Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignancies, and is also the leading cause of
cancer-associated mortalities (1,2).
Although progress has been achieved in the development of new
treatment strategies, HCC remains difficult to diagnose at an early
stage. Most HCC patients suffer of asymptomatic presentation at the
early stage, resulting in metastasis once diagnosis (3). The clinical outcome of advanced HCC
patients is still extremely poor. Most advanced HCC patients die as
a result of rapid tumor progression, and hepatic resection or
transplantation are the only potential curative therapy strategies
for patients with HCC (4). However,
the effective diagnostic and therapeutic targets are still unclear.
Carcinogenesis of HCC is a complex process involving multiple
factors, and multi-steps (5). To
improve the clinical outcome of HCC therapy, it is critical to
elucidate the molecular pathogenesis of HCC and investigate the
genes responsible for HCC development and progression.
MicroRNAs (miRNAs) are a group of small noncoding
RNAs (19–22 or 19–25 nucleotides) and play an important role in the
regulation of gene expression at the posttranscriptional level
(6). It was demonstrated that miRNA
plays a critical role in the regulation of a variety of
physiological and pathological processes, such as development, cell
proliferation, cell apoptosis, cell differentiation, cell fate
determination, and signal transduction (6–8). To
date, miRNAs control nearly 60% of human genes (7), and more than 1000 human miRNAs have
been identified. Increased evidence showed that miRNA act as
oncogenes or tumor suppressor genes, dysregulation of them in human
malignant tumors regulate the development and progression of cancer
via downregulation of their targeted tumor suppressor genes or
oncogene expression (9).
Recently, it was demonstrated miR-30e was
down-regulated in both plasma and breast cancer tissues (10), non-small cell lung cancer (11), as well as liver tumor tissues
(12–14). It was also demonstrated that
miR-147a is upregulated in hepatitis C virus-associated diffuse
large B-cell lymphoma, and in small cell lung cancer (15), human gastric cancer (16), squamous cell carcinoma of tongue
(17), and hepatocellular carcinoma
(18). Some studies demonstrate
that miR-147a plays critical effects on cell development,
migration, and invasion, but has no influence on apoptosis
(19,20). In gastric cancer, AKT2 and cyclin D1
were identified as direct targets in gastric cancer, contributing
to miR-147 strong inhibitory effect on G1/S transition (20). Hypoxia-induced HIF-1α increases the
expression of miR-147a via HNF4A, miR-147a induced cell
proliferation arrest under hypoxia (21). Therefore, each miRNA might target a
different gene to play distinct roles in the regulation of
fundamental cellular processes.
Herein, we show that miR-30e and miR-147a were
differentially expressed in HCC cells (HepG2, MHCC97H, HuH7, and
Bel-7402), and the liver cells L02. Two cell lines significantly
downregulated the miR-30e expression were selected to investigate
the effect of miR-30 on development and progression of HCC,
including cell proliferation, cell apoptosis, cell migration and
invasion. Mechanistically, we demonstrate that miR-30e target the
JAK1-STAT3-vimentin signaling pathway which could collectively
contribute to their efficient therapeutic significance, and that
IL-6 (agonist of the JAK1/STAT3 pathway) treatment could phenocopy
miR-30e downexpression and rescue the cell function induced by
miR-30e mimic transfection.
Materials and methods
Cell culture
The human hepatocellular carcinoma cell lines HepG2,
MHCC97H, HuH7, and Bel-7402, and the live cell line L-02, and human
embryonic kidney 293 (HEK-293) cells were purchased from Cell Bank
of Shanghai Institute of Biochemistry and Cell Biology, Chinese
Academy of Sciences. As previous described (22), cells were cultured in RPMI-1640
medium (Hyclone, Logan, UT, USA) and supplemented with 10% fetal
bovine serum (Gibco, Carlsbad, CA, USA), 1% penicillin/streptomycin
at 37°C in 5% CO2.
Cell transfection
The miR-30e mimics, small-interfering RNAs targeting
JAK1 (siJAK1) and their respective negative controls were obtained
from GenePharma (Shanghai GenePharma Co. Ltd., Shanghai, China).
The primers were as follows: miR-30e mimics: 5′- UGU AAA CAU CCU
UGA CUG GAAG-3′ (forward), and 5′-UGG UGU UAG UUG GUU GCG UUUU-3′
(reverse); mimic NC: 5′-UUC UCC GAA CGU GUC ACG UTT-3′ (forward),
5′-ACG UGA CACG UUC GGA GAATT-3′ (reverse); JAK1 siRNA: 5′-UUG UUU
UGU UUU GUU UGA GCC-3′ (forward), and 5′-CUC AAA CAA AAC AAA ACA
AAA-3′ (reverse). Cells were seeded into six-well plates, incubated
for 24 h before transfected with miR-30e mimics or siJAK1 by using
Lipofectamine-2000 (Invitrogen, Carlsbad, CA, USA) in according to
the manufacturer's instructions. At 48 h post-transfection, cells
were harvested, expression of miR-30e and JAK1 was tested, and
prepared for subsequent experiments.
Luciferase reporter assay
The fragment of wild-type JAK1 3′-UTR (wt 3′-UTR)
containing predicted miR-30e binding sites was amplified by PCR,
and mutant JAK1 3′-UTR (MUT 3′-UTR) was obtained by mutating the
conserved binding sites for miR-30e. The fragments including the wt
3′-UTR or MUT 3′-UTR regions of JAK1 were cloned into
XhoI/NotI-digested psiCHECK-2 vector (Promega,
Madison, WI, USA), which included both renilla and firefly
luciferase reporter genes. Then the psiCHECK-2 vectors with wt
3′-UTR or MUT 3′-UTR regions of JAK1 were transfected into HEK23
cells and transfected with miR-30e mimics or negative control
mimics, respectively. After 24 h, the firefly and renilla
luciferase activities in cells were determined with a
dual-luciferase reporter assay system (Promega) in accordance with
the manufacturer's instructions.
Quantitative real-time PCR
Total RNA in transfected cells was isolated using
the RNeasy Plus Mini kit (Qiagen) according to the manufacturer's
instructions. The expression level of miR-30e was determined using
Taqman miRNA assays (Applied Biosystems, Foster City, CA USA) with
miRNA-specific primers (forward primer: 5′-ACA CTC CAG CTG GGT GTA
AAC ATC CTTG-3′ and universal reverse primer: 5′-CTC AAC TGG TGT
CGT GGA GTC GGC AAT TCA GTT GAG CTT CCA GTC-3′). For data
normalization, U6 small nuclear RNA was used as an endogenous
control. The expression level of JAK1 mRNA was determined using
PrimeScript RT-PCR kits (Takara, Shiga, Japan) with primers
(forward primer: 5′-CGCTCTGGGAAATCTGCT-3′ and reverse primer:
5′-TGATGGCTCGGAAGAAAGG-3′), and β-actin was used as internal
control.
Western blotting
Total proteins were extracted from cells as previous
described (23) and separated by
10% SDS-PAGE electrophoresis and then electroblotted onto a
nitrocellulose membrane in 25 mM Tris base and 190 mM glycine at 50
V for 3 h at 4°C. The membranes were probed with primary antibodies
overnight, followed by incubation with horseradish
peroxidase-conjugated secondary antibodies. Protein was detected by
enhanced chemiluminescence kit (Amersham Life Science, Buckingham,
UK). All antibodies were purchased from Santa Cruz Biotechnology,
Santa Cruz, CA, USA.
Cell proliferation assay
Cell proliferation was determined by suing the
colorimetric water-soluble tetrazolium salt assay using a Cell
Counting Kit-8 (Beyotime, Haimen, China). In brief, cells with a
density of 2×103 cells per well were seeded in 96-well
plates, and then cell proliferation was documented at 24, 48, 72,
and 96 h. The number of viable cells was obtained by reading the
absorbance at 450 nm using a microplate reader Thermo Plate (Rayto
Life and Analytical Science C. Ltd., Wetzlar, Germany).
Cell apoptosis assay
Cell apoptosis was assessed by using an Annexin
V-FITC apoptosis detection kit (BD Pharmingen, Franklin Lakes, NJ,
USA). In brief, Annexin V-FITC (5 µl) and propidium iodide (5 µl)
were added in 100 µl of cells at concentration of 1×106
cells/ml and incubated in the dark for 15 min. Then, binding buffer
was added and apoptosis was analyzed by flow cytometry (BD
Biosciences, Franklin Lakes, NJ, USA).
Cell cycle assay
After transfection, cells were harvested after
trypsinization and were resuspended with concentration of
1×106 cells/ml and prepared using Cycle Test Plus DNA
Reagent kit (Becton Dickinson, San Jose, CA, USA) according to the
manufacturer's instructions. Cell cycle was analyzed by flow
cytometer using propidium iodide (PI) as a specific fluorescent dye
probe. The PI fluorescence intensity of 10,000 cells was measured
for each sample using a Becton-Dickinson FACSCalibur flow
cytometer.
Migration and invasion
Cell migration and invasion of HCC cells (HepG2 and
HuH7 cells) were performed by using a Transwell chamber (Millipore,
Billerica, MA, USA). For cell invasion, transwell chamber was
coated with 30 µl Matrigel. Cells were seeded into 24-well plate
and cultured at 37°C in RPMI-1640 medium with 2% serum, while 600
µl of 10% FBS RPMI-1640 was added to the lower chamber. After 48 h,
HCC cells were fixed with 100% methanol for 30 min and stained
using crystal violet for 20 min. Non-migrated cells were removed
using cotton swabs. Cell images were obtained under a
phase-contrast microscope (Olympus, Tokyo, Japan).
Statistical analysis
The results are expressed as the mean ± SD.
Statistical evaluation was performed using GraphPad Prism software
version 5.01 (GraphPad, Inc., La Jolla, CA, USA). The normal
distribution of variables was assessed prior to selecting the tests
to use for statistical analyses with ANOVA or student t-test. A
P-value <0.05 was considered significant.
Results
miR-30e is significantly downregulated
in HepG2 and HuH7 cells, whereas miR-147a is significantly
upregulated in HuH7 cells
According to the miRNA array analysis data (24,25),
we tested the expression of miR-30e, miR-147a in HepG2, MHCC97H,
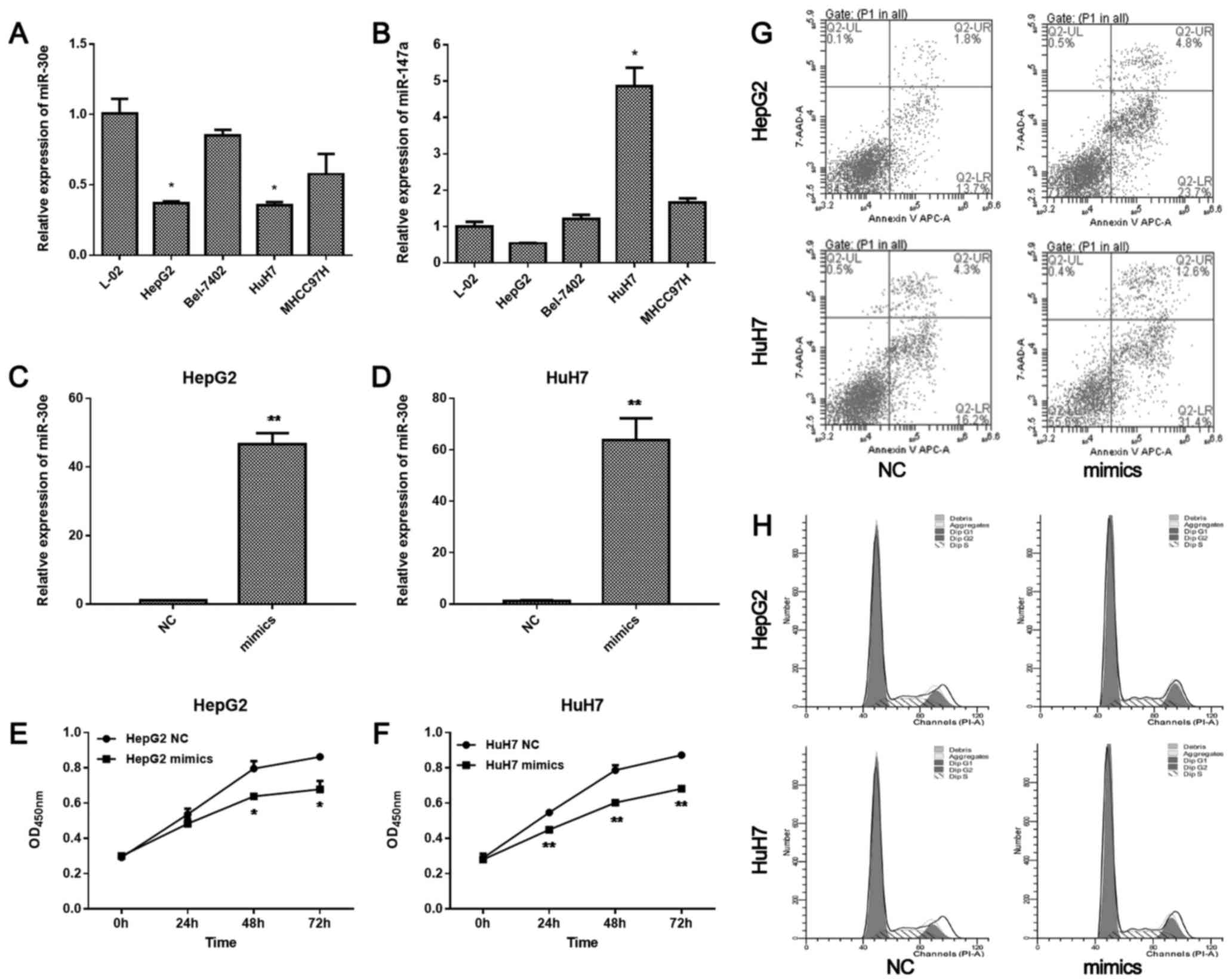
HuH7, and Bel-7402 hepatoma carcinoma cell lines, and L-02 live
cell line to screen the most pronounced miRNA by qRT-PCR (Fig. 1A and B). miR-30e was significantly
downregulated in both HepG2 and HuH7 cells, while it was not
significantly changed in L-02, Bel-7402, and MHCC97H cells.
miR-147a was significantly upregulated in HuH7 cells, but not in
other cell lines. Thus, we further examined the role of miR-30e in
HepG2 and HuH7 cell lines.
miR-30e mimics inhibit the
proliferation, migration, and invasion of HepG2 and HuH7 cells, and
promote cell apoptosis, but do not influence the cell cycle
After transfection of miR-30e mimics, miR-30e was
overexpressed in HepG2 cells (P<0.01; Fig. 1C) and HuH7 cells (P<0.01;
Fig. 1D). The proliferation of
HepG2 was significantly inhibited by miR-30e mimics after 48 h
(P<0.05; Fig. 1E). Similar
change in proliferation was observed in HuH7 cells. After 24 h, the
proliferation of HuH7 cells was significantly inhibited by miR-30e
mimics (P<0.05, 0.01 at 24, 48, 72 h, respectively; Fig. 1F). Furthermore, miR-30e mimics
promoted apoptosis of HepG2 and HuH7 cells (Fig. 1G). However, miR-30e mimics did not
significantly change the cell cycle (Fig. 1H). The migration and invasion of
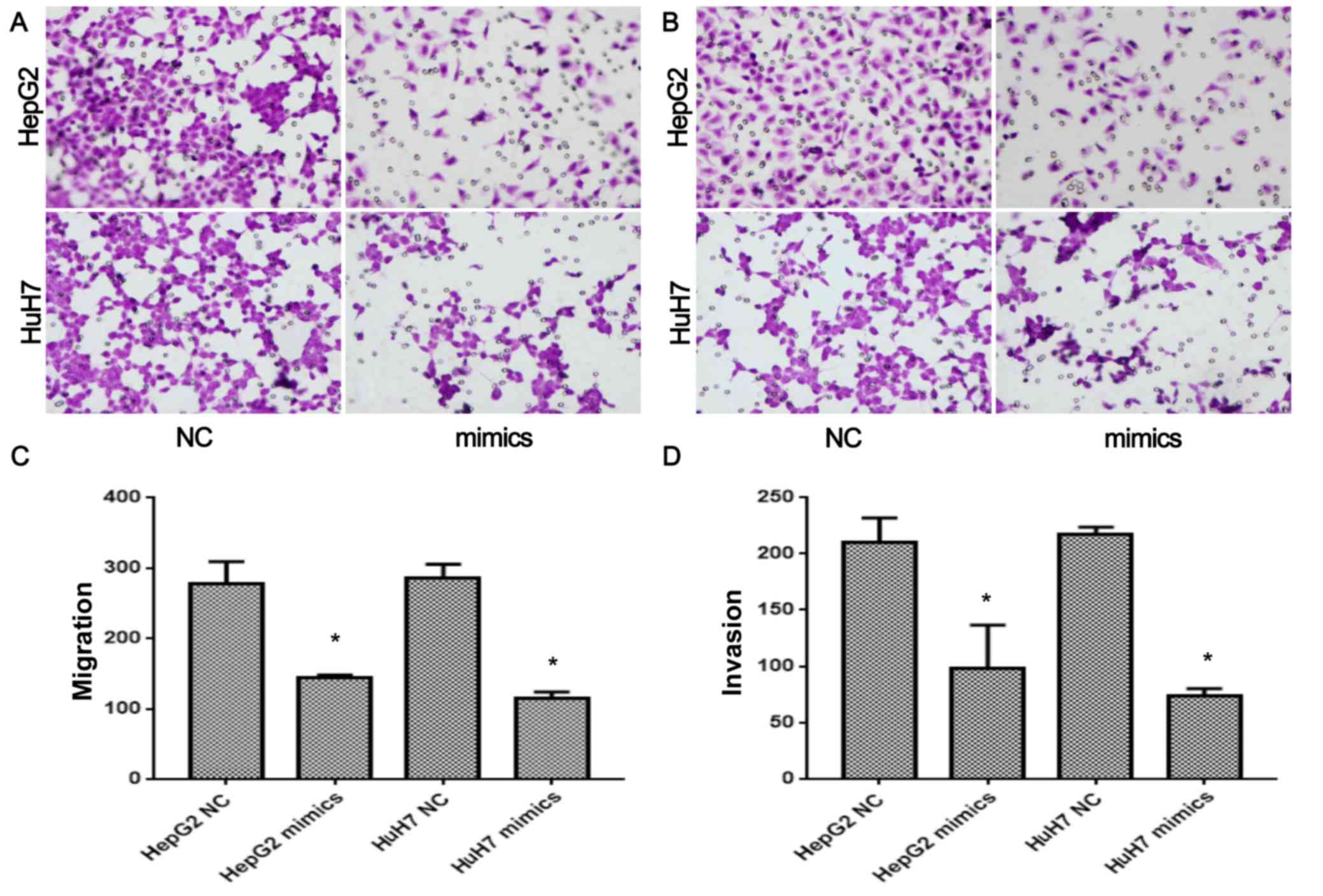
HepG2 and HuH7 cells after transfection of miR-30e were also
detected (Fig. 2A-D). miR-30e
mimics significantly inhibited the migration of HepG2 and HuH7
cells (P<0.05, Fig. 2A and C).
Similarly, the invasion of HepG2 and HuH7 cells after transfection
of miR-30e were also significantly inhibited (P<0.05, Fig. 2B and D). Thus, overexpression of
miR-30e significantly inhibited the proliferation, migration, and
invasion of HCC cells, and promoted cell apoptosis.
JAK1, not vimentin is a direct target
of miR-30e
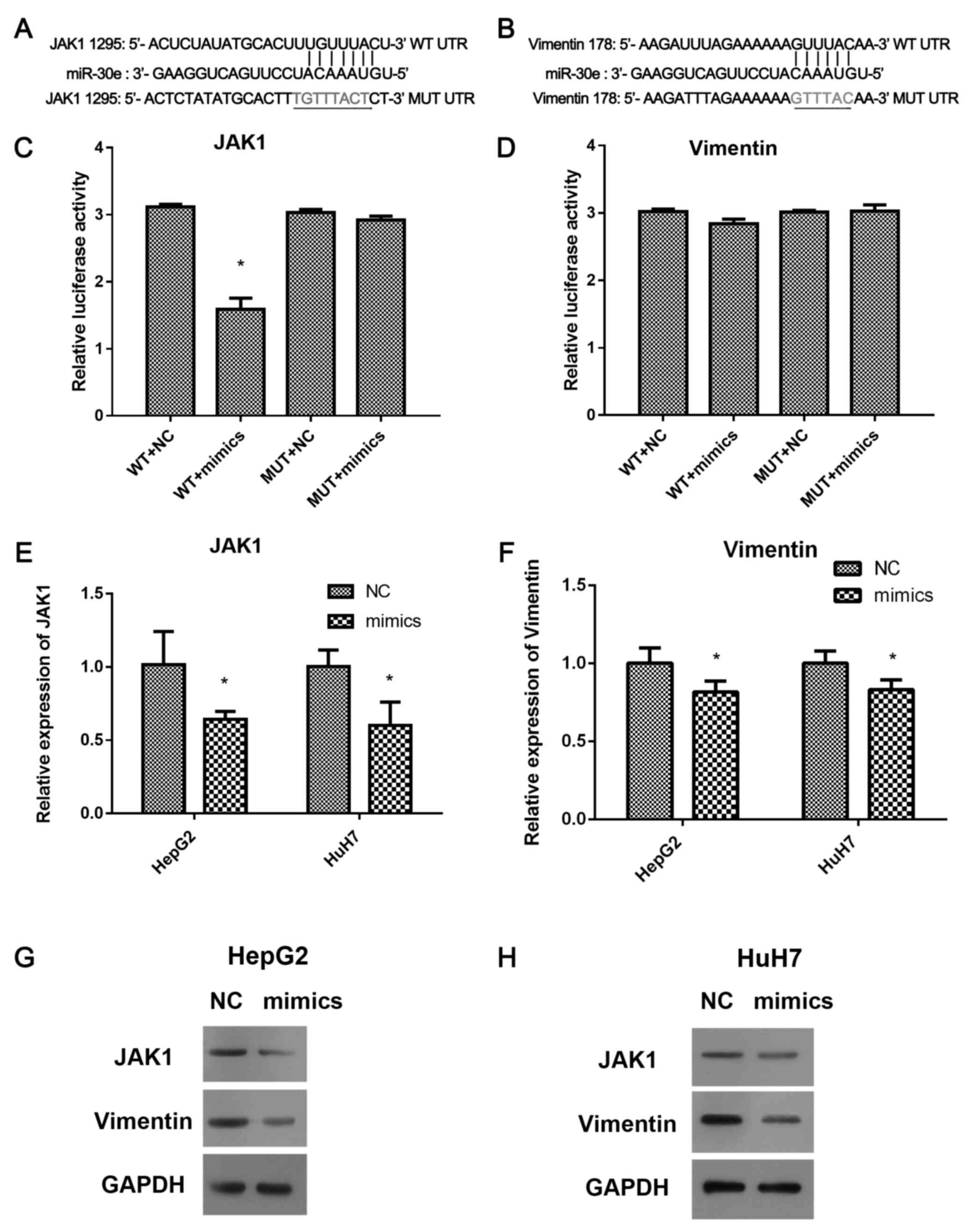
Luciferase reporter assay was used to validate the
target of miR-30e (Fig. 3A-D). The
Reporter luciferase vectors, wt-3′UTR of JAK1, vimentin containing
the predicted miR-30e binding sites and the corresponding mutated
vectors (Mut-3′UTR) were achieved (Fig.
3A and B) and transfected into HepG2 and HuH7 cells. The
luciferase activity was inhibited by miR-30e in the cells with
wt-3′UTR of JAK1 (P<0.05, Fig.
3C), but not changed in mut-3′UTR of JAK1, or wt-and Mut-3′UTR
of vimentin (Fig. 3D), suggesting
JAK1, not vimentin is a direct target of miR-30e. Furthermore, the
expression of JAK1 and vimentin in miR-30e mimics transfected-HepG2
and HuH7 cells were performed by qRT-PCR and western blotting
(Fig. 3C-H). miR-30e mimics
inhibited the expression of JAK1 and vimentin in mRNA and protein
levels in HepG2 and HuH7 cells (P<0.05, Fig. 3E and F). Thus, overexpression of
miR-30e significantly downregulated the expression of JAK1 and
vimentin, but only JAK1 was a direct target of miR-30e.
Silence of JAK1 inhibits cell
proliferation, migration and invasion of HCC cells
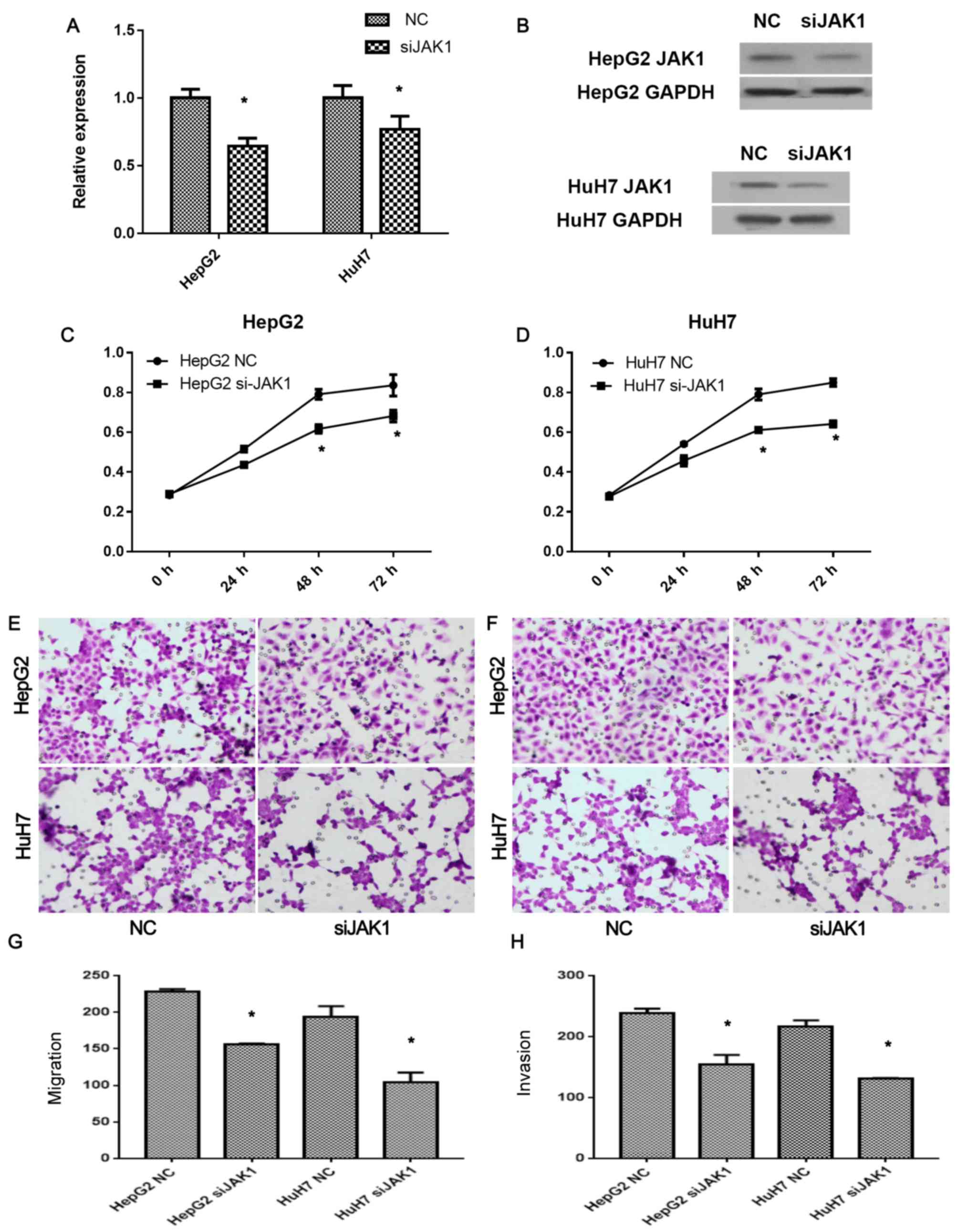
To further confirm whether miR-30e effects HCC cells
via JAK1, JAK1 siRNA (siJAK1) was transfected in the cells, and the
expression of JAK1 in mRNA and protein levels was detected by
qRT-PCR (Fig. 4A) and western
blotting (Fig. 4B), respectively.
Silence of JAK1 by JAK1 siRNA was confirmed by the lower expression
levels of JAK1 in both HepG2 and HuH7 cells (P<0.05; Fig. 4A and B), compared with the
non-transfected controls. siJAK1 significantly inhibited the
proliferation of HepG2 (P<0.05; Fig.
4C) and HuH7 (P<0.05; Fig.
4D) cells after 48 h. The migration and invasion of the
siJAK1-transfected HCC cells were also detected. After siJAK1
transfection, the migration of HepG2 (P<0.05; Fig. 4E and G) and HuH7 cells (P<0.05;
Fig. 4F and H) were significantly
inhibited. Similarly, the invasion of HepG2 (P<0.05; Fig. 4E and G) and HuH7 cells (P<0.05;
Fig. 4F and H) were also inhibited
by siJAK1, respectively. These results suggested that miR-30e
inhibited cell proliferation, migration, and invasion of HCC cells
via direct downregulation of JAK1.
JAK/STAK pathway mediates the effects
of miR-30e
It was demonstrated that vimentin is related to the
motility capacity of HCC cells (5,26) and
important molecule in JAK/STAK/vimentin signaling pathway (27). We further investigated whether
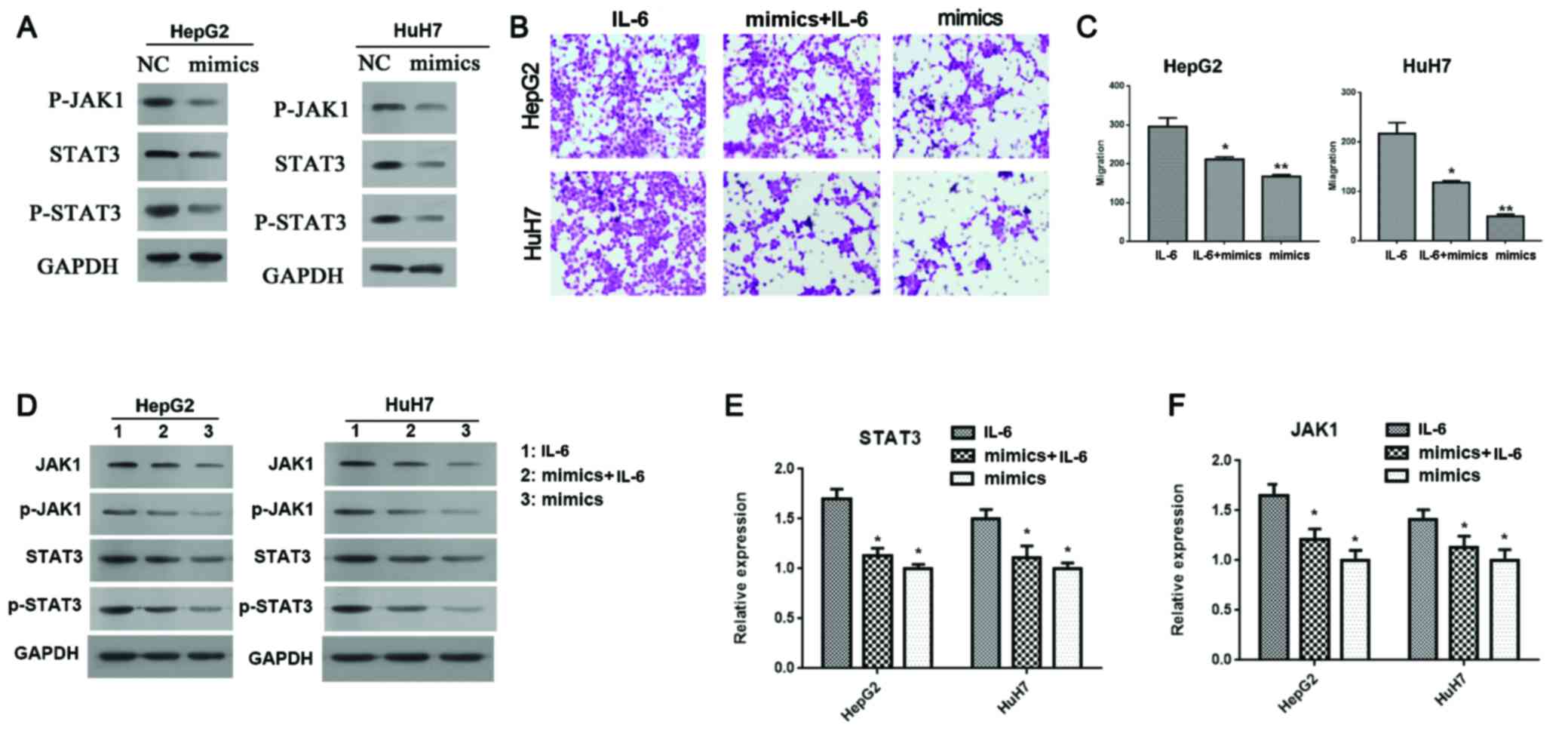
JAK/STAK pathway mediated the effect of miR-30e (Fig. 5). First, miR-30e mimics not only
significantly downregulated the expression of JAK1 (Fig. 3G and H) and STAT3 (Fig. 5A), but also significantly
downregulated phosphorylation levels of JAK1 and STAT3 (Fig. 5A). Second, IL-6, the agonist of
JAK/STAT3 pathway decreased the inhibition of cell migration of
HepG2 and HuH7 cells by miR-30e partly (P<0.05, Fig. 5B and C). Third, IL-6 reduced the
inhibition of expression (in both mRNA and protein levels) and
phosphorylation levels of JAK1 and STAT3 (Fig. 5D-F). Thus, JAK/STAT3 pathway
mediated the effect of miR-30e on cell migration.
Discussion
Hepatocellular carcinoma (HCC) ranks among the top
10 of the most common malignancies in the world, and is also the
leading cause of cancer-associated mortalities in China (1,2). HCC
remains difficult to diagnose at an early stage, and hepatic
resection or transplantation are the only potential curative
therapy strategies for patients with HCC (4). HCC patients with poor diagnosis
undergo metastasis fast and die as a result of the rapid tumor
progression. To improve the clinical outcome of HCC therapy, it is
critical to elucidate the molecular pathogenesis of HCC and
investigate the genes responsible for HCC development and
progression. Herein, we demonstrate that miR-30e and miR-147a are
differentially expressed in HCC cells (HepG2, MHCC97H, HuH7, and
Bel-7402), and liver cells L02. miR-30e mimics inhibited the
development and progression of HCC including inhibited the cell
proliferation, cell migration and invasion, promoted cell
apoptosis. Mechanistically, we demonstrate that miR-30e target the
JAK1-STAT3-vimentin signaling pathway which could collectively
contribute to their efficiently therapeutic significance, and that
IL-6 (agonists of the JAK1/STAT3 pathway) treatment could phenocopy
miR-30e downexpression and rescue the cell function induced by
miR-30e mimics transfection.
Recently, it was demonstrated miR-30e was
downregulated in both plasma and breast cancer tissues (10), in non-small cell lung cancer
(11), as well as in liver tumor
tissues in hepatocellular carcinoma (12–14).
It was also demonstrated that miR-147a was upregulated in hepatitis
C virus-associated diffuse large B-cell lymphoma, and in small cell
lung cancer (15), human gastric
cancer (16), squamous cell
carcinoma of tongue (17), and
hepatocellular carcinoma (18).
Consistently, our results demonstrated the downregulation of
miR-30e in HepG2 and HuH7 HCC cell lines, and upregulation of
miR-147a only in HuH7 cell lines. Thus, the HepG2 and HuH7 cell
lines were selected for investigation of the miR-30e role.
miR-30e was proved to be suppressor of human NK cell
cytotoxicity, and could directly target perforin (28). In breast cancer, it upregulated
three predicted targets of miR-30e including RAB11A, BNIP3L, and
RAB32 associated with downregulation of miR-30e (10). In addition, miR-30e targeted
3′-untranslated region (3′UTR) of prolyl 4-hydroxylase subunit
alpha-1 (P4HA1) mRNA, and reduced the expression of P4HA1 at the
levels of mRNA and protein (12).
Overexpression of miR-30e suppressed cell proliferation of HepG2
cells and reduced colony formation (12). Herein, we found JAK1 was also the
target of miR-30e by luciferase reporter gene assays. Enforced
expression of miR-30e inhibited cell proliferation, cell migration
and invasion, promoted cell apoptosis, but had no effect on the
cell cycle arrest. Silence of JAK1 also inhibited cell
proliferation, cell migration and invasion, suggesting miR-30e
inhibited the cell proliferation, cell migration and invasion
partly via JAK1. miR-23a suppressed the JAK1/STAT-6 pathway and
reduced production of M2 type cytokines by targeting JAK1 and
STAT-6 directly (29). In acute
erythroid leukemia, miR-23a, miR-27a and miR-24 formed a miRNA
cluster, synergistically targeting multiple members of the
oncogenic JAK1-STAT3 pathway, and thus reinforced their inhibition
on the cascade to regulate cell proliferation and apoptosis
(30). Exogenous miR-9 activated
JAK-STAT pathway in tumor angiogenesis (31). MiR-30c also targets JAK1 playing
important roles in porcine reproductive and respiratory syndrome
virus (32). Therefore, there is a
complex network between miRNAs and their targets. The investigation
on the correlation between miRNAs and their targets should be
helpful in clearing the mechanism.
miR-30e has no effect on the cell cycle. Some
studies demonstrate that miR-147a plays critical effects on cell
development, migration, and invasion, but has no influence on
apoptosis (19,20). In gastric cancer, AKT2 and cyclin D1
were identified as direct targets in gastric cancer, contributing
to miR-147 strong inhibitory effect on G1/S transition (20). Hypoxia-induced HIF-1α increases the
expression of miR-147a via HNF4A, miR-147a induced cell
proliferation arrest under hypoxia (21). Therefore, each miRNAs might target a
different gene to play distinct roles in the regulation of
fundamental cellular processes like development and proliferation,
cell fate determination and apoptosis.
In conclusion, we demonstrated that miR-30e targets
the JAK1-STAT3-vimentin signaling pathway playing critical roles in
inhibition of the cascade to regulate cell proliferation and
apoptosis, which could collectively contribute to their efficient
therapeutic significance.
Acknowledgements
This work is supported by the National Natural
Science Foundation Key Project of China (grant no. 81430041), the
National Natural Science Foundation of China (grant nos. 81271621,
81501561, 81620108017), Natural Science Foundation of Guangdong
Province (2014A030310043), the Science and Technology Planning
Project of Guangzhou Province (201604020098, 201610010006).
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yegin EG, Oymaci E, Karatay E and Coker A:
Progress in surgical and nonsurgical approaches for hepatocellular
carcinoma treatment. Hepatobiliary Pancreat Dis Int. 15:234–256.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Buendia MA and Neuveut C: Hepatocellular
carcinoma. Cold Spring Harb Perspect Med. 5:a0214442015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zeng YE, Yao XH, Yan ZP, Liu JX and Liu
XH: Potential signaling pathway involved in
sphingosine-1-phosphate-induced epithelial-mesenchymal transition
in cancer. Oncol Lett. 12:379–382. 2016.PubMed/NCBI
|
|
6
|
Kim VN: MicroRNA biogenesis: Coordinated
cropping and dicing. Nat Rev Mol Cell Biol. 6:376–385. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lages E, Ipas H, Guttin A, Nesr H, Berger
F and Issartel JP: MicroRNAs: Molecular features and role in
cancer. Front Biosci (Landmark Ed). 17:2508–2540. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin Z, Li JW, Wang Y, Chen T, Ren N, Yang
L, Xu W, He H, Jiang Y, Chen X, et al: Abnormal miRNA-30e
expression is associated with breast cancer progression. Clin Lab.
62:121–128. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Y, Chen J, Lin Z, Cao J, Huang H,
Jiang Y, He H, Yang L, Ren N and Liu G: Role of deregulated
microRNAs in non-small cell lung cancer progression using
fresh-frozen and formalin-fixed, paraffin-embedded samples. Oncol
Lett. 11:801–808. 2016.PubMed/NCBI
|
|
12
|
Feng G, Shi H, Li J, Yang Z, Fang R, Ye L,
Zhang W and Zhang X: MiR-30e suppresses proliferation of hepatoma
cells via targeting prolyl 4-hydroxylase subunit alpha-1 (P4HA1)
mRNA. Biochem Biophys Res Commun. 472:516–522. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wong CM, Wong CC, Lee JM, Fan DN, Au SL
and Ng IO: Sequential alterations of microRNA expression in
hepatocellular carcinoma development and venous metastasis.
Hepatology. 55:1453–1461. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bhattacharya S, Steele R, Shrivastava S,
Chakraborty S, Di Bisceglie AM and Ray RB: Serum miR-30e and
miR-223 as novel noninvasive biomarkers for hepatocellular
carcinoma. Am J Pathol. 186:242–247. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yi Z, Fu Y, Ji R, Li R and Guan Z: Altered
microRNA signatures in sputum of patients with active pulmonary
tuberculosis. PLoS One. 7:e431842012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni
L, Zhang WG, Nan KJ, Song TS and Huang C: MicroRNA profiling of
human gastric cancer. Mol Med Rep. 2:963–970. 2009.PubMed/NCBI
|
|
17
|
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP
and Wei WI: Mature miR-184 as potential oncogenic microRNA of
squamous cell carcinoma of tongue. Clin Cancer Res. 14:2588–2592.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Han ZB, Zhong L, Teng MJ, Fan JW, Tang HM,
Wu JY, Chen HY, Wang ZW, Qiu GQ and Peng ZH: Identification of
recurrence-related microRNAs in hepatocellular carcinoma following
liver transplantation. Mol Oncol. 6:445–457. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bertero T, Grosso S, Robbe-Sermesant K,
Lebrigand K, Hénaoui IS, Puisségur MP, Fourre S, Zaragosi LE,
Mazure NM, Ponzio G, et al: ‘Seed-Milarity’ confers to hsa-miR-210
and hsa-miR-147b similar functional activity. PLoS One.
7:e449192012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Uhlmann S, Mannsperger H, Zhang JD, Horvat
EÁ, Schmidt C, Küblbeck M, Henjes F, Ward A, Tschulena U, Zweig K,
et al: Global microRNA level regulation of EGFR-driven cell-cycle
protein network in breast cancer. Mol Syst Biol. 8:5702012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang F, Zhang H, Xu N, Huang N, Tian C, Ye
A, Hu G, He J and Zhang Y: A novel hypoxia-induced miR-147a
regulates cell proliferation through a positive feedback loop of
stabilizing HIF-1α. Cancer Biol Ther. 17:790–798. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng L, Deng CL, Wang L, Huang XB, You N,
Tang YC, Wu K, Liang P, Mi N and Li J: COMMD7 is correlated with a
novel NF-κB positive feedback loop in hepatocellular carcinoma.
Oncotarget. 7:32774–32784. 2016.PubMed/NCBI
|
|
23
|
Zeng Y, Sun HR, Yu C, Lai Y, Liu XJ, Wu J,
Chen HQ and Liu XH: CXCR1 and CXCR2 are novel mechano-sensors
mediating laminar shear stress-induced endothelial cell migration.
Cytokine. 53:42–51. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gramantieri L, Fornari F, Callegari E,
Sabbioni S, Lanza G, Croce CM, Bolondi L and Negrini M: MicroRNA
involvement in hepatocellular carcinoma. J Cell Mol Med.
12:2189–2204. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Budhu A, Jia HL, Forgues M, Liu CG,
Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, et al:
Identification of metastasis-related microRNAs in hepatocellular
carcinoma. Hepatology. 47:897–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin X, Yang Z, Zhang P, Liu Y and Shao G:
miR-154 inhibits migration and invasion of human non-small cell
lung cancer by targeting ZEB2. Oncol Lett. 12:301–306.
2016.PubMed/NCBI
|
|
27
|
Barcelona PF, Ortiz SG, Chiabrando GA and
Sánchez MC: Alpha2-macroglobulin induces glial fibrillary acidic
protein expression mediated by low-density lipoprotein
receptor-related protein 1 in Müller cells. Invest Ophthalmol Vis
Sci. 52:778–786. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang P, Gu Y, Zhang Q, Han Y, Hou J, Lin
L, Wu C, Bao Y, Su X, Jiang M, et al: Identification of resting and
type I IFN-activated human NK cell miRNomes reveals microRNA-378
and microRNA-30e as negative regulators of NK cell cytotoxicity. J
Immunol. 189:211–221. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ma S, Liu M, Xu Z, Li Y, Guo H, Ge Y, Liu
Y, Zheng D and Shi J: A double feedback loop mediated by
microRNA-23a/27a/24-2 regulates M1 versus M2 macrophage
polarization and thus regulates cancer progression. Oncotarget.
7:13502–13519. 2016.PubMed/NCBI
|
|
30
|
Su R, Dong L, Zou D, Zhao H, Ren Y, Li F,
Yi P, Li L, Zhu Y, Ma Y, et al: microRNA-23a, −27a and −24
synergistically regulate JAK1/Stat3 cascade and serve as novel
therapeutic targets in human acute erythroid leukemia. Oncogene.
35:6001–6014. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhuang G, Wu X, Jiang Z, Kasman I, Yao J,
Guan Y, Oeh J, Modrusan Z, Bais C, Sampath D, et al:
Tumour-secreted miR-9 promotes endothelial cell migration and
angiogenesis by activating the JAK-STAT pathway. EMBO J.
31:3513–3523. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang Q, Huang C, Yang Q, Gao L, Liu HC,
Tang J and Feng WH: MicroRNA-30c modulates type I IFN responses to
facilitate porcine reproductive and respiratory syndrome virus
infection by targeting JAK1. J Immunol. 196:2272–2282. 2016.
View Article : Google Scholar : PubMed/NCBI
|