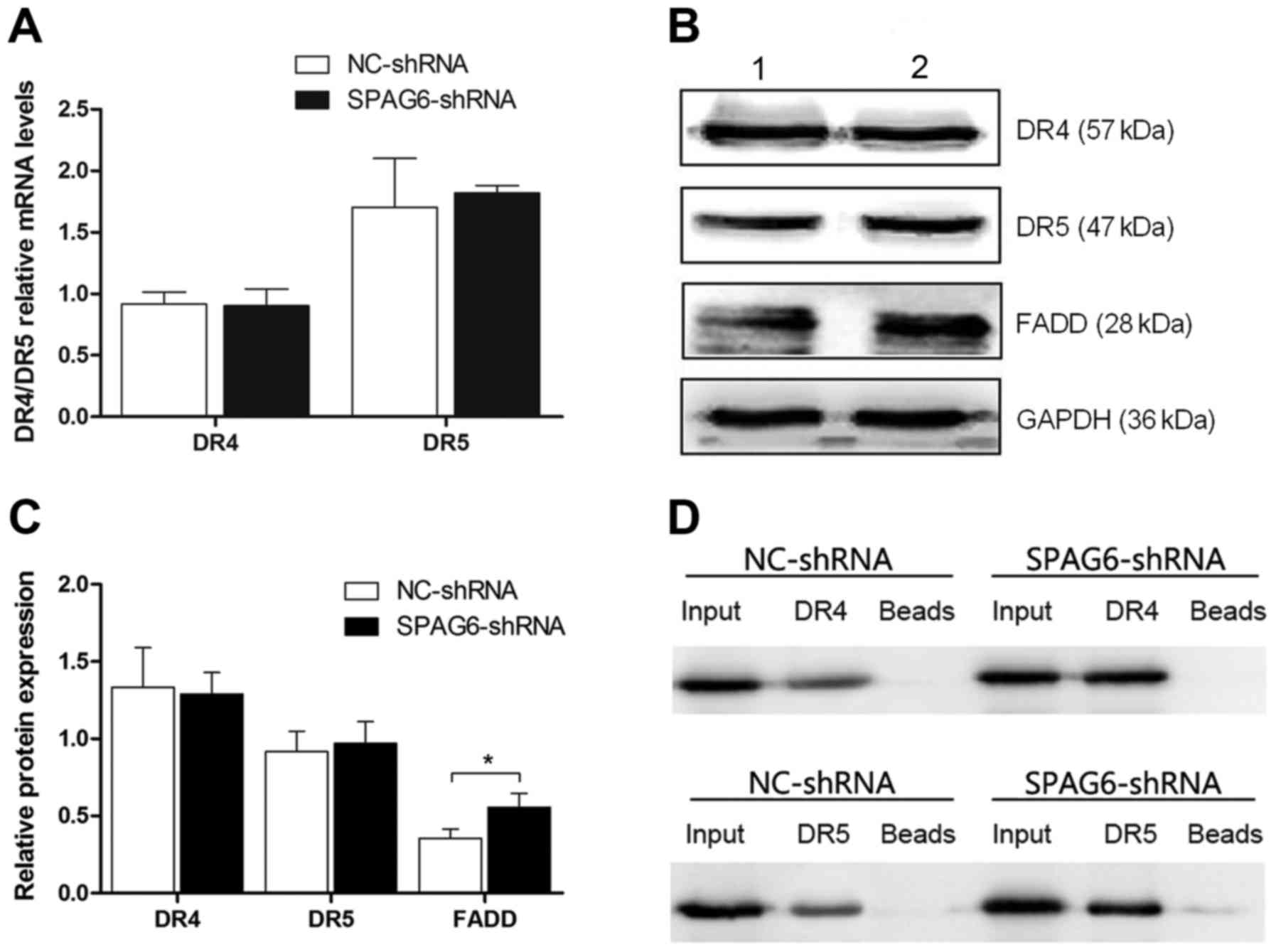
TRAIL induces apoptosis through DR4 (TNFRSF10A) and
DR5 (TNFRSF10B). Therefore, the degree of apoptosis induced by
TRAIL is closely related to the levels of DR4 and DR5. As
illustrated in the figure, both the mRNA and protein levels of the
two receptors were not changed (Fig.
4A-C). However, the expression of FADD was statistically
different (Fig. 4B and C),
indicating that the regulation of apoptosis may not be through DR4
and DR5 expression.
SPAG6 influences the interaction
between the FADD and TRAIL death receptors
As shown in the Fig.
4, the expressions of DR4 and DR5 showed no difference.
Therefore, immunoprecipitation was used to explore the relationship
between the FADD and TRAIL death receptors. FADD was used as the
bait protein to immunoprecipitate DR4 and DR5. Then, DR4 and DR5
were blotted. As shown in the figure, the interaction between FADD
and both death receptors increased in SKM-1 infected with SPAG6
shRNA-lentivirus as compared to the control group (Fig. 4D).
Discussion
Apoptosis is a process of programmed cell death,
which can occur in multicellular organisms in order to maintain
homeostasis. In brief, apoptosis is initiated through two pathways,
the extrinsic pathway and the intrinsic pathway mediated by
mitochondrion (17). Abnormal
apoptosis, including resistance to apoptosis, is regarded as an
important mechanism of tumorigenesis (18). MDS is characterized by ineffective
hematopoiesis leading to peripheral blood cytopenia, and one of the
possible mechanisms is excessive apoptosis in hematopoietic
precursors. It has been found that TRAIL and its receptors are
expressed at a low level in normal marrow, and the opposite occurs
in MDS marrow cells (19).
Furthermore, the expression of TRAIL signal inhibitor including
cFLIP, XIAP and the Bcl-2 family, is higher in patients with MDS in
advanced stages than those with MDS in the early stages (20–22).
The evidence indicates that the TRAIL signal pathway may play a
role in the development of MDS.
SPAG6 was first detected in human testis tissue. Its
major functions are participation in the maturation of reproductive
cells and maintaining sperm motility and fertility (10,23,24).
Recent studies have found that SPAG6 is upregulated in
CALM/AF10-positive leukemia and pediatric AML (11,12).
Additionally, a prospective multicenter study indicated that
patients with AML may obtain a better prognosis and relapse-free
survival (RFS) when SPAG6 is expressed at a low level (25). Furthermore, in lung and breast
cancer, SPAG6 is defined as a novel cancer-testis (CT) antigen,
which can be considered as a tumor marker and potential candidate
for use in cancer therapy (26),
implying that SPAG6 may lead to carcinogenesis and can be a
parameter for assessing their efficacy or prognosis. However, the
molecular mechanism of SPAG6 in hematologic malignancy has seldom
been studied.
Therefore, in the present study, we explored the
relationship between SPAG6 and the TRAIL signal pathway to
illustrate the possible mechanism of the regulation of apoptosis in
SKM-1. TRAIL can induce apoptosis in cells through its receptors at
the cell surface, DR4 and DR5 (27). After TRAIL binds to receptors, the
adapter protein FADD is recruited through cytoplasmic death
domains, and then, FADD interacts with pro-caspase-8 recruited to
form the death-inducing signaling complex (DISC). DISC then
mediates the activation of pro-caspase-8 (28). Caspase-8 directly activates
caspase-3 to cleave the substrates, such as PARP (29). Therefore, the activation of
caspase-8 and the cleavage of PARP are chosen as parameters to
measure apoptosis. Moreover, BID, a target of active caspase-8, is
cleaved to form truncated BID (tBID) and tBID then leads the
participation of mitochondrial apoptotic factors (30). As is shown in the results, after the
expression of SPAG6 was reduced by the lentivirus, the apoptosis
rates increased, and the TRAIL signal pathway was activated, and
both the intrinsic and extrinsic pathways were triggered. This
results suggests that SPAG6 may affect apoptosis through the TRAIL
signaling pathway.
A preliminary study indicated that a high expression
of SPAG6 is connected with apoptosis resistance in SKM-1 and K562
(13). In this study, when SKM-1
cells were treated with different concentrations of rTRAIL, the
apoptosis rates showed an increased tendency. However, given the
same concentration of rTRAIL, cells with high expression levels of
SPAG6 exhibited insensitivity to apoptosis induced by rTRAIL. In
addition, the expression of DR4 and DR5 showed no differences at
the mRNA and protein levels, but the expression of FADD increased.
These data imply that the regulation may not occur through the
receptors. In order to explore the possible mechanism,
immunoprecipitation was used to measure the interaction between
FADD and both DR4 and DR5. It was found that the interaction
between FADD and DR4 and that between FADD and DR5 both increased.
Moreover, p53 is the most commonly mutated gene in human cancer
(31), and large quantities of
studies have demonstrated its anticancer functions. Additionally,
some studies have shown that p53 may also participate in the
regulation of apoptosis induced by TRAIL, and the possible
approaches are to influence the expression of the TRAIL receptors
(32,33) or the BCL-2 family (34). In addition, TRAIL can induce not
only the initiation of apoptosis but also the non-apoptotic
pathways, such as nuclear factor-κB (NF-κB), phosphatidylinositol
3-kinase (PI3K) and Akt, and mitogen activated protein kinases
(MAPKs) (35). It is important to
note that these studies are performed on the cellular level, so an
in vivo study is needed to confirm the results. Furthermore,
there are limitations inherent in using a cell line in culture
versus using primary bone marrow cells from patients. Therefore,
further studies are needed to explore the possible mechanisms.
In conclusion, this study demonstrates that SPAG6
may regulate apoptosis through the TRAIL signal pathway by
inhibiting the expression of FADD and the interactions between FADD
and the TRAIL death receptors. TRAIL has been proven to possess
anticancer functions. In animal models, it has shown that the
growth of a TRAIL-sensitive tumor is suppressed by rTRAIL without
significant system toxicity (36,37).
MDS treatments have rapidly improved; however, there is still lack
of effective methods, and antileukemic drugs can enhance apoptosis
mediated by TRAIL (38). The above
may indicate a new therapy for MDS. Therefore, SPAG6 may be a
potential target in therapy for MDS, and it may also be worthwhile
to explore drugs against SPAG6 function.
Acknowledgements
We thank Mr. Nian Zhou, Ms. Xingyi Kuang and Mr.
Hongrong Zhang for their valuable assistance. We would like to
acknowledge the service provided by Laboratory of Lipid and Glucose
Metabolism, the First Affiliated Hospital of Chongqing Medical
University, Chongqing, China. This study is supported by the
Natural Science Foundation of Chongqing (no. CSTC2013jjB10020) and
the National Natural Sciences Foundation of China (no.
81570109).
References
|
1
|
Corey SJ, Minden MD, Barber DL, Kantarjian
H, Wang JC and Schimmer AD: Myelodysplastic syndromes: the
complexity of stem-cell diseases. Nat Rev Cancer. 7:118–129. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wouters BJ and Delwel R: Epigenetics and
approaches to targeted epigenetic therapy in acute myeloid
leukemia. Blood. 127:42–52. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shih AH, Abdel-Wahab O, Patel JP and
Levine RL: The role of mutations in epigenetic regulators in
myeloid malignancies. Nat Rev Cancer. 12:599–612. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Haase D, Germing U, Schanz J, Pfeilstöcker
M, Nösslinger T, Hildebrandt B, Kundgen A, Lübbert M, Kunzmann R,
Giagounidis AA, et al: New insights into the prognostic impact of
the karyotype in MDS and correlation with subtypes: evidence from a
core dataset of 2124 patients. Blood. 110:4385–4395. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bernasconi P, Klersy C, Boni M, Cavigliano
PM, Calatroni S, Giardini I, Rocca B, Zappatore R, Caresana M,
Dambruoso I, et al: World Health Organization classification in
combination with cytogenetic markers improves the prognostic
stratification of patients with de novo primary myelodysplastic
syndromes. Br J Haematol. 137:193–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schanz J, Steidl C, Fonatsch C,
Pfeilstöcker M, Nösslinger T, Tuechler H, Valent P, Hildebrandt B,
Giagounidis A, Aul C, et al: Coalesced multicentric analysis of
2,351 patients with myelodysplastic syndromes indicates an
underestimation of poor-risk cytogenetics of myelodysplastic
syndromes in the international prognostic scoring system. J Clin
Oncol. 29:1963–1970. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Maciejewski JP and Mufti GJ: Whole genome
scanning as a cytogenetic tool in hematologic malignancies. Blood.
112:965–974. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kralovics R: Genetic complexity of
myeloproliferative neoplasms. Leukemia. 22:1841–1848. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang L, Fidler C, Nadig N, Giagounidis A,
Della Porta MG, Malcovati L, Killick S, Gattermann N, Aul C,
Boultwood J, et al: Genome-wide analysis of copy number changes and
loss of heterozygosity in myelodysplastic syndrome with del(5q)
using high-density single nucleotide polymorphism arrays.
Haematologica. 93:994–1000. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sapiro R, Kostetskii I, Olds-Clarke P,
Gerton GL, Radice GL and Strauss JF III: Male infertility, impaired
sperm motility, and hydrocephalus in mice deficient in
sperm-associated antigen 6. Mol Cell Biol. 22:6298–6305. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Steinbach D, Schramm A, Eggert A, Onda M,
Dawczynski K, Rump A, Pastan I, Wittig S, Pfaffendorf N, Voigt A,
et al: Identification of a set of seven genes for the monitoring of
minimal residual disease in pediatric acute myeloid leukemia. Clin
Cancer Res. 12:2434–2441. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mulaw MA, Krause A, Deshpande AJ, Krause
LF, Rouhi A, La Starza R, Borkhardt A, Buske C, Mecucci C, Ludwig
WD, et al: CALM/AF10-positive leukemias show upregulation of
genes involved in chromatin assembly and DNA repair processes and
of genes adjacent to the breakpoint at 10p12. Leukemia.
26:1012–1019. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang B, Wang L, Luo X, Chen L, Yang Z and
Liu L: SPAG6 silencing inhibits the growth of the malignant myeloid
cell lines SKM-1 and K562 via activating p53 and caspase
activation-dependent apoptosis. Int J Oncol. 46:649–656.
2015.PubMed/NCBI
|
|
14
|
Kerbauy DB and Deeg HJ: Apoptosis and
antiapoptotic mechanisms in the progression of myelodysplastic
syndrome. Exp Hematol. 35:1739–1746. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Griffith TS and Lynch DH: TRAIL: a
molecule with multiple receptors and control mechanisms. Curr Opin
Immunol. 10:559–563. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kranz D and Boutros M: A synthetic lethal
screen identifies FAT1 as an antagonist of caspase-8 in extrinsic
apoptosis. EMBO J. 33:181–197. 2014.PubMed/NCBI
|
|
17
|
Igney FH and Krammer PH: Death and
anti-death: tumour resistance to apoptosis. Nat Rev Cancer.
2:277–288. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Martin GS: Cell signaling and cancer.
Cancer Cell. 4:167–174. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zang DY, Goodwin RG, Loken MR, Bryant E
and Deeg HJ: Expression of tumor necrosis factor-related
apoptosis-inducing ligand, Apo2L, and its receptors in
myelodysplastic syndrome: effects on in vitro hemopoiesis. Blood.
98:3058–3065. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Benesch M, Platzbecker U, Ward J, Deeg HJ
and Leisenring W: Expression of FLIP(Long) and FLIP(Short) in bone
marrow mononuclear and CD34+ cells in patients with
myelodysplastic syndrome: correlation with apoptosis. Leukemia.
17:2460–2466. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yamamoto K, Abe S, Nakagawa Y, Suzuki K,
Hasegawa M, Inoue M, Kurata M, Hirokawa K and Kitagawa M:
Expression of IAP family proteins in myelodysplastic syndromes
transforming to overt leukemia. Leuk Res. 28:1203–1211. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Parker JE, Mufti GJ, Rasool F, Mijovic A,
Devereux S and Pagliuca A: The role of apoptosis, proliferation,
and the Bcl-2-related proteins in the myelodysplastic syndromes and
acute myeloid leukemia secondary to MDS. Blood. 96:3932–3938.
2000.PubMed/NCBI
|
|
23
|
Zhang Z, Jones BH, Tang W, Moss SB, Wei Z,
Ho C, Pollack M, Horowitz E, Bennett J, Baker ME, et al: Dissecting
the axoneme interactome: the mammalian orthologue of
Chlamydomonas PF6 interacts with sperm-associated antigen 6,
the mammalian orthologue of Chlamydomonas PF16. Mol Cell
Proteomics. 4:914–923. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pearson CG, Giddings TH Jr and Winey M:
Basal body components exhibit differential protein dynamics during
nascent basal body assembly. Mol Biol Cell. 20:904–914. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Steinbach D, Bader P, Willasch A,
Bartholomae S, Debatin KM, Zimmermann M, Creutzig U, Reinhardt D
and Gruhn B: Prospective validation of a new method of monitoring
minimal residual disease in childhood acute myelogenous leukemia.
Clin Cancer Res. 21:1353–1359. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Silina K, Zayakin P, Kalnina Z, Ivanova L,
Meistere I, Endzelins E, Abols A, Stengrēvics A, Leja M, Ducena K,
et al: Sperm-associated antigens as targets for cancer
immunotherapy: expression pattern and humoral immune response in
cancer patients. J Immunother. 34:28–44. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ashkenazi A and Dixit VM: Death receptors:
signaling and modulation. Science. 281:1305–1308. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Scaffidi C, Kirchhoff S, Krammer PH and
Peter ME: Apoptosis signaling in lymphocytes. Curr Opin Immunol.
11:277–285. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gonzalvez F and Ashkenazi A: New insights
into apoptosis signaling by Apo2L/TRAIL. Oncogene. 29:4752–4765.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li H, Zhu H, Xu CJ and Yuan J: Cleavage of
BID by caspase 8 mediates the mitochondrial damage in the Fas
pathway of apoptosis. Cell. 94:491–501. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kandoth C, McLellan MD, Vandin F, Ye K,
Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al:
Mutational landscape and significance across 12 major cancer types.
Nature. 502:333–339. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu X, Yue P, Khuri FR and Sun SY: p53
upregulates death receptor 4 expression through an intronic p53
binding site. Cancer Res. 64:5078–5083. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Takimoto R and El-Deiry WS: Wild-type p53
transactivates the KILLER/DR5 gene through an intronic
sequence-specific DNA-binding site. Oncogene. 19:1735–1743. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Henry H, Thomas A, Shen Y and White E:
Regulation of the mitochondrial checkpoint in p53-mediated
apoptosis confers resistance to cell death. Oncogene. 21:748–760.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Falschlehner C, Emmerich CH, Gerlach B and
Walczak H: TRAIL signalling: decisions between life and death. Int
J Biochem Cell Biol. 39:1462–1475. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ashkenazi A, Pai RC, Fong S, Leung S,
Lawrence DA, Marsters SA, Blackie C, Chang L, McMurtrey AE, Hebert
A, et al: Safety and antitumor activity of recombinant soluble Apo2
ligand. J Clin Invest. 104:155–162. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Walczak H, Miller RE, Ariail K, Gliniak B,
Griffith TS, Kubin M, Chin W, Jones J, Woodward A, Le T, et al:
Tumoricidal activity of tumor necrosis factor-related
apoptosis-inducing ligand in vivo. Nat Med. 5:157–163. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wen J, Ramadevi N, Nguyen D, Perkins C,
Worthington E and Bhalla K: Antileukemic drugs increase death
receptor 5 levels and enhance Apo-2L-induced apoptosis of human
acute leukemia cells. Blood. 96:3900–3906. 2000.PubMed/NCBI
|