Introduction
Glioma is the most common, aggressive and lethal
type of malignant tumor of the central nervous system (1,2). Its
relapse and mortality rate are constantly increasing due to the
inefficiency of current treatment and this has led to its poor
prognosis despite the combination of multidisciplinary therapies
including surgical resection, chemotherapy and radiotherapy
(3,4). Although diverse therapeutic efforts
have been made, the median survival of glioma patients has not
changed markedly in the last few years (5,6). The
poor prognosis highlights the urgent need to elucidate the detailed
molecular mechanism for the development of novel therapeutic tools
against glioma and the identification of diagnostic and prognostic
markers of glioma (7).
MicroRNAs (miRNAs) are an endogenous group of
evolutionarily conserved, non-coding RNAs, which function as
negative modulators of gene expression at the post-transcriptional
level via binding to the 3′-untranslated (3′-UTR) regions of target
mRNAs through complementary binding, causing translational
suppression or mRNA degradation (8–10).
Accumulating evidence has confirmed that miRNAs are involved in
diverse fundamental biological processes including cell
differentiation, apoptosis, proliferation and invasion (11–13).
An increasing number of studies have demonstrated that aberrant
miRNAs play crucial roles in cancer pathogenesis and progression
(14). Recently, miR-142 was
reported to be dysregulated in multiple cancers (15–17).
For instance, it was significantly downregulated in cervical cancer
(13), osteosarcoma (18), lung (19) and colon cancer (20,21).
Previous studies revealed that miR-142 was identified as a
regulator of cell proliferation, migration and invasion in
different cancers. miR-142 inhibits cell proliferation and invasion
of cervical cancer cells by targeting FZD7 (13). miR-142 functions as a potential
tumor suppressor in human osteosarcoma by targeting HMGA1 (22). In addition, serum miR-142 is
associated with early relapse in operable lung adenocarcinoma
patients (23). However, the
expression of miR-142 is overexpressed in nasopharyngeal carcinoma
(24), testicular (25), and head and neck cancer (26), esophageal squamous cell (27) and renal cell carcinoma (28). miR-142 suppresses SOCS6 expression
and promotes cell proliferation in nasopharyngeal carcinoma
(24). Oncogenic miR-142 is
associated with cellular migration, proliferation and apoptosis in
renal cell carcinoma (28).
Therefore, it is suggested that the functional significance of
miR-142 in tumorigenesis and progression is cancer-type specific.
However, the functional role and the underlying molecular mechanism
by which miR-142 regulates the initiation and development of glioma
have yet to be elucidated.
In the present study, we investigated the expression
and biological function of miR-142 in glioma progression. We found
that miR-142 was significantly decreased in both glioma tissues and
cell lines. Its expression level was closely correlated with World
Health Organization (WHO) grading and poor survival outcome in
patients. miR-142 inhibited glioma cell migration and invasion
in vitro as determined by gain- and loss-of-function
experiments by directly targeting Rac1 thus leading to the
suppression of matrix metalloproteinases (MMPs). Collectively,
these data confirmed the underlying mechanism by which miR-142
inhibits migration and invasion of glioma and identified miR-142 as
a novel prognostic biomarker for glioma patients.
Materials and methods
Clinical tissues and cell lines
Ninety-seven glioma and 25 human normal brain
tissues were obtained from The First Affiliated Hospital of
Chongqing Medical University from January 2007 to December 2010.
None of the patients had received local or systemic treatment
before surgery. Informed consent was obtained from all the patients
and the present study was approved by the Ethics Committee of the
First Affiliated Hospital of Chongqing Medical University according
to the Declaration of Helsinki.
Human glioma cell lines (U87, U251, LN229 and U373)
and normal human astrocytes (NHA) were purchased from the Institute
of Biochemistry and Cell Biology (Chinese Academy of Sciences,
Shanghai, China) and were cultured in Dulbecco's modified Eagle's
medium/F12 (DMEM; HyClone, Logan, UT, USA) supplemented with 10%
fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and 1%
penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO, USA) at 37°C
with 5% CO2.
RNA extraction, quantitative real-time
PCR (qRT-PCR)
Total RNA was extracted from tissues and cells by
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer's instructions. RNA was reverse-transcribed using a
TaqMan Human MiRNA Assay kit (Applied Biosystems, Foster City, CA,
USA) and cDNA was then amplified with a SYBR® Premix Ex
Taq™ II (Perfect Real-Time) kit (Takara Bio Inc., Shiga, Japan) and
analyzed using an Applied Biosystems 7900 Real-Time PCR System.
Hsa-miR-142 primer (HmiRQP0185), snRNA U6 qPCR primer (HmiRQP9001)
and GAPDH (HQP006940) were purchased from GeneCopoeia (Guangzhou,
China). The primer for Rac1 was synthesized by GenePharma
(Shanghai, China).
Western blotting
Proteins were isolated in RIPA buffer (Beyotime,
Haimen, China) and the protein concentration was determined using a
Bicinchoninic Acid (BCA) Protein Assay kit (Thermo Fischer
Scientific, Rockford, IL, USA). Protein (40 µg) was separated on
10% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF)
membranes (Millipore, Billerica, MA, USA). The membranes were
blocked with 5% non-fat milk in Tris-buffered saline with Tween-20
(TBST) and then incubated at 4°C overnight with primary antibodies:
Rac1, MMP2, MMP9 and GAPDH (1:1,000; Cell Signaling Technology,
Inc., Danvers, MA, USA). After being washed with TBST, the
membranes were incubated with an appropriate peroxidase-conjugated
secondary antibody for 2 h at room temperature (ZSGB-BIO, Beijing,
China). Protein bands were visualized using an enhanced
chemiluminescence kit (Amersham, Little Chalfont, UK).
Immunohistochemical staining analysis
(IHC)
IHC was performed on formalin-fixed paraffin
sections. MMP2 and MMP9 (Cell Signaling Technology, Inc.) (1:100)
antibodies were used in immunohistochemical staining with the
streptavidin peroxidase-conjugated (SP-IHC) method. The percentage
of positive cells was expressed as the following grades: 0, <5%;
1, 6–25%; 2, 26–50%; 3, 51–75%; and 4, >75%.
Cell transfection
The cells were seeded into 6-well plates and
transfected with vectors using Lipofectamine 2000 reagent
(Invitrogen Life Technologies) in accordance with the
manufacturer's protocol. miRNA expression vectors, including
miR-142 expression vector (HmiR0282), the control vector
(CmiR0001-MR04) were obtained from GeneCopoeia. The Rac1 expression
plasmid was produced by GenePharma.
Wound-healing migration assay
To confirm the migratory ability of glioma cells, we
performed a wound-healing assay in each group. Cells were seeded
into 6-well plates and when confluence reached 80–90%, a 200 µl tip
was used to create a wound across the cell plates. Subsequently the
cells were washed twice with phosphate-buffered saline (PBS) to
remove the detached cells. The cells were then cultured in DMEM in
a common incubator for 48 h. Finally, the images were captured
using phase-contrast microscope.
Transwell invasion assays
Cell invasion assays were determined by 8-µM
pore-sized Transwell inserts (Nalge Nunc International Corp.,
Naperville, IL, USA). The upper chambers were pre-coated with
Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). The cells
(2.5×104) in 150 µl of serum-free DMEM were loaded into
the top chamber, and 750 µl of DMEM with 10% FBS were placed in the
bottom chamber. After 24 h of incubation, the cells were fixed in
4% paraformaldehyde and stained with 0.3% crystal violet. The cells
remaining in the top layer were swabbed carefully and the bottom
cells were counted under microscope.
Luciferase reporter assay
Luciferase activity was determined using Dual
Luciferase Assay (Promega, Madison, WI, USA) according to the
manufacturer's protocols. The wild-type 3′-UTR of Rac1 containing
the putative miR-142 target site was amplified and cloned into the
pGL3-control luciferase reporter vector (Promega). The mutant
constructs were generated using a QuickChange site-directed
mutagenesis kit (Stratagene, La Jolla, CA, USA). The experiment was
performed in duplicate in three independent experiments.
Statistical analysis
Data are presented as the mean ± SD from at least
three independent replicates. SPSS software 16.0 (SPSS, Inc.,
Chicago, IL, USA) and GraphPad Prism 6.0 (GraphPad, San Diego, CA,
USA CA, USA) were used for a two-tailed Students t-test, Pearson's
correlation analysis, Kaplan-Meier method and the log-rank test to
evaluate statistical significance. Differences were defined as
P<0.05.
Results
Decreased expression of miR-142 in
human glioma tissues and cells
We performed qRT-PCR to investigate the expression
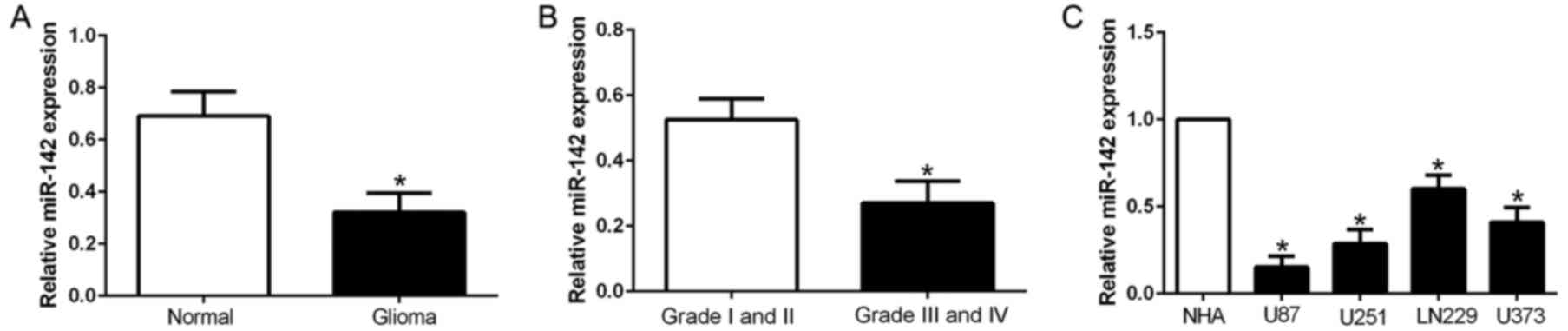
level of miR-142 in clinical tissues. As shown in Fig. 1A, the expression of miR-142 was
significantly decreased in the glioma tissues compared with the
normal brain tissues (Fig. 1A;
P<0.05). According to the pathological diagnosis, the expression
of miR-142 in the high grade glioma (III and IV) were lower than
that in the low grade glioma tissues (I and II), which indicated
that miR-142 expression was associated with glioma malignancy
(Fig. 1B; P<0.05). Consistently,
all glioma cell lines had significantly decreased expression of
miR-142 than normal astrocytes (Fig.
1C; P<0.05). These data revealed that miR-142 may act as a
tumor suppressor in glioma.
Decreased expression of miR-142
predicts a poor prognosis
To assess the clinical significance of miR-142 in
glioma, we determined the median expression value as a cut-off to
divide glioma patients into groups of high or low expression levels
of miR-142. As shown in Table I,
low expression of miR-142 was prominently associated with the
histological grade of the glioma (P=0.001). Hence, these findings
revealed that decreased expression of miR-142 was involved in the
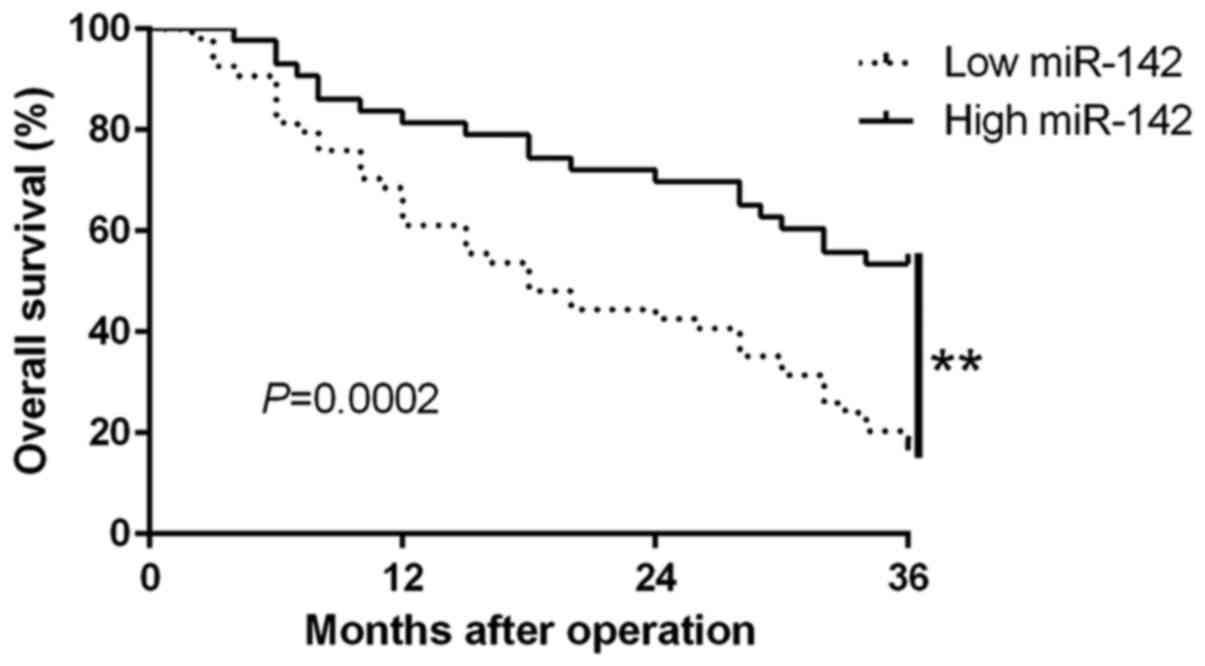
progression of glioma. Moreover, survival analysis revealed that
the downregulation of miR-142 was significantly correlated with
shorter overall survival (P=0.0002; Fig. 2) in glioma patients. Furthermore,
miR-142 expression was an independent prognostic factor for
predicting overall survival in glioma patients (P=0.001; 0.001,
respectively; Table II). These
results revealed that miR-142 may possibly serve as a potential
prognostic biomarker in glioma patients.
 | Table I.Association of miR-142 expression with
clinicopathological characteristics of glioma patients. |
Table I.
Association of miR-142 expression with
clinicopathological characteristics of glioma patients.
|
|
| miR-142 expression
level |
|
|---|
|
|
|
|
|
|---|
| Clinical
parameters | Cases (n) | High (n=43) | Low (n=54) | P-value
(aP<0.05) |
|---|
| Age (years) |
|
|
| 0.936 |
|
<60 | 65 | 29 | 36 |
|
|
≥60 | 32 | 14 | 18 |
|
| Sex |
|
|
| 0.629 |
|
Male | 59 | 25 | 34 |
|
|
Female | 38 | 18 | 20 |
|
| WHO grade |
|
|
| 0.001a |
|
I+II | 34 | 24 | 10 |
|
|
III+IV | 63 | 19 | 44 |
|
| Location |
|
|
| 0.791 |
|
Supratentorial | 69 | 30 | 39 |
|
|
Subtentorial | 28 | 13 | 15 |
|
 | Table II.Univariate and multivariate analysis
of prognostic factors in glioma patients. |
Table II.
Univariate and multivariate analysis
of prognostic factors in glioma patients.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (years) | 1.205 | 0.563–2.273 | 0.458 | 1.180 | 0.397–1.709 | 0.436 |
| Sex | 1.538 | 0.558–2.783 | 0.523 | 1.039 | 0.482–1.927 | 0.329 |
| WHO grade | 4.327 | 1.872–5.674 | 0.001a | 2.512 | 1.287–3.165 | 0.002a |
| miR-142 | 3.965 | 1.583–7.382 | 0.001a | 3.218 | 1.237–6.962 | 0.001a |
miR-142 inhibits cell migration and
invasion in human glioma cells
To explore the biological function of miR-142 in
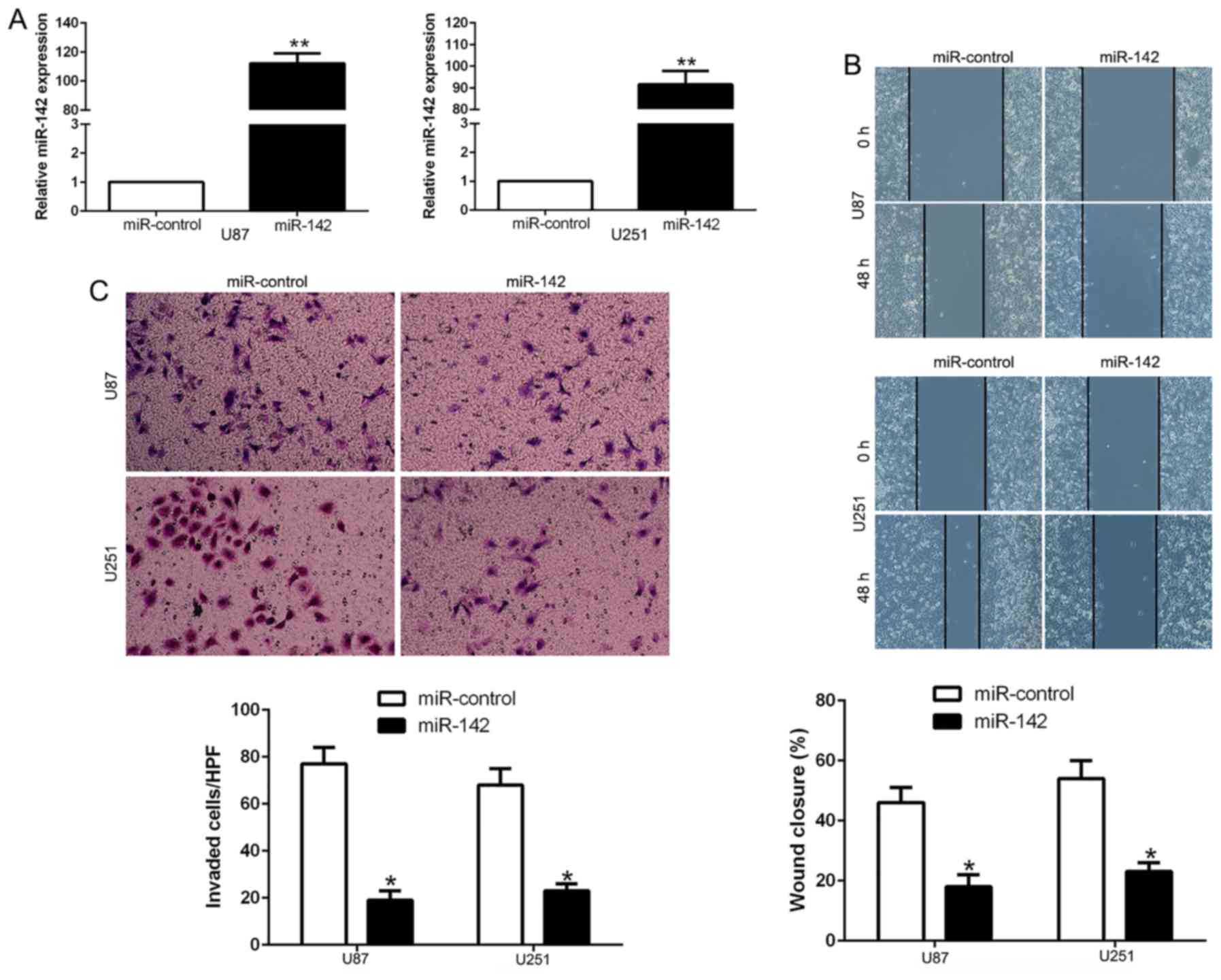
glioma, we transfected miR-142 mimics to ectopically express this
miRNA in U87 and U251 cells. The results revealed that the
expression of miR-142 was significantly increased by appropriate
vectors in both cell lines as determined by qRT-PCR (Fig. 3A; P<0.01). Wound-healing assay
demonstrated that the overexpression of miR-142 significantly
inhibited the migration ability of U87 and U251 cells (Fig. 3B; P<0.05). Consistently,
Transwell assay revealed that the ectopic expression of miR-142
suppressed the invasive number of glioma cells (Fig. 3C; P<0.05). These results
demonstrated that miR-142 regulates the migration and invasion of
glioma cells.
Overexpression of miR-142 inhibits
invasion-related genes
MMP2 and MMP9 are important members of the MMP
family, which degrade all essential components of the extracellular
matrix and play critical roles in cancer metastasis, including
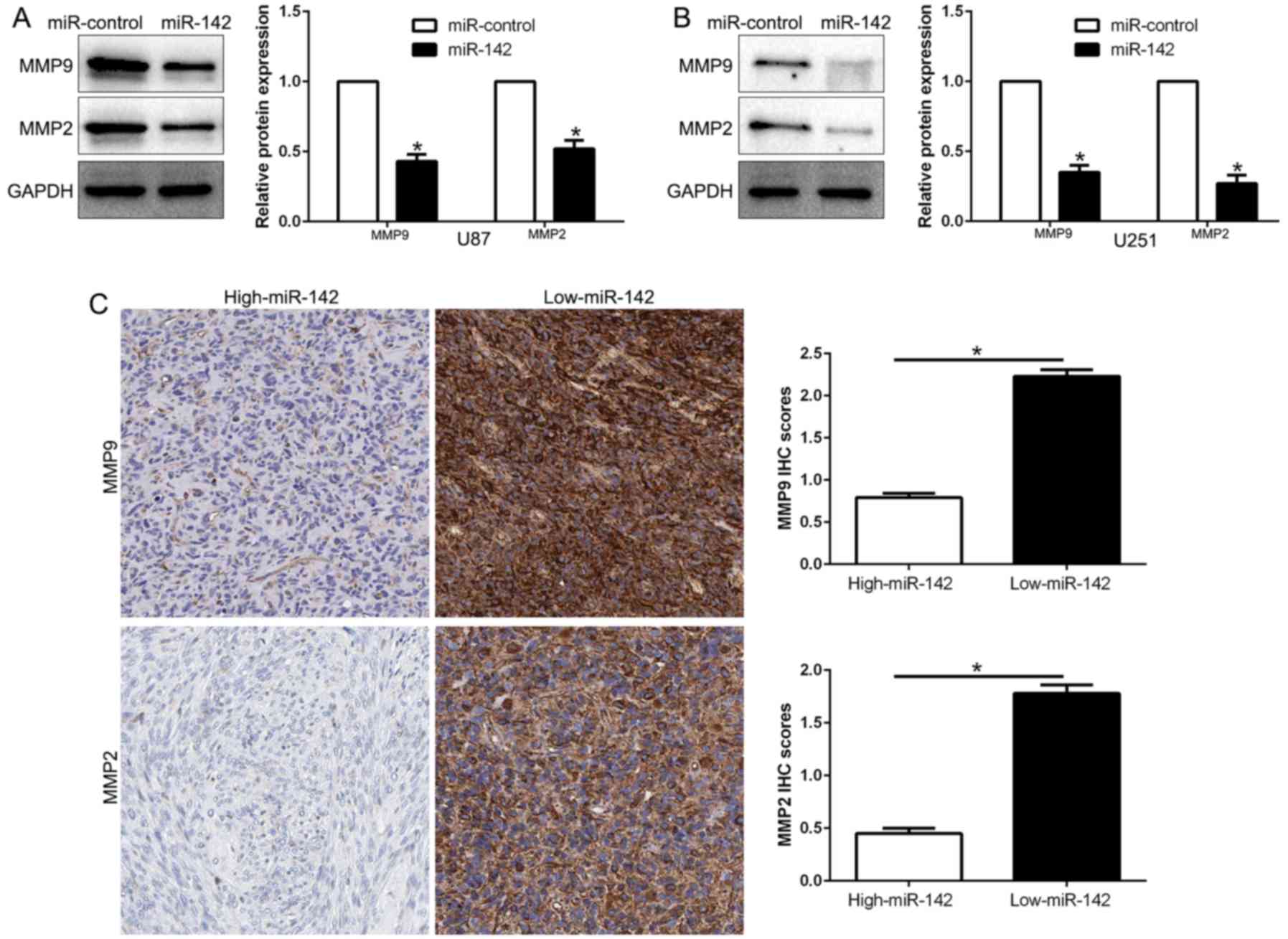
glioma. We disclosed whether miR-142 modulated these
invasion-related genes. Our results revealed that miR-142
overexpression significantly decreased the expression of MMP2 and
MMP9 in both U87 and U251 cells (P<0.05; Fig. 4A and B). Moreover, we investigated
the correlation between miR-142 expression and MMPs in glioma
tissues. We found that MMP2 and MMP9 expression in the high miR-142
expression group was significantly lower than that in the low
expression miR-142 group (P<0.05; Fig. 4C). In conclusion, these results
revealed that miR-142 is critical for glioma cell invasion by
regulating the expression of MMPs.
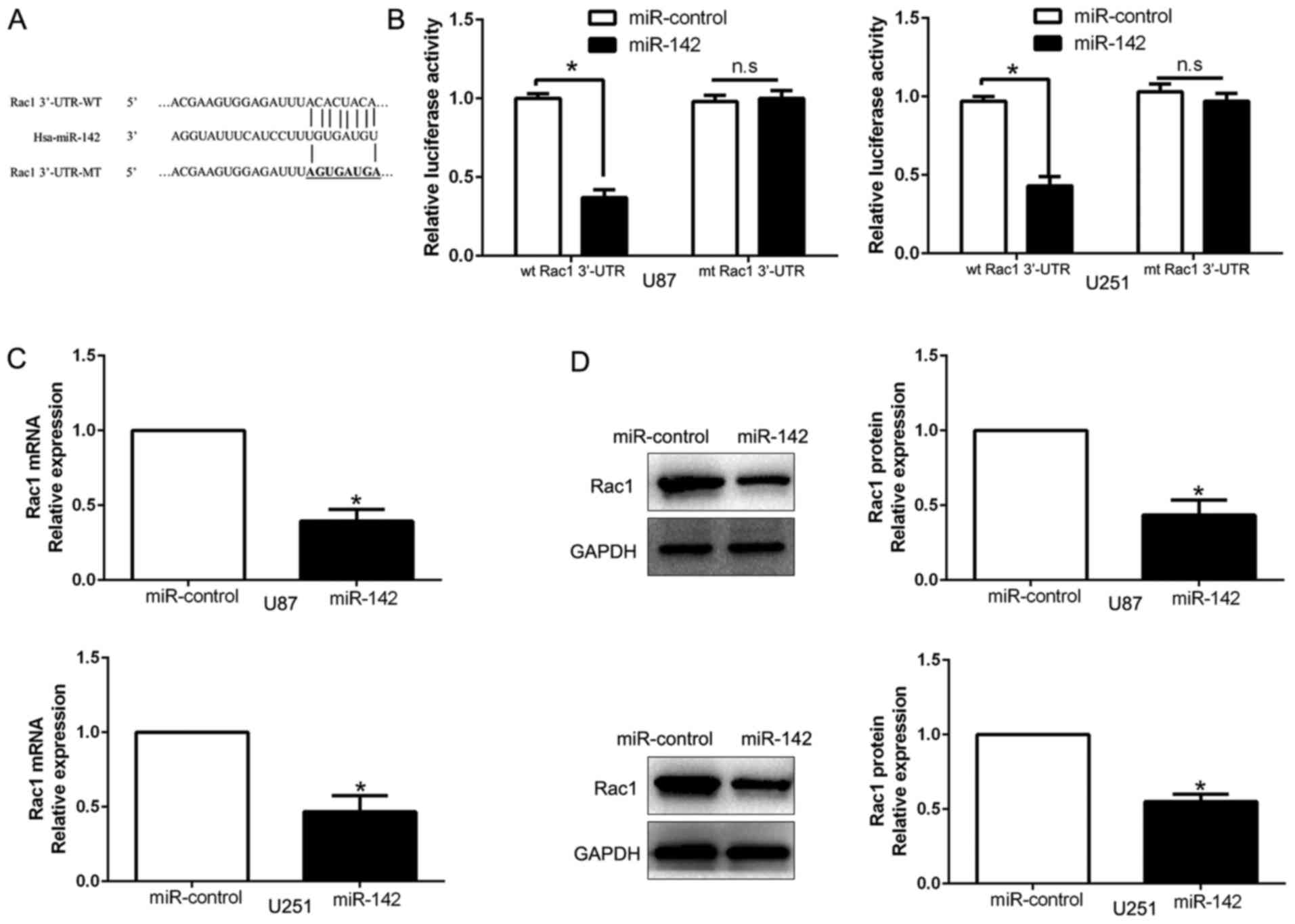
Rac1 is the direct downstream target
of miR-142
We searched for candidate target genes of miR-142 by
exploring different algorithm programs (TargetScan and miRanda) and
predicted that the 3′-UTR of Rac1 contains a putative binding site
for miR-142 (Fig. 5A). A luciferase
assay was performed to investigate whether miR-142 directly targets
Rac1. Our data demonstrated that ectopic expression of miR-142
induced a clear decrease of the luciferase activity of the
wild-type (wt) Rac1 3′-UTR, but did not inhibit the activity of
Rac1 with a mutant (mt) 3′-UTR (Fig.
5B; P<0.05). In addition, miR-142 overexpression obviously
suppressed Rac1 mRNA (Fig. 5C;
P<0.05) and protein (Fig. 5D;
P<0.05) expression in U87 and U251 cells. These results
indicated that miR-142 directly targeted Rac1 in glioma cells
through interaction with its 3′-UTR.
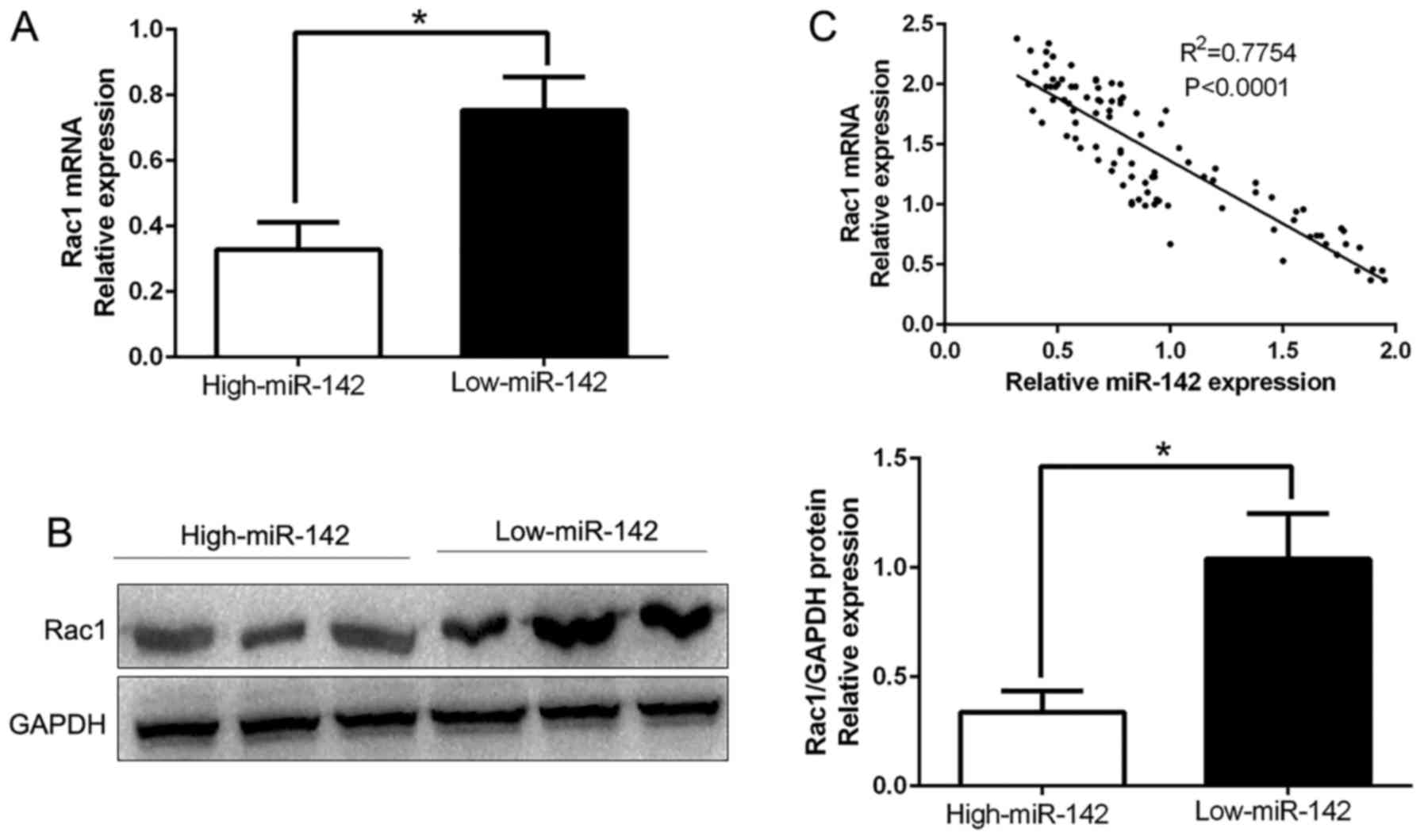
Rac1 levels are inversely correlated
with miR-142 levels in glioma tissues
To further confirm the aforementioned findings, we
detected the expression of Rac1 in human glioma tissues. Our data
demonstrated that Rac1 mRNA and protein levels were significantly
lower in the high miR-142 expression group than that in the low
miR-142 expression group in glioma (P<0.05; Fig. 6A and B). In addition, the results
revealed that the mRNA level of Rac1 in glioma tissues was
inversely correlated with miR-142 expression (R2=0.7754;
P<0.0001; Fig. 6C). In
conclusion, these data revealed that Rac1 was a direct downstream
target of miR-142 in glioma.
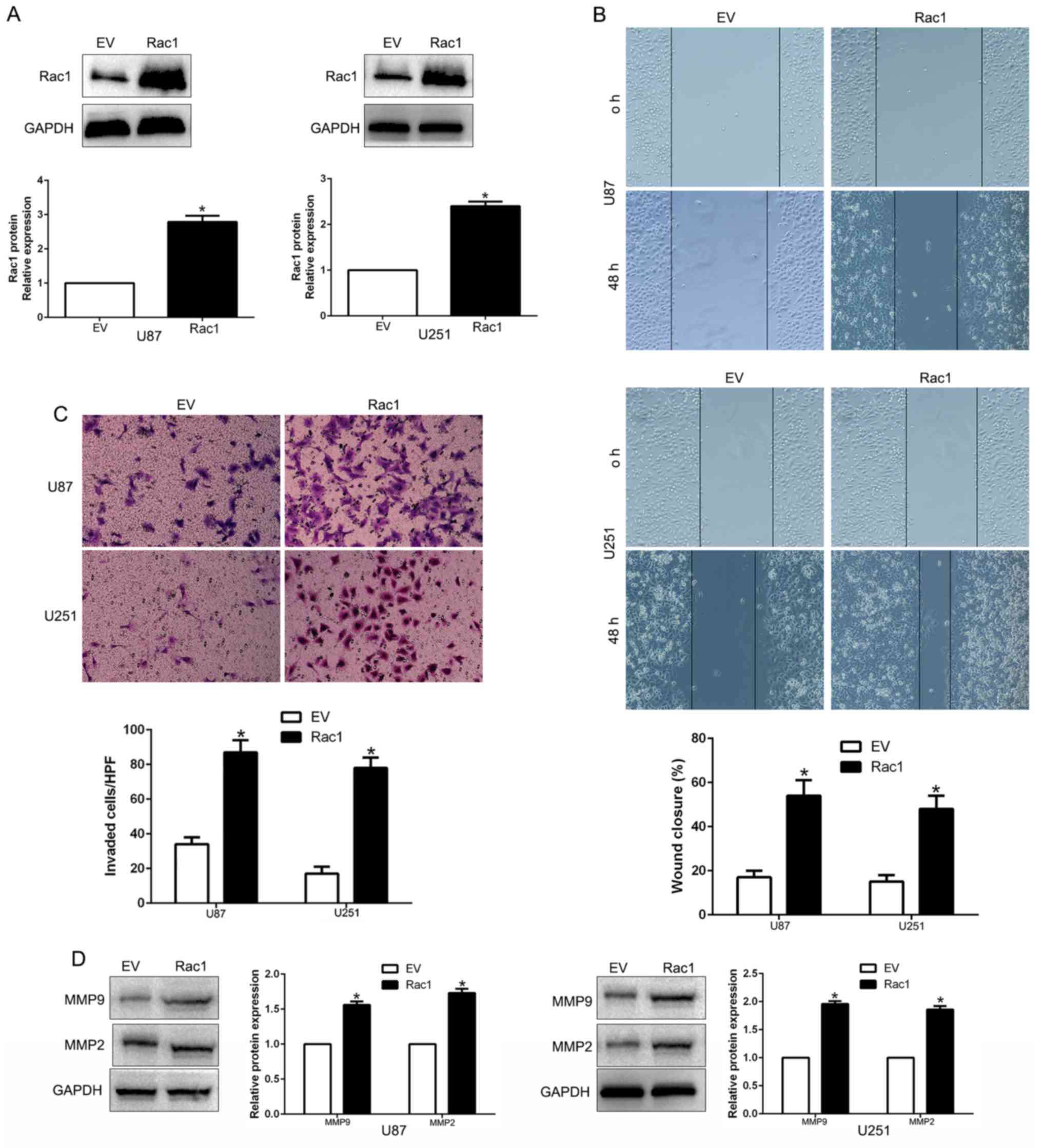
Overexpression of Rac1 expression
counteracted the suppressive effects of miR-142 expression
To further confirm whether Rac1 was involved in
miR-142-mediated inhibition of migration, invasion and
invasion-related genes, we restored Rac1 expression in U87-miR-142
and U251-miR-142 cells by transfecting Rac1 expression plasmid
(P<0.05; Fig. 7A). The results
revealed that Rac1 overexpression promoted cell migration
(P<0.05; Fig. 7B), invasion
(P<0.05; Fig. 7C) and
invasion-related MMP expression (P<0.05; Fig. 7D). These data demonstrated that Rac1
is a downstream mediator in the function of miR-142 in glioma.
Discussion
Recently, accumulating evidence has confirmed that
aberrant miRNAs are involved in the initiation, development and
progression of human malignancies via modulation of their targeting
oncogenes or tumor-suppressor genes, which could potentially serve
as biomarkers for the prediction and prognosis of diverse cancers,
including glioma. Therefore, identification of cancer-specific
miRNAs is critical for elucidating their molecular mechanism and
vital for developing novel therapeutic targets. In previous
studies, miRNA-142 negatively regulated canonical Wnt/β-catenin
signaling, which plays pivotal roles in cancer progression
(29). Schwickert et al
demonstrated that miRNA-142 inhibits breast cancer cell
invasiveness by synchronous targeting of WASL, integrin αV and
additional cytoskeletal elements (30). miR-142 functions as a potential
tumor suppressor directly targeting HMGB1 in non-small cell lung
carcinoma (31). However,
conversely, miR-142 could promote cell proliferation in
nasopharyngeal carcinoma (24).
Moreover, miR-142 plays an oncogenic role in cell migration,
proliferation and apoptosis in renal cell carcinoma (28). These data indicated that the
expression level and biological function of miR-142 was cancer
type-dependent.
In the present study, we confirmed that the
expression of miR-142 was significantly downregulated in clinical
specimens and cell lines. Moreover, our data revealed that
decreased expression of miR-142 was associated with advanced WHO
grade in glioma. In addition, miR-142 expression was an independent
prognostic indicator in the prediction of the 5-year overall
survival of glioma patients. Collectively, these data revealed that
miR-142 may play a critical role in the development of glioma and
the decreased expression of miR-142 confers a worse prognosis and
could be identified as a prognostic indicator. Mechanically,
overexpression of miR-142 suppressed the cell migration and
invasion capacity of glioma U87 and U251 cells. Moreover, miR-142
inhibited the invasion-related MMP gene expression. Furthermore, we
demonstrated that Rac1 was a direct target of miR-142 in glioma,
which was consistent with studies on hepatocellular carcinoma and
osteosarcoma (18,32). We also observed that miR-142
overexpression suppressed the expression of Rac1 mRNA and protein.
Furthermore, we found an inverse correlation between miR-142
expression and Rac1 in glioma tissues. Additionally, the effects of
miR-142 alteration on cell migration and invasion of glioma cells
were abolished by Rac1 modulation. Thus, these results indicated
that miR-142 inhibited migration and invasion in glioma cells by
directly blocking the Rac1 pathway.
Emerging evidence has revealed that Rac1-dependent
signaling is essential for tumor initiation, development and
progression (33). The expression
of Rac1 has been demonstrated to be much higher in several human
cancers, including glioma (34).
The high expression of Rac1 induced glioma cell proliferation and
inhibited apoptosis. Moreover, previous studies confirmed that Rac1
could promote migration and invasion by inducing
epithelial-to-mesenchymal transition (EMT) and MMP expression
(35), which have been identified
as key regulators in the invasion and metastasis of diverse
cancers. In the present study, we found miR-142 inhibited MMP
expression by targeting Rac1. Rac1 restoration abrogated the
suppressive effect of miR-142. Collectively, these data
demonstrated the suppressive effect of miR-142 was mediated by
targeting Rac1 to inhibit MMP expression in glioma.
In conclusion, we demonstrated that miR-142 was
downregulated in glioma tissues and cell lines, and its decreased
expression was associated with advanced clinicopathological
features. Furthermore, we confirmed that miR-142 inhibited cell
migration, invasion and MMP expression by inhibiting Rac1. These
results revealed that miR-142 is a potential metastasis-associated
tumor suppressor in glioma. Collectively, decreased miR-142
expression may play an important role in tumor metastasis and may
be a novel prognostic factor and potential therapeutic target for
glioma.
Acknowledgements
The present study was supported by a grant from the
National Natural Scientific Foundation of China (no. 81403243).
References
|
1
|
Vredenburgh JJ, Desjardins A, Reardon DA
and Friedman HS: Experience with irinotecan for the treatment of
malignant glioma. Neuro Oncol. 11:80–91. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xie Q, Mittal S and Berens ME: Targeting
adaptive glioblastoma: An overview of proliferation and invasion.
Neuro Oncol. 16:1575–1584. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Preusser M, Haberler C and Hainfellner JA:
Malignant glioma: Neuropathology and neurobiology. Wien Med
Wochenschr. 156:332–337. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vigneswaran K, Neill S and Hadjipanayis
CG: Beyond the World Health Organization grading of infiltrating
gliomas: Advances in the molecular genetics of glioma
classification. Ann Transl Med. 3:952015.PubMed/NCBI
|
|
5
|
Cohen AL and Colman H: Glioma biology and
molecular markers. Cancer Treat Res. 163:15–30. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: European Organisation for Research and Treatment of
Cancer Brain Tumor and Radiotherapy Groups; National Cancer
Institute of Canada Clinical Trials Group: Radiotherapy plus
concomitant and adjuvant temozolomide for glioblastoma. N Engl J
Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Davis FG and McCarthy BJ: Current
epidemiological trends and surveillance issues in brain tumors.
Expert Rev Anticancer Ther. 1:395–401. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Makeyev EV and Maniatis T: Multilevel
regulation of gene expression by microRNAs. Science. 319:1789–1790.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartels CL and Tsongalis GJ: MicroRNAs:
Novel biomarkers for human cancer. Clin Chem. 55:623–631. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu Z, Dou C, Yao B, Xu M, Ding L, Wang Y,
Jia Y, Li Q, Zhang H, Tu K, et al: Methylation-mediated repression
of microRNA-129-2 suppresses cell aggressiveness by inhibiting high
mobility group box 1 in human hepatocellular carcinoma. Oncotarget.
7:36909–36923. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu J, Cui H, Zhu Z and Wang L:
MicroRNA-200b-3p suppresses epithelial-mesenchymal transition and
inhibits tumor growth of glioma through down-regulation of ERK5.
Biochem Biophys Res Commun. 478:1158–1164. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Deng B, Zhang Y, Zhang S, Wen F, Miao Y
and Guo K: MicroRNA-142-3p inhibits cell proliferation and invasion
of cervical cancer cells by targeting FZD7. Tumour Biol.
36:8065–8073. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Malzkorn B, Wolter M, Liesenberg F,
Grzendowski M, Stühler K, Meyer HE and Reifenberger G:
Identification and functional characterization of microRNAs
involved in the malignant progression of gliomas. Brain Pathol.
20:539–550. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cao XC, Yu Y, Hou LK, Sun XH, Ge J, Zhang
B and Wang X: miR-142-3p inhibits cancer cell proliferation by
targeting CDC25C. Cell Prolif. 49:58–68. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pan D, Du Y, Ren Z, Chen Y, Li X, Wang J
and Hu B: Radiation induces premature chromatid separation via the
miR-142-3p/Bod1 pathway in carcinoma cells. Oncotarget.
7:60432–60445. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma Z, Liu T, Huang W, Liu H, Zhang HM, Li
Q, Chen Z and Guo AY: MicroRNA regulatory pathway analysis
identifies miR-142-5p as a negative regulator of TGF-β pathway via
targeting SMAD3. Oncotarget. 7:71504–71513. 2016.PubMed/NCBI
|
|
18
|
Zheng Z, Ding M, Ni J, Song D, Huang J and
Wang J: miR-142 acts as a tumor suppressor in osteosarcoma cell
lines by targeting Rac1. Oncol Rep. 33:1291–1299. 2015.PubMed/NCBI
|
|
19
|
Su YH, Zhou Z, Yang KP, Wang XG, Zhu Y and
Fa XE: MIR-142-5p and miR-9 may be involved in squamous lung cancer
by regulating cell cycle related genes. Eur Rev Med Pharmacol Sci.
17:3213–3220. 2013.PubMed/NCBI
|
|
20
|
Yin Y, Song M, Gu B, Qi X, Hu Y, Feng Y,
Liu H, Zhou L, Bian Z, Zhang J, et al: Systematic analysis of key
miRNAs and related signaling pathways in colorectal tumorigenesis.
Gene. 578:177–184. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shen WW, Zeng Z, Zhu WX and Fu GH:
MiR-142-3p functions as a tumor suppressor by targeting CD133,
ABCG2, and Lgr5 in colon cancer cells. J Mol Med. 91:989–1000.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu G, Wang J, Jia Y, Shen F, Han W and
Kang Y: MiR-142-3p functions as a potential tumor suppressor in
human osteosarcoma by targeting HMGA1. Cell Physiol Biochem.
33:1329–1339. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kaduthanam S, Gade S, Meister M, Brase JC,
Johannes M, Dienemann H, Warth A, Schnabel PA, Herth FJ, Sültmann
H, et al: Serum miR-142-3p is associated with early relapse in
operable lung adenocarcinoma patients. Lung Cancer. 80:223–227.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qi X, Li J, Zhou C, Lv C and Tian M:
MiR-142-3p suppresses SOCS6 expression and promotes cell
proliferation in nasopharyngeal carcinoma. Cell Physiol Biochem.
36:1743–1752. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pelloni M, Coltrinari G, Paoli D, Pallotti
F, Lombardo F, Lenzi A and Gandini L: Differential expression of
miRNAs in the seminal plasma and serum of testicular cancer
patients. Endocrine. Oct 28–2016.(Epub ahead of print). https://doi.org/10.1007/s12020-016-1150-z
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Summerer I, Unger K, Braselmann H,
Schuettrumpf L, Maihoefer C, Baumeister P, Kirchner T, Niyazi M,
Sage E, Specht HM, et al: Circulating microRNAs as prognostic
therapy biomarkers in head and neck cancer patients. Br J Cancer.
113:76–82. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin RJ, Xiao DW, Liao LD, Chen T, Xie ZF,
Huang WZ, Wang WS, Jiang TF, Wu BL, Li EM, et al: MiR-142-3p as a
potential prognostic biomarker for esophageal squamous cell
carcinoma. J Surg Oncol. 105:175–182. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Y, Chen D, Jin LU, Liu J, Li Y, Su Z,
Qi Z, Shi M, Jiang Z, Yang S, et al: Oncogenic microRNA-142-3p is
associated with cellular migration, proliferation and apoptosis in
renal cell carcinoma. Oncol Lett. 11:1235–1241. 2016.PubMed/NCBI
|
|
29
|
Isobe T, Hisamori S, Hogan DJ, Zabala M,
Hendrickson DG, Dalerba P, Cai S1, Scheeren F, Kuo AH1, Sikandar
SS, et al: miR-142 regulates the tumorigenicity of human breast
cancer stem cells through the canonical WNT signaling pathway.
Elife. 3:doi: 10.7554/eLife.01977. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Schwickert A, Weghake E, Brüggemann K,
Engbers A, Brinkmann BF, Kemper B, Seggewiß J, Stock C, Ebnet K,
Kiesel L, et al: microRNA miR-142-3p inhibits breast cancer cell
invasiveness by synchronous targeting of WASL, integrin alpha V,
and additional cytoskeletal elements. PLoS One. 10:e01439932015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xiao P and Liu WL: MiR-142-3p functions as
a potential tumor suppressor directly targeting HMGB1 in
non-small-cell lung carcinoma. Int J Clin Exp Pathol.
8:10800–10807. 2015.PubMed/NCBI
|
|
32
|
Wu L, Cai C, Wang X, Liu M, Li X and Tang
H: MicroRNA-142-3p, a new regulator of RAC1, suppresses the
migration and invasion of hepatocellular carcinoma cells. FEBS
Lett. 585:1322–1330. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yoon CH, Hyun KH, Kim RK, Lee H, Lim EJ,
Chung HY, An S, Park MJ, Suh Y, Kim MJ, et al: The small GTPase
Rac1 is involved in the maintenance of stemness and malignancies in
glioma stem-like cells. FEBS Lett. 585:2331–2338. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhong J, Bach CT, Shum MS and O'Neill GM:
NEDD9 regulates 3D migratory activity independent of the Rac1
morphology switch in glioma and neuroblastoma. Mol Cancer Res.
12:264–273. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Man J, Shoemake J, Zhou W, Fang X, Wu Q,
Rizzo A, Prayson R, Bao S, Rich JN and Yu JS: Sema3C promotes the
survival and tumorigenicity of glioma stem cells through Rac1
activation. Cell Rep. 9:1812–1826. 2014. View Article : Google Scholar : PubMed/NCBI
|