Introduction
Colon cancer is a commonly occurring malignant tumor
in the world. It is considered as the fourth most commonly
diagnosed cancer and the second leading cause of cancer-related
death in China. Furthermore, due to environmental and lifestyle
changes, the incidence of colon cancer has gradually increased
(1,2). Nowadays, 5-fluorouracil (5-FU) and
cisplatin (DDP) are widely used in treating colon cancer. However,
drug resistance and rapid growth of tumors are the major
obstructions in the treatment of colon cancer. Therefore, the
combination of anticancer drugs and other regimens is often
practiced to strengthen the efficacy of chemotherapy and suppress
cancer proliferation. Previous research has proved that microRNAs
(miRNAs) could restrain cancer cell growth and sensitize the
pharmacological action of chemotherapy drugs (3–5).
miRNAs are a class of small noncoding RNA molecules
that contain approximately 22 nucleotides. miRNAs are widely known
to regulate gene expression at the post-transcriptional level and
play an important role in diverse biological processes of cells,
such as proliferation, differentiation, and apoptosis (6). Accumulated research indicates that
aberrant miRNA expression is closely related to various types of
human cancers (7). It has been
reported that 50% of miRNAs are located in the chromosomal regions,
which often increase or decrease in human cancer cells (8). Altered miRNA expression profiles are
reported in many types of cancers, including liver, gastric, and
prostate cancers (9–11). Moreover, recent studies revealed
that miRNAs could function as tumor suppressors or oncogenes in
human colon cancer (12,13).
Quantitative reverse transcription polymerase chain
reaction (qRT-PCR) was used in this study to detect the expression
of miR-101 in colon cancer tissues and adjacent normal tissues.
Recombinant adenovirus Ad-miR-101 was used to upregulate the
expression of miR-101 in HT-29 and RKO colon cancer cells, and the
effects of miR-101 on cell proliferation, colony formation,
migration, and invasion abilities were analyzed. Moreover, the
effect of miR-101 on the drug sensitivity of colon cancer cells to
5-FU and DDP was explored. This investigation provided insight into
the function of miR-101 in colon cancer and indicates the potential
application of miR-101 in cancer therapy.
Materials and methods
Patients
Forty-two pairs of matched colon tumorous and
adjacent non-tumorous mucosal tissues (>5 cm from the edge of
the tumor) were collected from the patients with colon cancer at
the Zhejiang Provincial People's Hospital between April 2015 and
December 2015. The cohort comprised 20 patients aged <60 years
and 22 patients aged ≥60 years. The patient age ranged from 43 to
87 years; 17 had left colon tumor and 25 had right colon cancer.
Furthermore, 13 had adenocarcinoma and 29 had mucinous carcinoma.
The number of patients classified by TNM stage were as follows: 36
were at TNM stages I–III and 6 were at TNM stage IV. Informed
consent from all the patients, before surgery was obtained prior to
the use of their tissues. The use of all specimens was approved by
the ethics committee of Zhejiang Provincial People's Hospital. None
of the patients received radiotherapy or chemotherapy prior to the
operation. After surgical removal, the tissues were immediately
frozen in liquid nitrogen and stored at −80°C until further
use.
Cell culture
Human colon cell lines HT-29 and RKO were purchased
from Shanghai Institute of Digestive Surgery (Shanghai, China) and
cultured in Dulbecco's modified Eagle's medium (DMEM) containing
10% fetal bovine serum, 50 U/ml penicillin, and 50 µg/ml
streptomycin at 37°C under an atmosphere of 5% CO2.
Medium renewal was performed every 3–4 days, and subculturing was
done when the cells reached 80–90% confluence.
Reagents
TRIzol reagent was purchased from Invitrogen
(Carlsbad, CA, USA; 15596–018). One-Step Prime Script miRNA cDNA
synthesis kit and SYBR Premix Ex Taq II quantitative PCR reagent
box were purchased from Takara, Japan. Recombinant adenovirus
Ad-miR-101 and Ad-EGFP were constructed by the key laboratory of
gastroenterology of Zhejiang Province.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT),
acridine orange (AO), and ethidium bromide (EB) were obtained from
Sigma (St. Louis, MO, USA; CA1140). Migration kit (Transwell
chamber) assays were obtained from BD Biosciences (Franklin Lakes,
NJ, USA; PIEP12R48); invasion kit (Boyden chamber) assays were
purchased from Millipore Corp. (Billerica, MA, USA;
QIA129-1KIT).
Detection of miR-101 in colorectal
cancer tissues and adjacent tissues
Total RNA extraction and reverse
transcription
Total RNA was extracted according to the RNA
Extraction kit protocol. cDNA was synthesized by a reverse
transcription reaction: 2X miRNA reaction buffer mix (10 µl), 0.1%
phosphate-buffered saline (PBS) (2 µl), miRNA Prime Script RT
enzyme mix (2 µl), total RNA (1 µl), and RNase-free dH2O
(5 µl); reaction conditions: 37°C for 60 min and 85°C for 5
sec.
qRT-PCR assay for miR-101
miR-101 positive specific primer sequence:
5′-TACAGTACTGTGATAACTGAA-3′, reference U6B specific forward primer
sequence: 5′-ACGCAAATTCGTGAAGCGTT-3′; the reverse primer was the
universal primer provided by the reverse transcription kit. PCR
reaction system: SYBR Premix 10 µl, ROX 0.4 µl, reverse primer 0.5
µl, template 1 µl, and dH2O 7.6 µl; reaction conditions:
94°C for 4 min, 40 cycles of 94°C for 5 sec, 55°C for 20 sec, and
72°C for 20 sec; melting curve program: 95°C for 1 min, 55°C for 30
sec, and 95°C for 30 sec, 1 cycle. The relative expression of
miR-101 was calculated by the 2−∆Ct method: ∆Ct =
Ct(miR-101) - Ct(U6B).
Recombinant adenovirus infection
Recombinant adenovirus Ad-miR-101 was used to infect
HT-29 and RKO cells at a multiplicity of infection of 100 to
upregulate the expression level of miR-101. Recombinant adenovirus
Ad-EGFP was used as the control. The infected cells were harvested
and used for the following assays 24 h after infection. The miR-101
levels of infected cells were detected by qRT-PCR.
MTT assay and colony formation assay
for cell proliferation
MTT assay for cell proliferation
Ad-mir-101- and Ad-EGFP-infected HT-29 and RKO cells
were added to 96-well plates at a concentration of 2×103
cells/well in sextuple and placed at 37°C in a 5% CO2
incubator with saturated humidity. The MTT reagent (5 mg/ml, 20 µl)
was added to each well and further incubated for 4 h after 24, 48,
and 72 h. Dimethyl sulfoxide (150 µl) was added to quench the
reaction, and the absorbance (A) was measured at 570 and 630 nm
using a microplate reader. The relative cell proliferation rate was
calculated by the following formula:
Relativeproliferationrate(%)=(A570nm–A630nm)studygroup(A570nm–A630nm)controlgroupx100%
Colony formation assay
A total of 200 cells from the experimental and
control groups were cultured in a 6-well plate; the medium was
replenished every 5 days. The colonies were fixed with methanol for
15 min and stained with Giemsa stain after 2 weeks.
In vitro migration and invasion
assay
Migration assay
A 24-well Transwell chamber kit was used for the
assay. DMEM culture medium (200 µl, without any bovine serum)
containing 4×104 cells from the experimental and control
groups was added to the upper chamber, and 10% bovine serum was
added to the lower chamber. The cells were removed from the chamber
with a cotton swab 24 h post-culture. The cells adhering below the
surface of a membrane were fixed in methanol and stained. Five
randomly selected fields of view (x200) were observed under the
microscope to count the cells and calculate the mean.
Invasion assay
The Matrigel invasion chamber kit was used for the
assay. DMEM culture medium (300 µl) without any bovine serum was
added to the upper chamber, and 5×105 cells from each
group were suspended in it. The lower chamber consisted of DMEM
with 10% bovine serum. The cells were fixed and stained after 48-h
incubation; the cells in five randomly selected fields of view
(x200) were counted under a microscope, and the average was
calculated.
MTT assay and AO/EB staining analysis
of the sensitivity to chemotherapeutics
MTT assay for sensitivity to
chemotherapeutics
MTT assay was performed to analyze the synergism
between miR-101 and 5-FU or DDP in cell proliferation. Ad-miR-101-
and Ad-EGFP-infected HT-29 and RKO cells were seeded in 96-well
plates at 2×103 cells per well overnight. Then, the
cells were treated with PBS or 5-FU (5 µg/ml) or DDP (1 µg/ml). At
the end of 24-, 48-, and 72-h incubation, the MTT assay was
performed as described earlier. Ad-EGFP-infected cells in PBS
served as the negative control.
Relativeproliferationrate(%)=(A570nm–A630nm)studygroup(A570nm–A630nm)negativecontrolgroupx100%
AO/EB double staining for detecting sensitivity
to chemotherapeutics
AO/EB staining was performed to analyze the
synergism between miR-101 and 5-FU or DDP in cell apoptosis.
Ad-miR-101- and Ad-EGFP-infected HT-29 and RKO cells were seeded in
24-well plates at 2×104 cells per well overnight. Then,
PBS or 5-FU (5 µg/ml) or DDP (1 µg/ml) was added to respective
wells. The culture medium in each well was replaced with 100 µl of
PBS 24 h later. Then, the cells were stained with dual fluorescent
staining solution (2 µl) containing 100 µg/ml AO and 100 µg/ml EB,
and viewed under an epifluorescence microscope. Multiple
photographs at ×400 magnification were taken for analyzing the
results.
Statistic analyses
SPSS18.0 statistical software was used for data
analysis. Measurement data were expressed as mean ± standard
deviation. Paired data were analyzed using paired-samples t-test,
and ranked data using χ2 or Fisher's exact test.
Statistical significance was set at P<0.05.
Results
Decreased expression of miR-101 in
colon cancer tissues
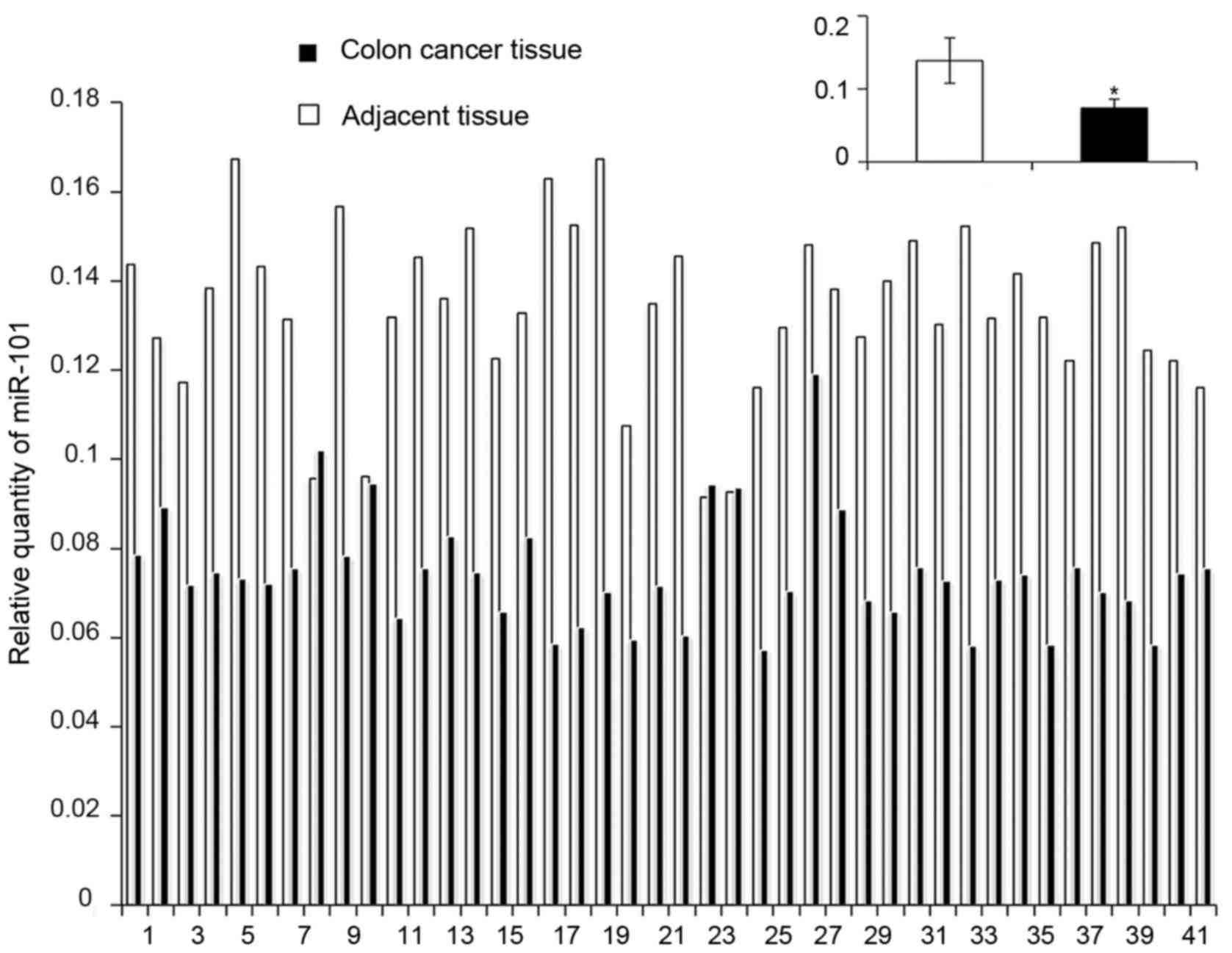
The relative expression of miR-101 in colon cancer
tissues was significantly lower than that in the adjacent tissues
(7.40×10−2±1.22×10−2 vs.
1.31×10-1±3.11×10−2, P<0.05) (Fig. 1). Moreover, the expression of
miR-101 was negatively correlated with the pathological type of
colon cancer in patients (P<0.05), but it was not correlated
with sex, age, tumor location, TNM staging, and differentiation
(P>0.05) (Table I).
 | Table I.Relationship between miR-101
expression and clinicopathological features of colon cancer. |
Table I.
Relationship between miR-101
expression and clinicopathological features of colon cancer.
| Clinical feature | n | Relative expression
of miR-101 | P-value
(χ2 test) |
|---|
| Sex |
|
| 0.350 |
|
Male | 26 |
7.51×10−2±1.21×10−2 |
|
|
Female | 16 |
7.35×10−2±1.45×10−2 |
|
| Age (year) |
|
| 0.720 |
|
<60 | 20 |
7.53×10−2±1.11×10−2 |
|
|
≥60 | 22 |
7.38×10−2±1.45×10−2 |
|
| Tumor location |
|
| 0.261 |
| Left
hemicolon | 17 |
7.72×10−2±1.08×10−2 |
|
| Right
hemicolon | 25 |
7.26×10−2±1.41×10−2 |
|
| Pathological
type |
|
| 0.0378 |
|
Adenocarcinoma | 29 |
7.69×10−2±1.43×10−2 |
|
|
Mucinous carcinoma | 13 |
6.92×10−2±6.93×10−3 |
|
| Differentiation
degree |
|
| 0.0990 |
| Low
differentiation | 10 |
7.94×10−2±9.94×10−3 |
|
| High
and medium differentiation | 32 |
7.32×10−2±1.34×10−2 |
|
| TNM staging |
|
| 0.200 |
|
I–III | 36 |
7.52×10−2±1.36×10−2 |
|
| IV | 6 |
7.03×10−2±6.62×10−3 |
|
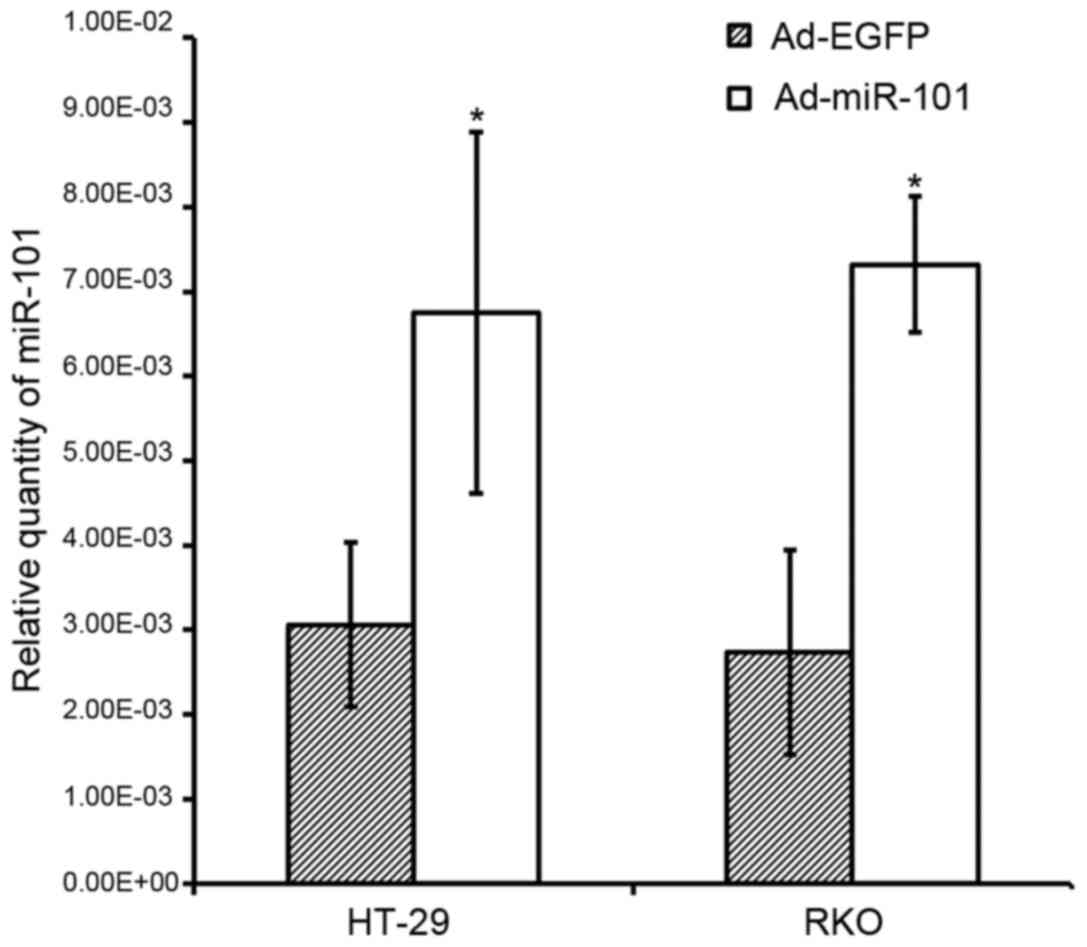
miR-101 expression is upregulated 24 h
after infection
The results showed that the expression level of
miR-101 of Ad-miR-101-infected HT-29 and RKO cells
(6.75×10−2±2.14×10−3 and
7.32×10−3±8.01×10−4) was much higher than
that of the Ad-EGFP-infected cells
(3.06×10−3±9.70×10−4 and
2.73×10−3±1.21×10−3, P<0.05, Fig. 2).
In summary, the expression of miR-101 of HT-29 and
RKO cell lines was significantly higher than that of the negative
control group after Ad-miR-101 infection. These results suggested
that Ad-miR-101 could effectively upregulate the expression of
miR-101 in colorectal cancer cell lines.
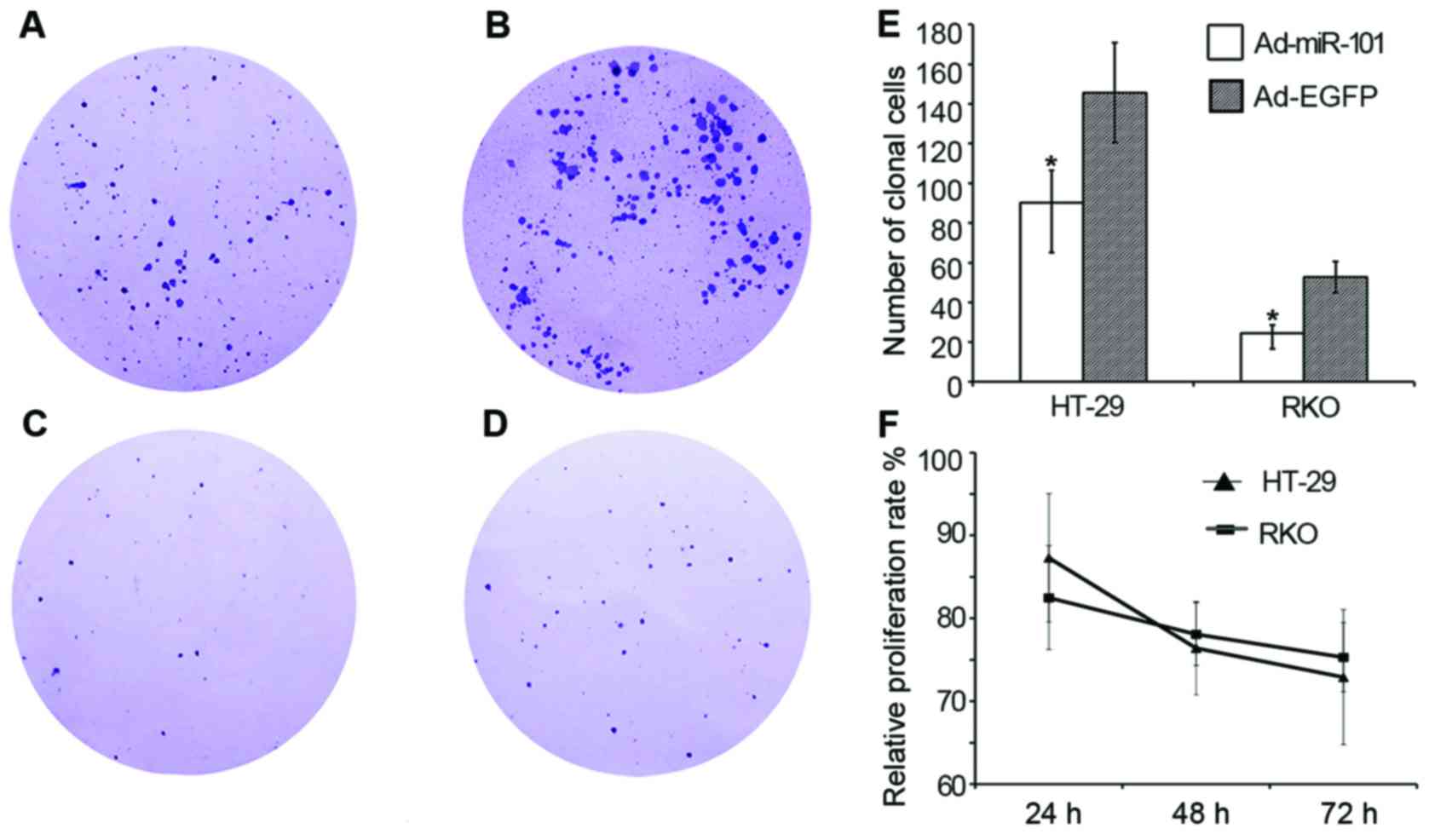
Effect of miR-101 on the proliferation
of HT-29 and RKO cells
MTT assay results (Fig.
3) showed that the relative proliferation rate of HT-29 cells
in the experimental group was 87.3±7.74%, 76.4±5.63%, and
72.9±8.15% after 24-, 48-, and 72-h cultures, respectively
(P<0.05), compared with the negative control group. The relative
proliferation rate of RKO cells in the experimental group was
82.5±6.26%, 78.1±3.78%, and 75.3±4.17% (P<0.05). It indicated
that the upregulation of miR-101 expression could inhibit the
proliferation of colon cancer cell lines HT-29 and RKO.
Consequently, the cloning formation assay was used
after 2 weeks to detect the cell proliferation ability. HT-29 and
RKO cell lines with overexpressed miR-101 demonstrated the clone
formation ability (90.3±16.4 and 24.5±4.2, respectively), and the
clone formation ability of the negative control group was
145.7±25.2 and 52.8±7.9, respectively. The clone formation ability
of the experimental group decreased significantly compared with the
negative control group (P<0.05), as shown in Fig. 3. The results suggested that the
upregulation of miR-101 had a significant inhibitory effect on the
proliferation of colon cancer cell lines HT-29 and RKO.
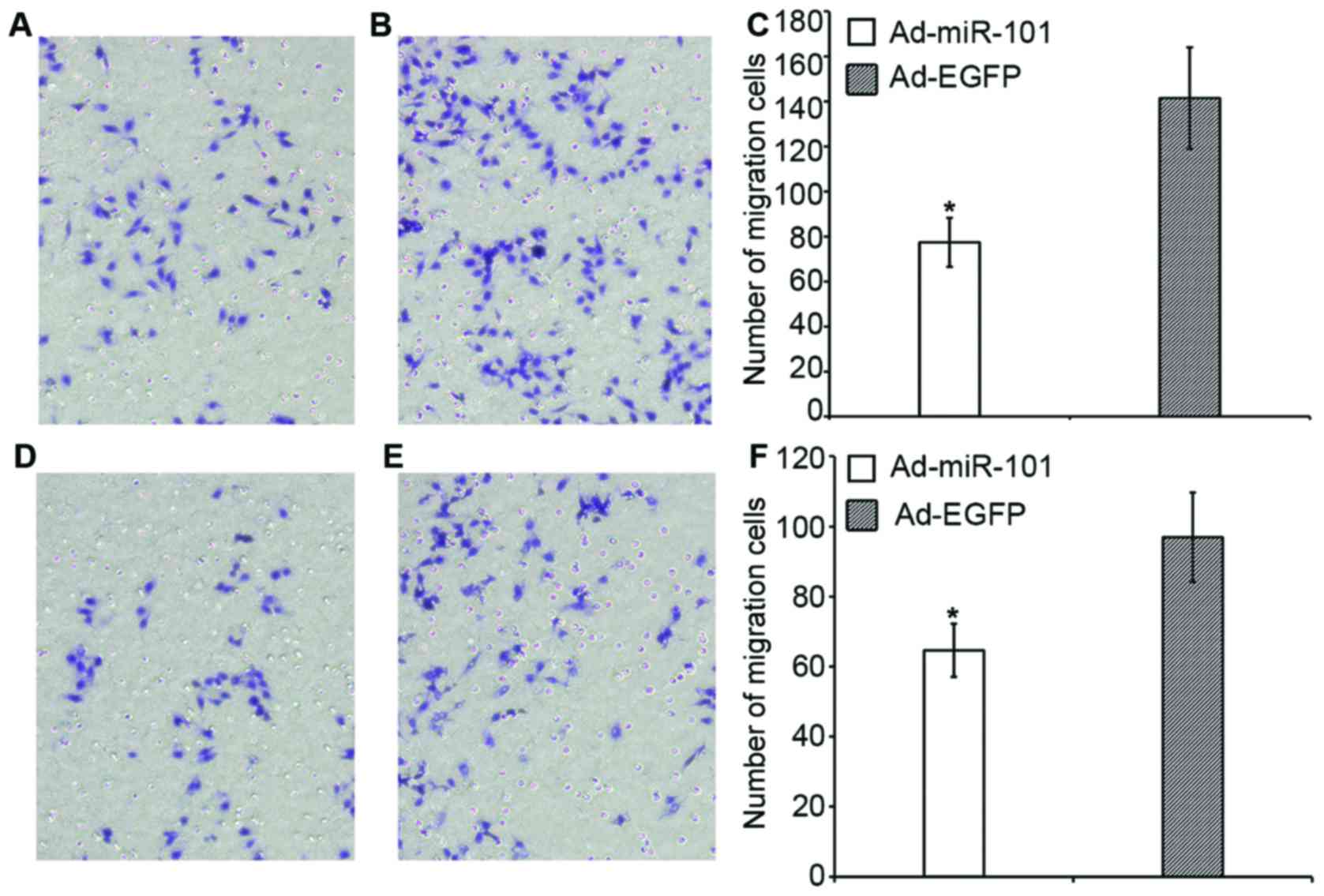
Effect of miR-101 on the migration and
invasion of colon cancer cells
The migration and invasion abilities of
overexpressed miR-101 cells were significantly lower than those of
the negative control group. Fig. 4
shows that in the migration assay, the number of HT-29 cells in the
experimental and negative control groups was 77.4±10.9 and
141.5±22.7, respectively; the invasion capacity decreased
approximately 45.3% (P<0.05). The number of RKO cells in the
experimental and negative control groups was 64.6±7.6 and
96.9±12.8, respectively; the migration ability decreased
approximately 33.3% (P<0.05).
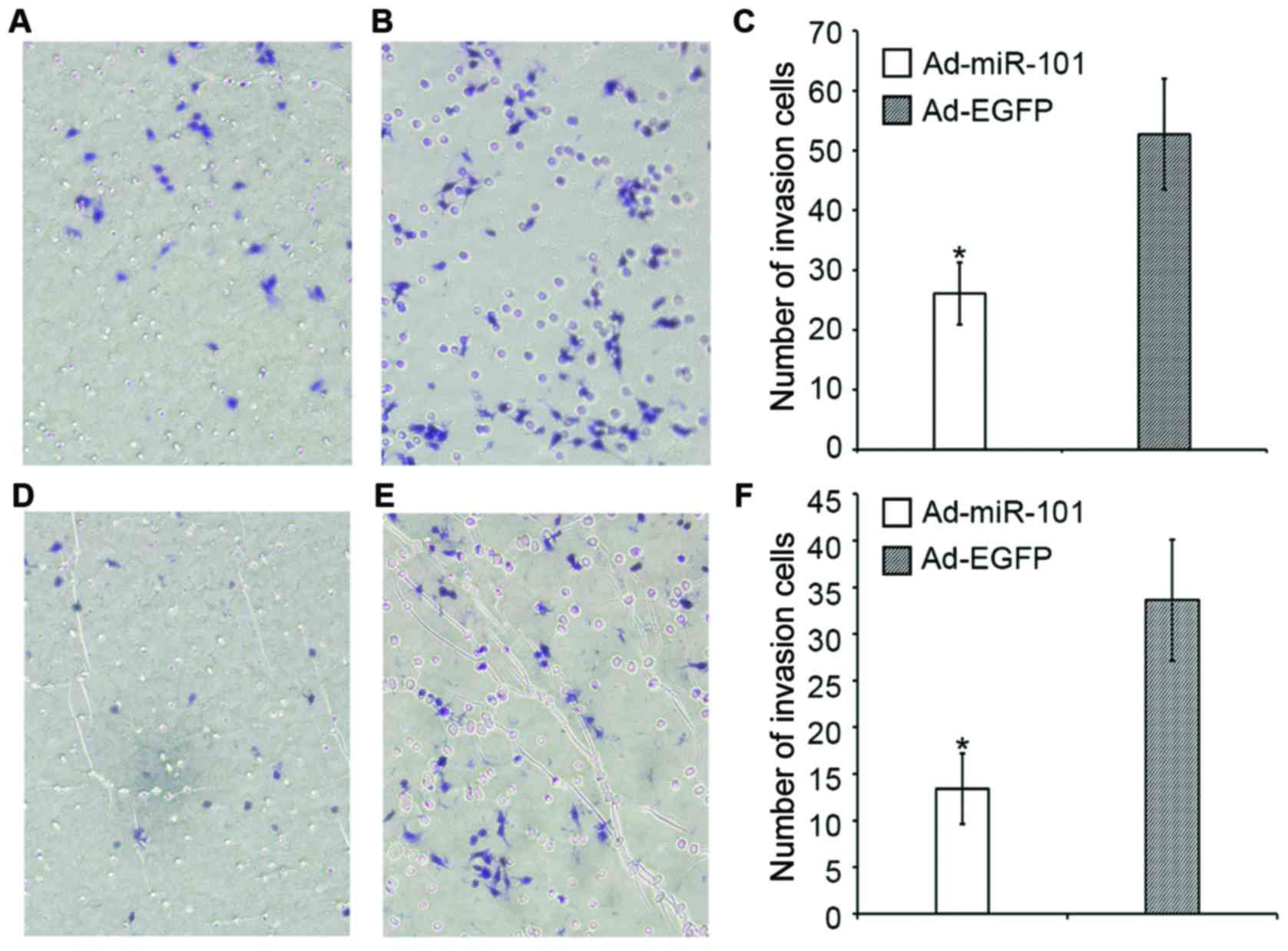
Fig. 5 shows that in
the invasion assay, the number of HT-29 cells in the experimental
and negative control groups was 26.1±5.2 and 52.7±9.3,
respectively; the invasion capacity decreased approximately 50.4%
(P<0.05). The number of RKO cells in the experimental and
negative control groups was 13.4±3.8 and 33.6±6.5, respectively;
the invasion ability decreased approximately 60.1% (P<0.05).
miR-101 could enhance the sensitivity
to chemotherapeutics
MTT assays showed that miR-101 increased the
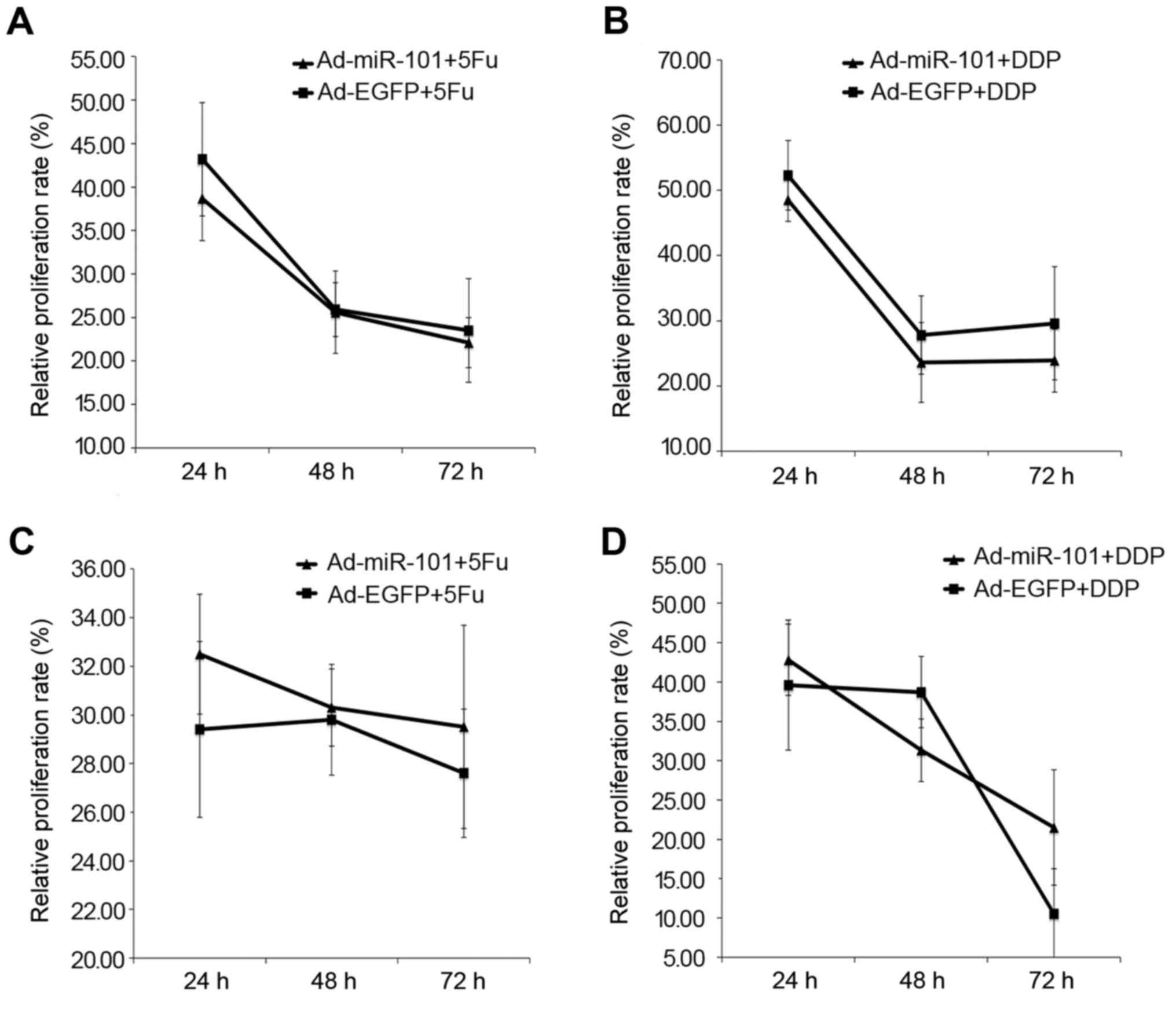
inhibitory effect of 5-FU and DDP on HT-29 cells. Fig. 6A shows that the relative
proliferation rate of HT-29 cells in the experimental group was
38.7±6.53%, 25.6±3.11%, and 22.1±5.97%, respectively, 24, 48, and
72 h after FU treatment (P<0.05) compared with the negative
control group; the relative proliferation rate of Ad-EGFP-infected
HT-29 cells was 43.2±4.89%, 25.9±4.73%, and 23.5±2.87%,
respectively (P<0.05). Therefore, the relative proliferation
rate of Ad-miR-101-infected cells was less than that of
Ad-EGFP-infected cells (P<0.05), implying that miR-101 could
improve the chemotherapy of HT-29 cells by 5-FU.
Fig. 6B shows that
the relative proliferation rate of Ad-miR-101-infected HT-29 cells
was 48.5±5.32%, 23.6±5.99%, and 23.9±8.67%, respectively, 24, 48,
and 72 h after DDP treatment (P<0.05) compared with the negative
control group; the relative proliferation rate of HT-29 cells in
the negative control group was 52.3±3.26%, 27.8±6.12%, and
29.6±4.84%, respectively (P<0.05). Therefore, the relative
proliferation rate of Ad-miR-101-infected cells was less than that
of Ad-EGFP-infected cells (P<0.05), implying that miR-101
enhanced the cytotoxic effect of DDP in HT-29 cells. However, the
sensitivity to chemotherapy was not increased by the overexpression
of miR-101 in RKO cells.
Fig. 6C shows that
the relative proliferation rate of HT-29 cells in the experimental
and negative control groups was 32.5±2.47%, 30.3±1.59%, and
29.5±4.18%, and 29.4±3.62%, 29.8±2.28%, and 27.6±2.64%,
respectively, 24, 48, and 72 h after 5-FU treatment (P>0.05).
The difference between the two groups was not statistically
significant. It did not imply that miR-101 could improve the
killing effect of 5-FU on RKO cells.
Fig. 6D shows that
the 24-, 48-, and 72-h relative proliferation rate of RKO cells in
the experimental group was 42.8±4.53%, 31.3±3.97%, and 21.5±7.33%,
and the relative proliferation rate of RKO cells in the negative
control group was 39.6±8.26%, 38.7±4.52%, and 10.5±5.74%,
respectively, on DDP treatment. No significant difference was
observed between the two groups (both P>0.05). It did not imply
that miR-101 could improve the killing effect of cisplatin on RKO
cells.
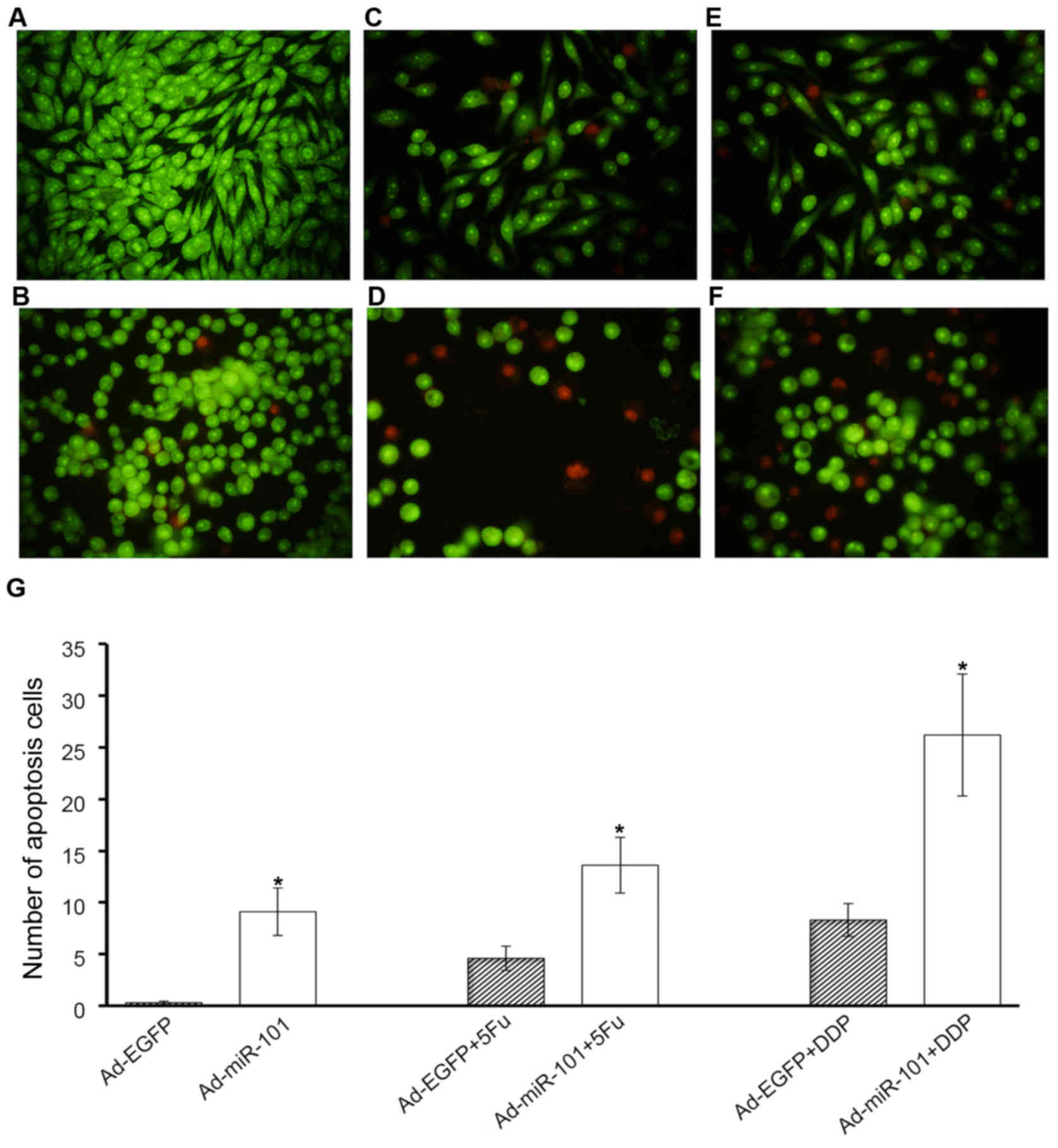
AO/EB staining indicated that miR-101 could
sensitize HT-29 cells to apoptosis induced by 5-FU and DDP. In
negative control groups, a small number of apoptotic HT-29 cells
were stained (0.3±0.17). The overexpression of miR-101 could
increase the apoptosis of HT-29 cells (9.1±2.3). Moreover, miR-101
could increase the number of apoptotic cells induced by 5-FU
significantly (4.6±1.2 vs. 13.6±2.7). The number of apoptotic cells
increased from 8.3±1.6 to 26.2±5.9, 24 h after low-dose DDP
treatment. It indicated that miR-101 could improve the
chemotherapeutic effect of 5-FU and DDP in HT-29 cells (Fig. 7).
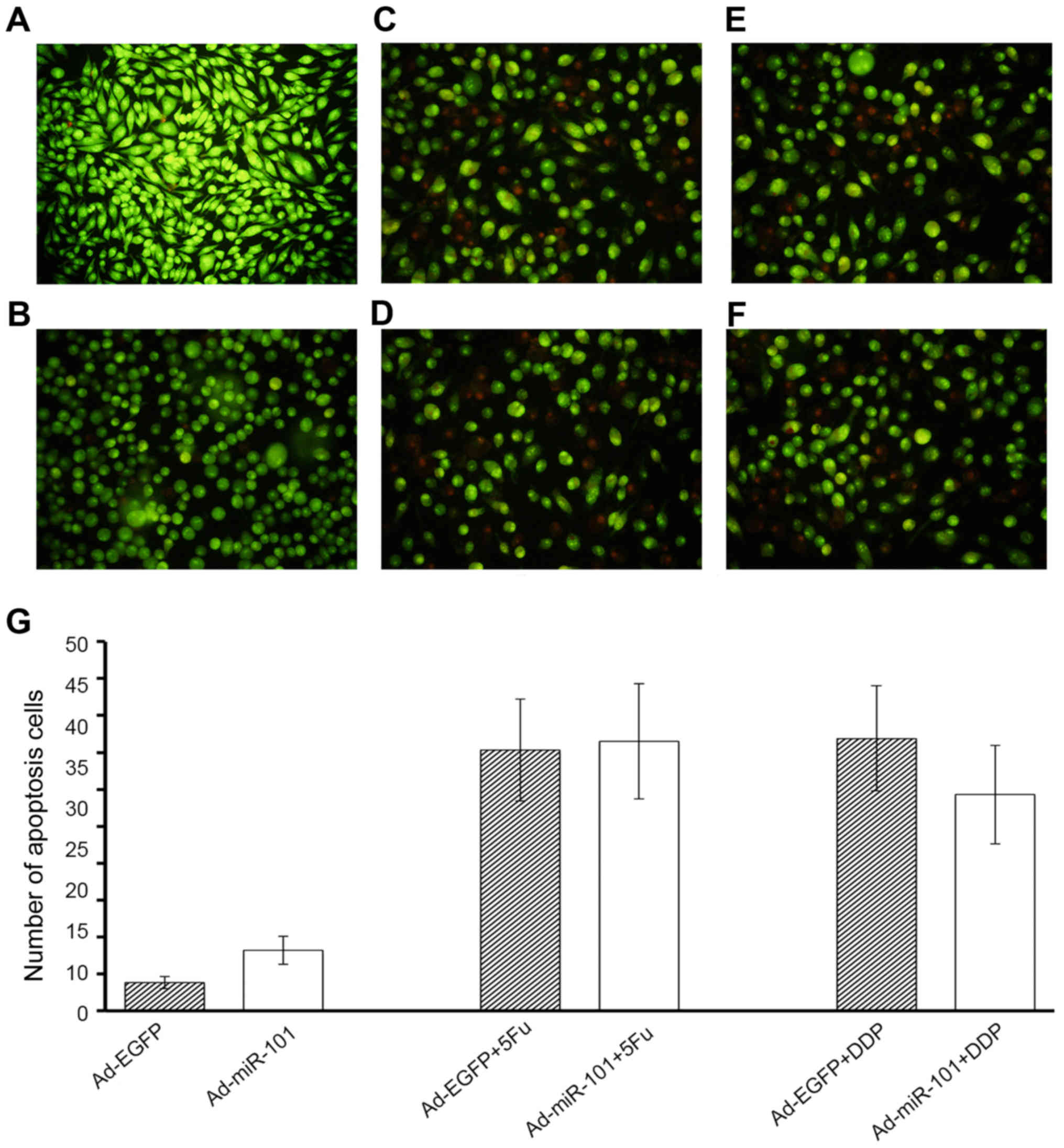
Despite exhibiting antitumor properties in RKO
cells, miR-101 failed to consolidate the inhibitory roles of 5-FU
and DDP. It was observed that overexpression of miR-101 could
increase the apoptosis of RKO cells (3.8±0.8 vs. 8.2±1.9). However,
the differences in the number of apoptotic cells between the
experimental and negative control groups were not statistically
significant on treatment with 5-FU or DDP, as shown in Fig. 8 (35.3±6.9 vs. 36.5±7.8; 36.9±7.1 vs.
29.3±6.7). It did not imply that miR-101 and antitumor drugs had a
synergistic effect in RKO cells.
Discussion
Dysregulation of miRNAs has frequently been observed
in human cancers, yet the molecular mechanisms of the involvement
of miRNAs in tumorigenesis and the behavior of cancer cells remain
to be explored (6). Recently, many
studies have indicated the participation of miRNAs in the
occurrence and development of colon cancer as oncogene or tumor
suppressor gene (7). Previous
studies (14) have indicated that
the expression of miR-200c decreases in colon cancer tissues and
blood circulation in patients. Therefore, it may be used as a tumor
marker in the diagnosis of colon cancer. Another study (15) found that miR-373 could stimulate
abnormal mitosis and morphological changes in the cells (epithelial
polarity deletion, restructuring of cells, and functional chaos),
leading to the occurrence and development of cancer; miR-205 could
promote the secretion of KLF4, MUC2, and TGFβ1, resulting in
resistance to chemotherapy. When these two kinds of miRNAs were
overexpressed, the tumor progression was rapid and the prognosis
was poor. The above research suggested that the ectopic expression
of miRNAs could be used in the diagnosis and treatment of various
malignant tumors. We investigated the abnormal expression of miRNA
in colorectal cancer and its role in tumorigenesis and development,
which could provide a theoretical basis for the eradication of
tumors. Therefore, in this study we analyzed not only the
expression of miR-101 in colon cancer tissues and adjacent normal
tissues, but also the relationship between the expression and the
clinicopathological characteristics of colon cancer. Ad-miR-101 was
used to upregulate the miR-101 level in HT-29 and RKO cells, and
the biological role of miR-101 in cell proliferation, invasion,
migration, and chemotherapeutic sensitivity was explored.
Recent studies have reported miR-101, whose whole
sequence is TACAGTACTGTGATAACTGAA, as an important tumor
suppressor. Its expression decreased or was missing in various
human cancers, such as embryonal rhabdomyosarcoma (eRMS),
endometrial cancer, and hepatocellular carcinoma (16–18).
However, not many studies show the relationship between miR-101 and
colon tumor. Vella et al (16) found that miR-101 could restrain the
migratory potential of eRMS cells by repressing enhancer of zeste
homolog 2 (EZH2). In aggressive endometrial cancer cells,
re-expression of miR-101 not only inhibited cell proliferation,
migration, and invasion but also induced cell apoptosis and
enhanced chemosensitivity to paclitaxel. This is similar to our
experimental results. Our experiments in vitro showed that
miR-101 expression was decreased in colon cancer tissues and
negatively correlated with the pathological type, but not with the
patient's sex, age, tumor location, TNM stage, or degree of
differentiation.
Another study in vivo found that the
upregulated expression of miR-101 suppressed proliferation,
migration, and invasion and induced apoptosis in HT-29 and RKO cell
lines. These results indicate that miR-101 is potentially an
anti-oncogene with negative regulation on the occurrence and
development of colon cancer. The ectopic overexpression of miR-101
markedly repressed proliferation, invasion, colony formation, and
cell cycle progression in human hepatocellular carcinoma cells and
suppressed tumorigenicity in vivo. Furthermore, miR-101
inhibited autophagy and synergized with either doxorubicin or
fluorouracil to induce apoptosis in tumor cells. Consistent with
previous studies, our present study revealed that miR-101 could
boost the apoptosis of HT-29 cells induced by 5-FU and DDP. This
means miR-101 played an additional role in colon cancer
chemotherapy. To summarize, this study confirmed the tumor
suppressive role of miR-101 in colon cancer and provided evidence
for the potential uses of miR-101 in miRNA-based cancer
therapy.
Regardless of the latest advances in research, drug
resistance remains a heavy burden in colon cancer therapy and
patient prognosis. One way to solve this problem is to find novel
therapeutic compounds for cancer treatment. The potential processes
of chemotherapy regulated by miRNAs remain to be evaluated, and
their merits of prognosis have not been well identified. However,
the role of miRNAs in oncotherapy has been a hot topic recently.
For instance, the overexpression of miR-21 increased the resistance
of prostate cancer cells to docetaxel, while the decreased
expression of miR-34a, miR-143, miR-148a, and miR-200 family was
involved in resistance to anticancer drugs by the inhibition of
apoptosis and the activation of signaling pathways (19). Vanas et al (20) found that ectopic expression of
miR-21 resulted in weakened resistance toward cisplatin while the
reduction in miRNA-21 activity showed the opposite effect in
osteosarcoma-derived cell lines. Zhao et al (21) suggested that miR-770-5p expression
decreased in platinum-resistant patients and could predict the
response to cisplatin treatment. Endogenous miR-770-5p might
function as an antioncogene by negatively regulating ERCC2 and
restored chemosensitivity to cisplatin due to the inhibition of DNA
repair.
Hu et al (22) found that the ectopic expression of
miR-205 led to an increase in apoptosis and resensitization of both
drug-resistant cell lines to doxorubicin and Taxol in breast
cancer. They further showed that miR-205 levels were negatively
correlated with the expression of VEGFA and FGF2 mRNA in patients
with breast cancer. Besides, these phenomena were also observed in
mouse tumor xenografts. The present study revealed that miR-101
increased the sensitivity of HT-29 colon cancer cells to
chemotherapy through strengthening the inhabitation of cell
proliferation and accelerating induced apoptosis. However, no
statistically significant interactions between miR-101 and
anticancer drugs were found in RKO cells. As we known from ATCC,
the HT-29 line is originated from tumor epithelium, which could
form well differentiated adenocarcinoma in nude mice. While RKO
containing wild-type p53 is a poorly differentiated colon carcinoma
cell line. The different characteristics of the two cell lines is
the cause of their differential expression of miR-101. The
infection efficiency of HT-29 and RKO cell lines was also
different. These distinctions might attribute to the different
experimental results. The variations of the research data might
also be related to the differences in cell biological behavior,
drug influx and efflux, metabolism of the drug, drug action
mechanisms, epithelial-to-mesenchymal transition, DNA damage
response, and other factors, which needs further validation. The
present results provided evidence that miR-101 might be a potent
therapeutic agent in the treatment of colorectal cancer.
In summary, this study provides new insights into
the role of miR-101 in human colon cancer. It showed that the
expression of miR-101 decreased in colon cancer tissues compared
with adjacent nontumor tissues. It was negatively correlated with
the pathological type, but not with the patient's sex, age, tumor
location, TNM stage, and degree of differentiation. The upregulated
expression of miR-101 suppressed cell proliferation and inhibited
cell migration and invasion in HT-29 and RKO colon cancer cell
lines. This study confirmed the tumor suppressive role of miR-101
in colon cancer and provided evidence for the potential uses of
miR-101 in miRNA-based cancer therapy.
Acknowledgements
This work was supported by Zhejiang Province medical
and health research project (2015114978).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sjo OH, Berg M, Merok MA, Kolberg M,
Svindland A, Lothe RA and Nesbakken A: Peritoneal carcinomatosis of
colon cancer origin: Highest incidence in women and in patients
with right-sided tumors. J Surg Oncol. 104:792–797. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mori F, Ferraiuolo M, Santoro R, Sacconi
A, Goeman F, Pallocca M, Pulito C, Korita E, Fanciulli M, Muti P,
et al: Multitargeting activity of miR-24 inhibits long-term
melatonin anticancer effects. Oncotarget. 7:20532–20548.
2016.PubMed/NCBI
|
|
4
|
Patel Y, Shah N, Lee JS, Markoutsa E, Jie
C, Liu S, Botbyl R, Reisman D, Xu P and Chen H: A novel
double-negative feedback loop between miR-489 and the
HER2-SHP2-MAPK signaling axis regulates breast cancer cell
proliferation and tumor growth. Oncotarget. 7:18295–18308.
2016.PubMed/NCBI
|
|
5
|
Konishi H, Fujiya M, Ueno N, Moriichi K,
Sasajima J, Ikuta K, Tanabe H, Tanaka H and Kohgo Y: microRNA-26a
and −584 inhibit the colorectal cancer progression through
inhibition of the binding of hnRNP A1-CDK6 mRNA. Biochem Biophys
Res Commun. 467:847–852. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M,
et al: Human microRNA genes are frequently located at fragile sites
and genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chauhan R and Lahiri N: Tissue- and
serum-associated biomarkers of hepatocellular carcinoma. Biomark
Cancer. 8 Suppl 1:37–55. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Suárez-Arriaga MC, Ribas-Aparicio RM and
Ruiz-Tachiquín ME: MicroRNAs in hereditary diffuse gastric cancer.
Biomed Rep. 5:151–154. 2016.PubMed/NCBI
|
|
11
|
Mohamad M, Wahab NA, Yunus R, Murad NA,
Zainuddin ZM, Sundaram M and Mokhtar NM: Roles of microRNA21 and
microRNA29a in regulating cell adhesion related genes in bone
metastasis secondary to prostate cancer. Asian Pac J Cancer Prev.
17:3437–3445. 2016.PubMed/NCBI
|
|
12
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mukohyama J, Shimono Y, Yamashita K, Sumi
Y, Mukohara T, Minami H and Kakeji Y: Effect of xenotransplantation
site on microRNA expression of human colon cancer stem cells.
Anticancer Res. 36:3679–3686. 2016.PubMed/NCBI
|
|
14
|
Kumar S, Nag A and Mandal CC: A
comprehensive review on miR-200c, a promising cancer biomarker with
therapeutic potential. Curr Drug Targets. 16:1381–1403. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Eyking A, Reis H, Frank M, Gerken G,
Schmid KW and Cario E: MiR-205 and miR-373 are associated with
aggressive human mucinous colorectal cancer. PLoS One.
11:e01568712016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vella S, Pomella S, Leoncini PP, Colletti
M, Conti B, Marquez VE, Strillacci A, Roma J, Gallego S, Milano GM,
et al: MicroRNA-101 is repressed by EZH2 and its restoration
inhibits tumorigenic features in embryonal rhabdomyosarcoma. Clin
Epigenetics. 7:822015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Konno Y, Dong P, Xiong Y, Suzuki F, Lu J,
Cai M, Watari H, Mitamura T, Hosaka M, Hanley SJ, et al:
MicroRNA-101 targets EZH2, MCL-1 and FOS to suppress proliferation,
invasion and stem cell-like phenotype of aggressive endometrial
cancer cells. Oncotarget. 5:6049–6062. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu L, Beckebaum S, Iacob S, Wu G, Kaiser
GM, Radtke A, Liu C, Kabar I, Schmidt HH, Zhang X, et al:
MicroRNA-101 inhibits human hepatocellular carcinoma progression
through EZH2 downregulation and increased cytostatic drug
sensitivity. J Hepatol. 60:590–598. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kopczyńska E: Role of microRNAs in the
resistance of prostate cancer to docetaxel and paclitaxel. Contemp
Oncol (Pozn). 19:423–427. 2015.PubMed/NCBI
|
|
20
|
Vanas V, Haigl B, Stockhammer V and
Sutterlüty-Fall H: MicroRNA-21 increases proliferation and
cisplatin sensitivity of osteosarcoma-derived cells. PLoS One.
11:e01610232016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao H, Yu X, Ding Y, Zhao J, Wang G, Wu
X, Jiang J, Peng C, Guo GZ and Cui S: MiR-770-5p inhibits cisplatin
chemoresistance in human ovarian cancer by targeting ERCC2.
Oncotarget. 7:53254–53268. 2016.PubMed/NCBI
|
|
22
|
Hu Y, Qiu Y, Yagüe E, Ji W, Liu J and
Zhang J: miRNA-205 targets VEGFA and FGF2 and regulates resistance
to chemotherapeutics in breast cancer. Cell Death Dis. 7:e22912016.
View Article : Google Scholar : PubMed/NCBI
|