Introduction
Glioblastoma (GBM) is one of the most common and
aggressive forms of primary brain tumour, with a median patient
survival of 9–12 months. Despite therapeutic advances and new
biological insights, ~10,000 new patients with high-grade or
malignant glioma suffer from tumour recurrence each year (1,2). The
lethality and poor prognosis of the disease are related to the
highly invasive/migratory capacity of glioma cells, making complete
surgical resection impossible (3,4).
Therefore, it is essential to investigate novel and effective
therapeutic approaches for glioma.
MicroRNAs (miRNAs) are a class of small non-coding
RNAs which regulate gene expression post-transcriptionally causing
target mRNA deadenylation and degradation in diverse biological
processes, such as proliferation, differentiation, invasion and
apoptosis (5). There is evidence
suggesting that various miRNAs can potentially regulate hundreds of
mRNAs and indicated that miRNAs are involved in tumourigenic
processes (6). It has been reported
that miRNAs are involved in the regulation of the expression of 1/3
of human protein-coding genes, accounting for ~1% of all the
expressed human genes (7,8). Recent studies have shown that a number
of miRNAs may function as oncogenes or tumour suppressor genes in
glioma, including miR-21, miR-221/222, miR-124 and miR-27b
(9–12). miR-216b has been demonstrated to
function as a regulator in hepatocellular carcinoma (13). However, the function of miR-216b in
gliomas remains unclear.
Forkhead box protein M1 (FoxM1, previously known as
HFH-11, INS-1, WIN, MPP2/MPHOSPH2 or Trident/FKHL16) plays a vital
role in animal development (14).
Overexpression of FoxM1 is common in many types of cancers
including glioma (15), and is
involved in cell cycle progression, cell differentiation, DNA
damage repair, angiogenesis and invasion. Recently, there is
evidence showing that FoxM1 is a key regulator in cancer drug
sensitivity and resistance (16).
The Ki-67 protein (also known as MKi-67) is a marker of cell
proliferation (17). In addition,
it is associated with ribosomal RNA transcription. MMP-2 and MMP-9
are members of the matrix metalloproteinase (MMP) family. The major
function of MMPs in cancer progression is their role in ECM
degradation, which allows cancer cells to migrate out of the
primary tumour to form metastases (18).
In high-grade glioma tissues and cells, miR-216b was
found to be downregulated compared to normal brain tissues and
normal human astrocyte (NHA) cells. In the present study, we found
that FoxM1 is a direct target of miR-216b. Transient transfection
of miR-216b mimics into glioma cell line suppressed cell
proliferation and invasion. Overexpression of FoxM1 partially
impeded the effects of miR-216b on the glioma biological
behaviours. An in vivo study demonstrated that ectopic
expression of miR-216b retarded the proliferation of U87 xenograft
tumours in BALB/c mice. In addition, the percentage of
Ki-67-positive tumour cells and FoxM1 protein expression had a
significant decline compared to the control tumours. These results
indicate that miR-216b may play a critical role in the regulation
of the proliferation and invasion of glioma cells, suggesting that
miR-216b could be a promising therapeutic target for glioblastoma
(GBM).
Materials and methods
Cells and tissues
Human glioma cell lines (U87, T98G, LN229 and U251)
were obtained from the Cell Bank of the Chinese Academy of Sciences
(Shanghai, China). NHA cells were purchased from the American Type
Culture Collection (ATCC; Rockville, MD, USA). All cells were
maintained in a 37°C/5% carbon dioxide incubator in Dulbecco's
modified Eagle's medium (DMEM; HyClone, Logan, UT, USA)
supplemented with 10% foetal-bovine serum (FBS) (Gibco, Los
Angeles, CA, USA).
Human GBM and adjacent normal brain tissues were
collected from 36 patients with histologically confirmed GBM who
underwent tumour resection at Xiangya Hospital of Central South
University between May 2010 and December 2014. No patients had
received any anticancer treatment. All tissues were pathologically
confirmed and immediately snap-frozen in liquid nitrogen and stored
at −80°C until RNA extraction. Written informed consent for
research purposes was obtained from each patient. All procedures
were subjected to the Declaration of Helsinki. In addition, all
applicable international, national, and/or institutional guidelines
for the care and use of animals were followed. The present study
was approved by the Ethical Committee of Central South University
(Changsha, China).
Real-time quantitative polymerase
chain reaction (RT-qPCR)
Total RNA was extracted from tissues and cultured
cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA)
according to the manufacturer's instructions. RT-qPCR chain
reactions were performed in triplicate in an ABI 7500HT Fast
Real-Time PCR System (Applied Biosystems, Foster City, CA, USA)
according to the manufacturers protocols. The miR-216b levels were
detected using the TaqMan MicroRNA Assay kit, and endogenous mRNA
levels of FoxM1 were measured using SYBR-Green PCR Master Mix kit
(Applied Biosystems). The following primer sequences were used in
the present study: FOXM1 F, ATACGTGGATTGAGGACCACT and R, TCCA
ATGTCAAGTAGCGGTTG; miR-216b, 5′-AAATCTCTGCAGGCAAATGTGA-3′; U6, F,
TGTGGGCATCAATGGATT TGG and R, ACACCATGTATTCCGGGTCAAT; FoxM1-Si F,
5′-GGACCACUUUCCCUACUUUUU-3′ and R, 5′-UUAAAGUAGGGAAAGUGGUCC-3′;
control F, 5′-AACAGUCGCGUUUGCGACUGUU-3′ and R,
5′-UUGUCAGCGCAAACGCUGACC-3′; GAPDH F, 5′-CCATGTTCGTCATGGTGTG-3′ and
R, 5′-GGTGCTAAGCAGTTGGTGGTG-3′. The cycling conditions were as
follows: 95°C for 5 min followed by 40 cycles of 95°C for 15 sec
and 60°C for 40 sec. U6 was used as a control to normalize the
miR-216b expression. Relative expression levels of miRNA and mRNA
expression in fresh tissues and cells were determined using the
2−ΔΔCt method.
Plasmids, oligonucleotides and cell
transfection
miR-216b mimics and negative control (NC)
oligonucleotides were purchased from GenePharma (Shanghai, China).
miR-216b mimics or NC were obtained from GeneChem (Shanghai,
China). Cells were transfected with miR-216b mimics, FoxM1 siRNA
and NC (200 pmol each) using Lipofectamine 2000 (Invitrogen)
according to the manufacturer's instructions. The FoxM1 open
reading frame without the 3′UTR region was amplified by PCR with
human FoxM1 cDNA as a template and inserted into the pcDNA3.1(+)
expression vector (Invitrogen). The human FoxM1 3′UTR-Luc reporter
was created by the ligation of FoxM1 3′UTR PCR product into the
XbaI site of the pcDNA3.1-control vector (Promega, Madison,
WI, USA), to generate the plasmid pcDNA3.1-WT-FoxM1-3′UTR
(FoxM1-wild). The mutant reporter was generated from
pcDNA3.1-WT-FoxM1 3′UTR-Luc by replacing the binding site of
miR-216b with a restriction enzyme cutting site GGUGACUC.
Luciferase reporter assay
For the luciferase reporter assay, U87 and U251
cells were co-transected with luciferase reporter vectors and
miR-216b using Lipofectamine 2000. Cells were transfected with
pRL-TK (Promega). The Renilla luciferase activity was
utilized as an internal control. Luciferase activity was analysed
48 h after transfection with a Dual-Luciferase Reporter Assay
System (Promega).
Cell proliferation and colony
formation assays
GBM cells were seeded into 96-well plates at 2,000
cells/well. Following transfection with miR-216b mimics, the MTT
assay was used to measure the cell viability of human glioma cells,
as previously described (9). Each
experiment was performed in triplicate. The optical density was
detected at the wavelength of 490 nm. The data are presented as the
mean ± standard error of the mean. All assays were repeated as
independent experiments at least 3 times. Cells were plated in
6-well plates (300 cells/well) and incubated for 2 weeks. Then,
cells were fixed with methanol and stained with violet (Sigma, St.
Louis, MO, USA). The colonies (>50 cells) were counted.
Wound healing and Transwell
assays
U87 and U251 cells were plated into 6-well plates.
When the cell confluence reached ~90%, ~24 h, after transfection, a
scratch was made using a sterile pipette tip on the monolayer.
After wounding, debris was removed by washing cells with
phosphate-buffered saline (PBS). Images of the scratched area were
captured at 0 h and after 48 h at a magnification of ×200 under a
light microscope. The number of cells that had migrated into the
wounded area or cells with extended protrusions from the wound
border were counted and averaged. Experiments were repeated 3
times.
The Transwell filters (Costar, Cambridge, MA, USA)
coated with Matrigel (Becton-Dickinson, Franklin Lakes, NJ, USA)
were used to quantify in vitro glioma cell invasion.
Transfected cells were cultured at 5×104/well into the
upper compartment of the chamber in serum-free medium. Medium
containing 20% FBS was added to the lower chamber. After 24 h of
incubation at 37°C with 5% carbon dioxide, the medium was removed
from the upper chamber. The non-invading cells were scraped off
with a cotton swab while the bottom cells were fixed with 3%
paraformaldehyde, stained with 0.1% crystal violet and photographed
in 3 independent 10x fields for each well. The fold-change in
migration was calculated relative to the blank control. Data
represent the mean ± SE of 3 independent experiments.
Western blotting
Glioma tissues and cells were scraped in Thermo
Scientific RIPA buffer (Pierce, Rockford, IL, USA) with protease
inhibitors. The Pierce BCA protein assay kit was used to determine
the protein concentration. The membranes were blocked with 5%
non-fat dry milk (w/v) at room temperature for 1 h and incubated
separately with rabbit anti-human FoxM1 (1:1,000), mouse anti-human
MMP-2, MMP-9 (1:1,000), rabbit anti-human Ki-67 (1:1,000) (all from
Cell Signaling Technology, Danvers, MA, USA) and mouse anti-human
β-actin (1:500; Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA). Then, they were incubated with horseradish
peroxidase-conjugated goat anti-rabbit secondary antibody
(1:10,000; ab150077; Abcam, Cambridge, MA, USA) for 1 h. The
relative protein expression levels on the polyvinylidene fluoride
(PVDF) membrane were scanned with the enhanced chemiluminescence
(ECL) system and quantified using Gel Doc 2000 (Bio-Rad, Hercules,
CA, USA).
Tumour xenograft model and Ki-67
immunohistochemical staining
miR-216b-overexpressing and control U87 glioma cells
(5×105 cells/mouse in 3 µl, pretreated with miR-216b and
NC mimics) were injected into the intracranial space of 5-week-old
female nude mice (n=14; Cancer Institute of the Chinese Academy of
Medical Science) using a stereotactic instrument. Bioluminescence
imaging was used to measure intracranial tumour growth. The mice
were anesthetized, injected intraperitoneally with D-luciferin at
50 mg/ml and imaged with Bruker In-Vivo FX PRO Imaging
System. At the end of the experiment (40 days after injection),
mice were sacrificed and fixed with 4% paraformaldehyde. Tumours
were processed for immunohistochemistry.
Paraffin-embedded tissue sections (4 µm) were
deparaffinized and subjected to heat-mediated antigen retrieval.
Sections were incubated with 3% H2O2 to
quench endogenous peroxidase activity. After washing, sections were
incubated with rabbit anti-Ki-67 and FoxM1 antibody (1:200; Cell
Signaling Technology). Then, the sections were incubated with
biotin-conjugated goat anti-rabbit IgG (Vector Laboratories,
Burlingame, CA, USA). After washing, the sections were developed
with a Vectastain ABC (avidin-biotin complex) peroxidase kit
(Vector Laboratories) and a 3,3-diaminobenzidine (DAB) substrate
(Sigma). The section was then counterstained with haematoxylin and
the percentage of Ki-67- and FoxM1-positive cells were estimated
under a microscope.
Statistical analysis
The data are expressed as the mean ± SD of 3
independent experiments. Statistics were estimated with Student's
t-test. All differences were considered to be statistically
significant at the level of P<0.05. Differences between groups
were analysed using a one-way ANOVA or χ2 test. The
results were analysed using GraphPad Prism 5 (GraphPad Software,
Inc., La Jolla, CA, USA). Statistics were performed using the SPSS
Graduate Pack, version 17.0, statistical software (SPSS, Inc.,
Chicago, IL, USA).
Results
miR-216b was downreglated in glioma
tissues and cell lines
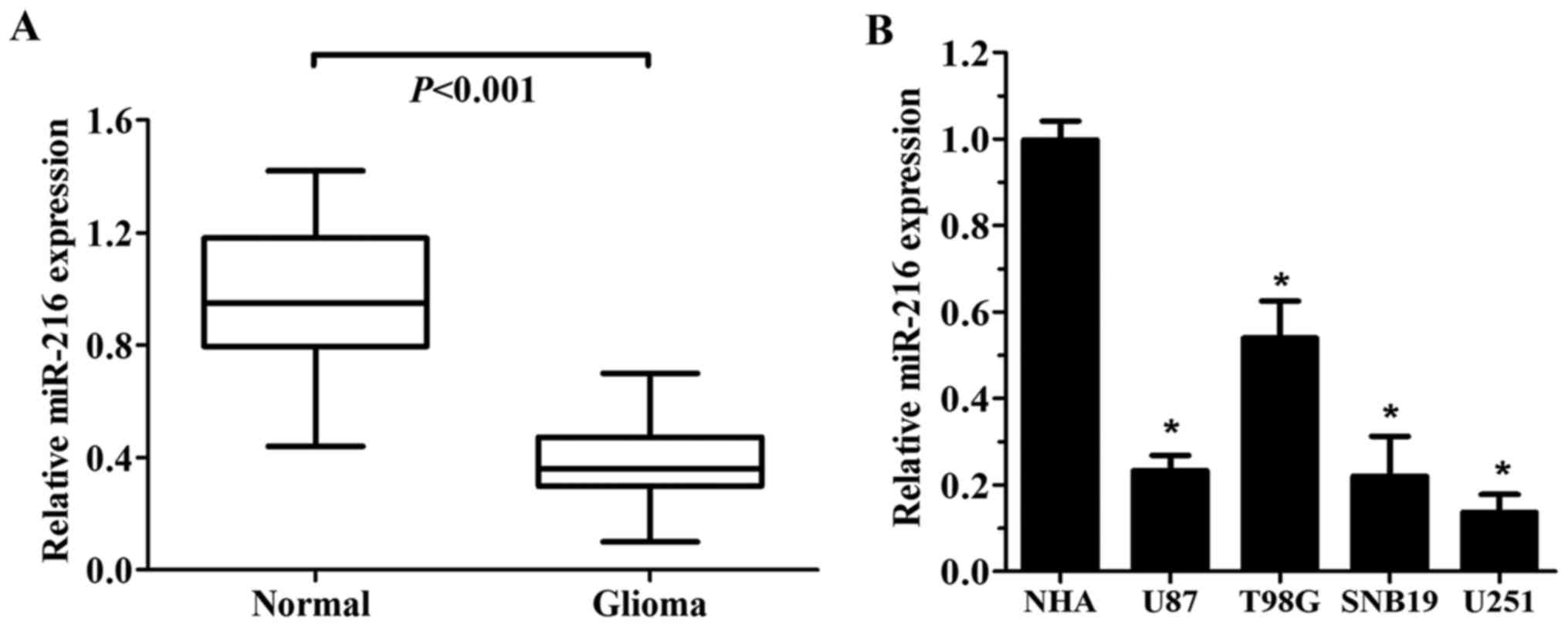
The expression of miR-216b in glioma and normal
brain tissues was examined using RT-qPCR analysis. As shown in
Fig. 1A, glioma tissues exhibited a
significantly lower level of miR-216b compared to that noted in the
adjacent normal brain tissues (P<0.001). miR-216b was
consistently observed to decrease in the GBM cell lines when
compared with the level noted in the NHA cell line (Fig. 1B). In addition, low expression of
miR-216b was significantly associated with KPS score and mean
tumour diameter (Table I)
(P<0.05).
 | Table I.Clinical characteristics of the glioma
patients according to miR-216b level in tissue. |
Table I.
Clinical characteristics of the glioma
patients according to miR-216b level in tissue.
|
|
| miR-216b
expression |
|
|---|
|
|
|
|
|
|---|
| Variables | No. of cases | Low | High | P-value |
|---|
| Age, years |
|
|
| 0.127 |
|
<60 | 19 | 10 | 9 |
|
|
≥60 | 17 | 13 | 4 |
|
| Sex |
|
|
| 0.374 |
|
Male | 22 | 15 | 7 |
|
|
Female | 14 | 8 | 6 |
|
| KPS |
|
|
|
0.029a |
|
<60 | 25 | 19 | 6 |
|
|
≥60 | 11 | 4 | 7 |
|
| Mean tumor diameter
(cm) |
|
|
|
0.015a |
|
<5 | 15 | 6 | 9 |
|
| ≥5 | 21 | 17 | 4 |
|
| Necrosis on
MRI |
|
|
| 0.087 |
|
Yes | 21 | 11 | 10 |
|
| No | 15 | 12 | 3 |
|
| Seizure |
|
|
| 0.273 |
|
Yes | 12 | 9 |
|
|
| No | 24 | 14 | 10 |
|
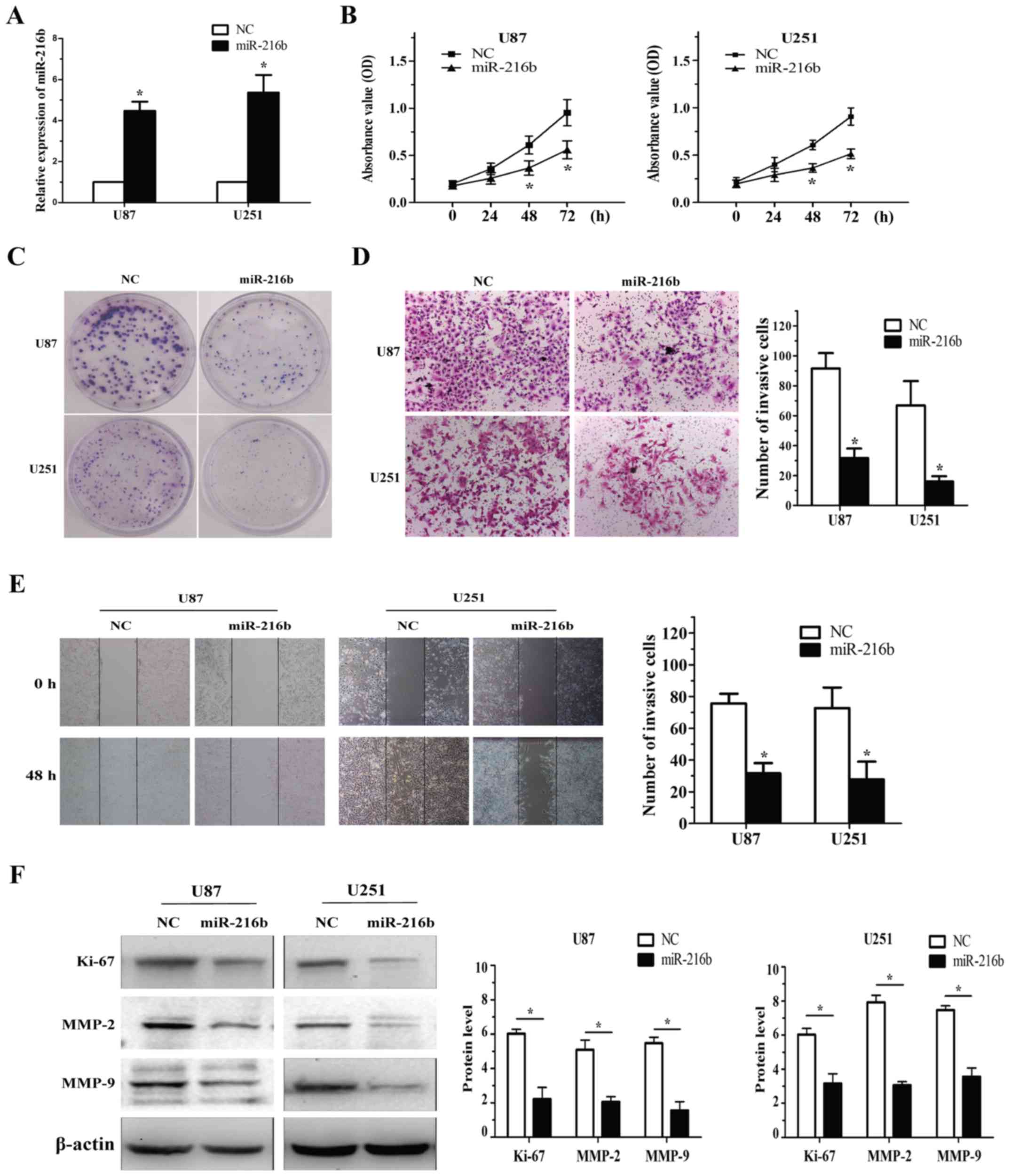
miR-216b inhibits glioma cell
proliferation, migration and invasion in glioma cells
To determine the biological function of miR-216b
downregulation in glioma, glioma cells were transfected with the
miR-216b plasmid and evaluated for proliferation and invasion using
the MTT, colony formation and Transwell assays. miR-216b expression
was confirmed by RT-qPCR in the U87 and U251 cells (Fig. 2A). The result showed that miR-216b
plays a vital role in suppressing the proliferation of glioma cells
(Fig. 2B and C).
Cell invasion was analysed with the Transwell
assays. After treatment with miR-216b, the number of invasive cells
was significantly decreased when compared with this number in the
NC group in the U87 and U251 cells. The experiment demonstrated
that miR-216b also markedly (P<0.05) inhibited the migration
ability of glioma cells (Fig. 2D and
E). To validate that miR-216b influences cell proliferation and
invasion, western blot analysis showed increased expression levels
of Ki-67, MMP-2 and MMP-9, specific markers of cell proliferation
and invasion, in the U87 and U251 cells following miR-216b
deregulation (Fig. 2F). These
results demonstrate that miR-216b inhibits cell proliferation and
invasion of glioma cells.
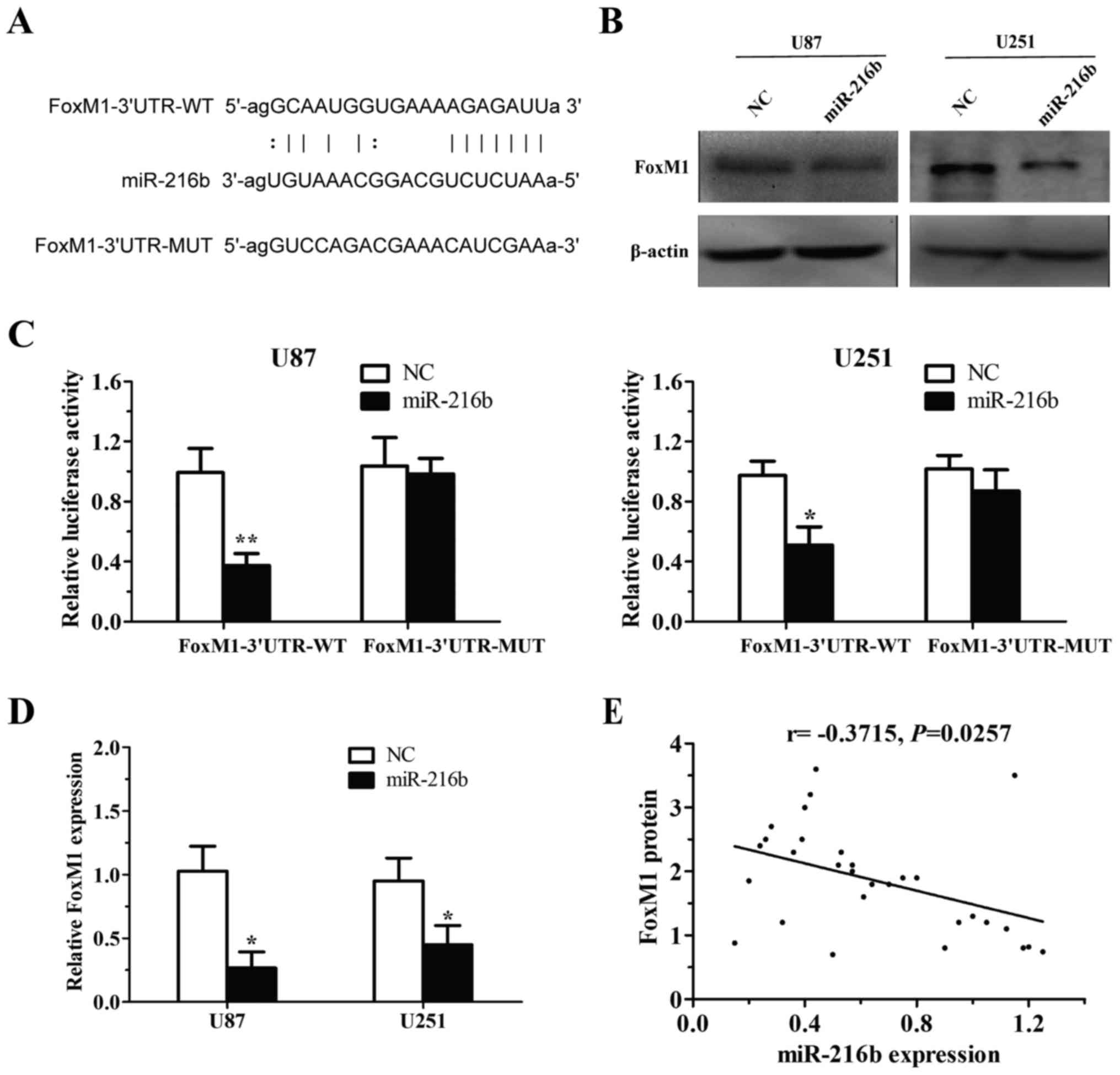
FoxM1 is a direct target of miR-216b
in glioma cells
To identify the mechanisms by which miR-216b
suppresses glioma cell growth and invasion, we searched the Sanger
microRNA database. Based on algorithm prediction, we observed that
the ‘seed sequence’ of miR-216b matched the 3′UTR of FoxM1 mRNA
(Fig. 3A). To test whether FoxM1 is
regulated by miR-216b, we performed western blot and RT-qPCR
analyses in U87 and U251 cells, and found that endogenous FoxM1
expression was decreased in accordance with miR-216b upregulation
(Fig. 3B and D). To determine
whether FoxM1 is a direct target of miR-216b, we then constructed
FoxM1-wild and FoxM1-mut luciferase reporter vectors. Then, we
co-transfected the miR-216b overexpression plasmid with FoxM1-wild
or FoxM1-mut constructs into the U87 and U251 cells. The assays
showed that the luciferase activity of wild-type 3′UTR of FoxM1
significantly decreased in cells tansfected with miR-216b, and was
recovered by FoxM1-mut (Fig. 3C).
Additionally, a significant negative correlation between miR-216b
expression and FoxM1 protein expression was observed in glioma
tissues (r=−0.3715, P=0.0257; Fig.
3E). Taken together, we conclude that miR-216b directly
modulates FoxM1 expression via binding the 3′UTR of FoxM1.
Expression of FoxM1 overrides the
effects of miR-216b in inhibiting glioma cell proliferation,
migration and invasion
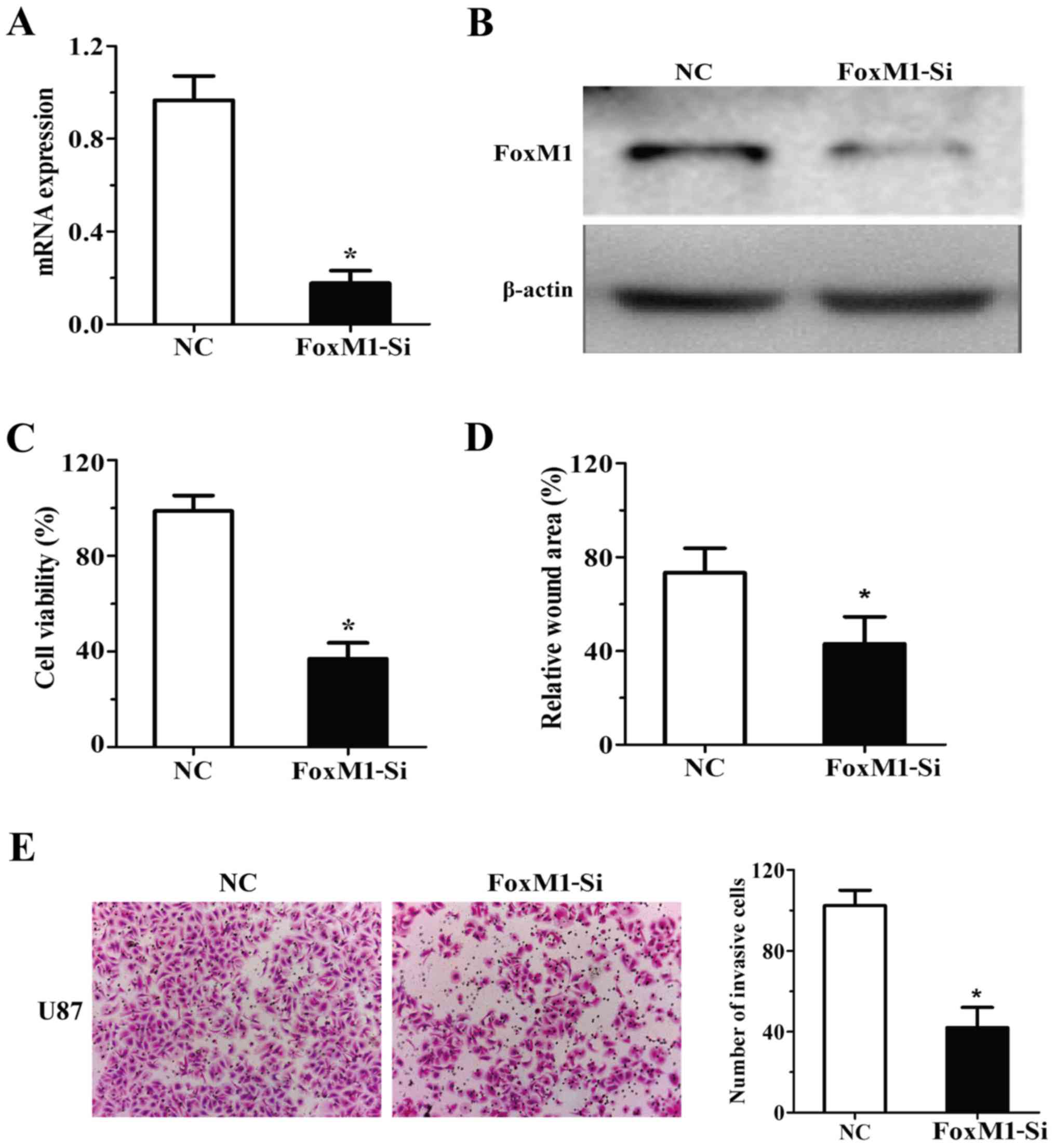
To determine whether FoxM1 is involved in the
antitumour effects of miR-216b, the expression of FoxM1 was
silenced by RNAi in the U87 cells (Fig.
4A and B). As shown in Fig.
4C-E, specific knockdown of FoxM1 by RNAi suppressed the
ability of proliferaion, migration and invasion in the U87
cells.
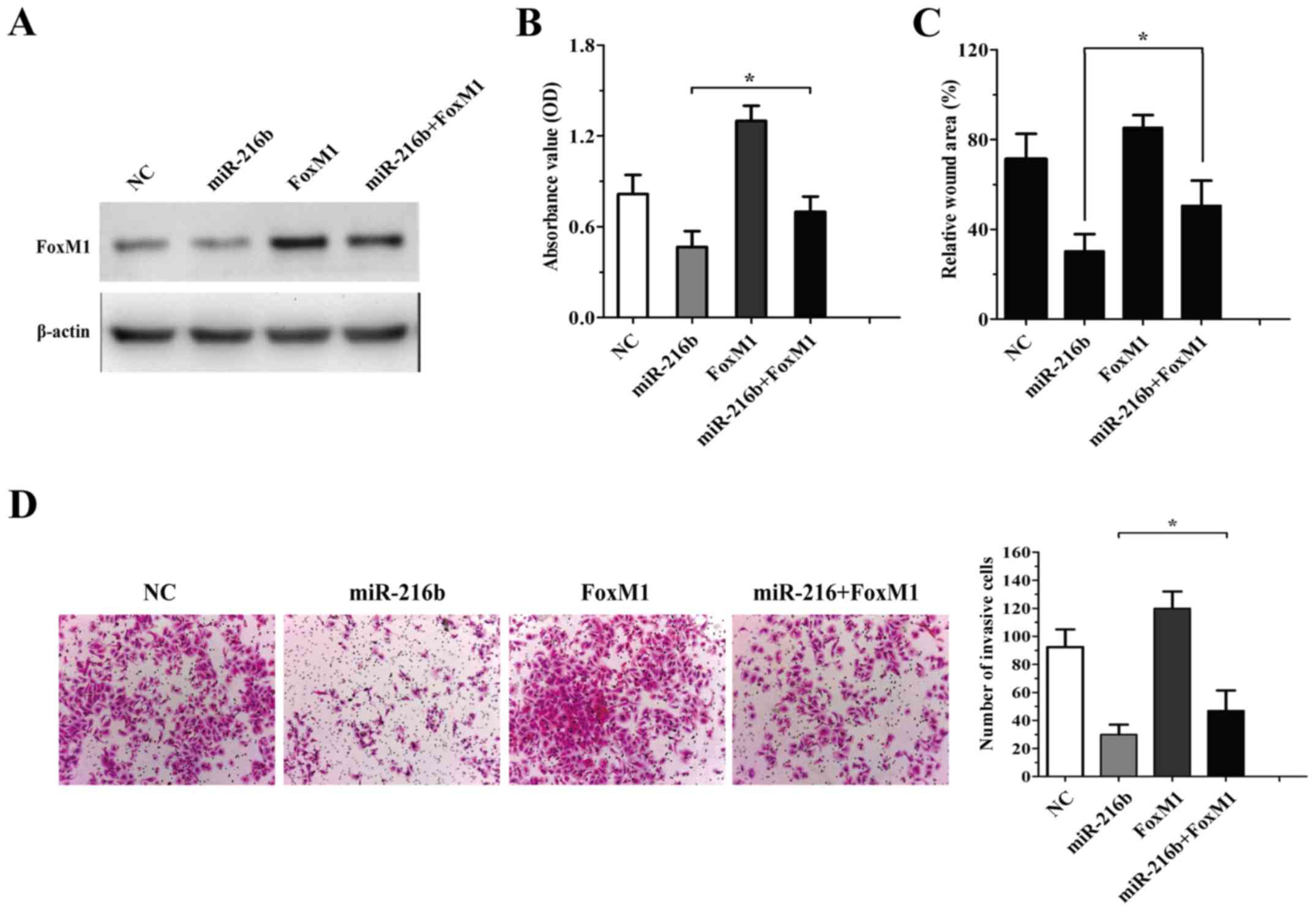
To define the importance of FoxM1 in
miR-216b-mediated cell survival and invasion, we constructed a
FoxM1-expression plasmid lacking the 3′UTR region. Rescue
experiments showed that co-transfection of FoxM1 lacking 3′UTR
significantly abolished miR-216b-induced inhibition of glioma cell
growth (Fig. 5B). U87 glioma cells
were transfected with NC, FxoM1, miR-216b mimics or miR-216b plus
FoxM1, and the expression of FoxM1 was detected by western blot
assay (Fig. 5A). Meanwhile,
miR-216b-mediated inhibition of cell migration and invasion was
impeded by FoxM1 restoration in the U87 glioma cells transfected
with miR-216b and FoxM1 (Fig. 5C and
D). These results indicate that the effects of miR-216b on
glioma cell biology were partially mediated by FoxM1.
miR-216b impairs glioma tumourigenesis
in vivo
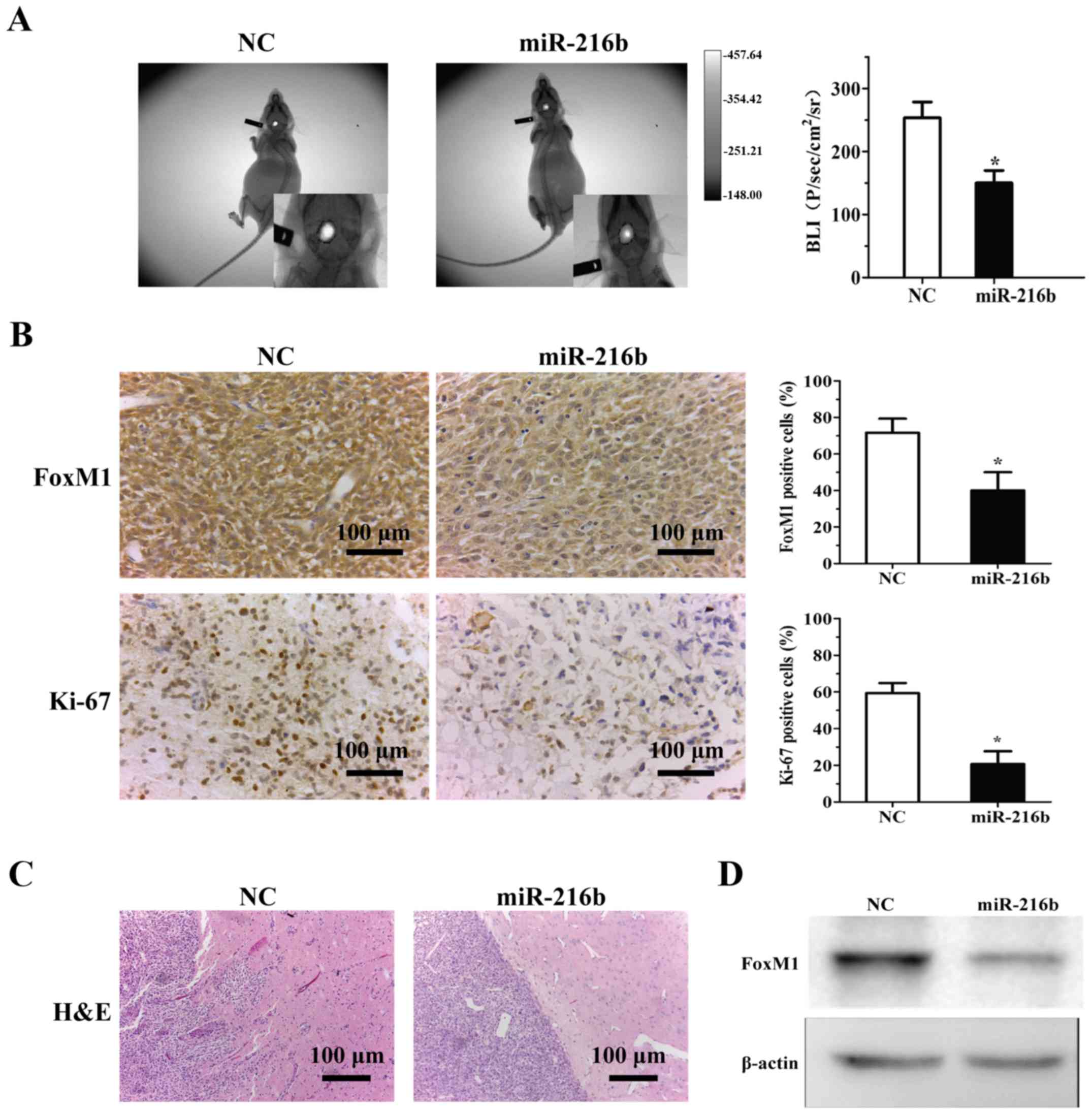
To further confirm the biological role of miR-216b
in vivo, an intracranial xenograft tumour model was
implanted intracranially into BALB/c nude mice. At the end of the
experiment (40 days), compared to the NC sequence-transfected
group, miR-216b significantly decreased tumour growth (Fig. 6A). Immunohistochemical analysis with
Ki-67 staining showed that the percentage of Ki-67-positive cells
was ~50% lower in the miR-216b tumours than that in the control
group (Fig. 6B). Western blot and
IHC analyses confirmed the downregulation of FoxM1 in the
miR-216b-transfected tumours in vivo (Fig. 6B-D). These results reveal that
miR-216b impedes xenograft glioma growth.
Discussion
miRNAs have been identified to play a vital role in
tumourigenesis since they can participate in many biological
processes, such as cell proliferation, differentiation and invasion
in a variety of cancer types. The loss of miR-216b expression has
been reported in liver and breast cancer, pancreatic ductal
adenocarcinoma, nasopharyngeal carcinoma and colorectal cancer
(19–23). It was reported that miR-216b
inhibited cell proliferation, metastasis and invasion by regulating
PKC-α and KRAS in nasopharyngeal carcinoma (24), and HBx-miR-216b-IGF2BP2 signaling
pathway in hepatocellular carcinoma (22). These results recognized miR-216b as
a new tumour regulatory molecule.
However, the mechanism involved in the regulation of
the growth of glioma by miR-216b remains unknown. In the present
study, we first detected the downregulation of miR-216b in glioma
tissues and cells and then confirmed the inhibitory effect on
proliferation and invasion by miR-216b in glioma U87 and U251 cells
by in vitro assays. These results provide us elementary
evidence that miR-216b may exert a tumour-suppressor function in
glioma.
FoxM1 is a protein involved in proliferation and
cell cycle as a proto-oncogene (16), which is overexpressed in many human
cancers, including glial tumours (25). FoxM1 belongs to the Forkhead Box
superfamily of transcription factors, which regulates cell
proliferation, differentiation, apoptosis and invasion (26). Upregulated FoxM1 expression is found
in several types of cancers and plays a key role in G1/S and G2/M
transition as a cell cycle-regulatory protein (27). Recently a study showed that FoxM1
regulates glucose metabolism in epithelial ovarian cells by
targeting GLUT1 and HK2 transcription (28). In renal cell cancer cells, FoxM1 was
found to regulate the cell cycle by activating PLK1 (29), and promoted colorectal cancer cell
invasion and migration by regulating HSPA5 trans-activation
(30).
MMP-2 plays a vital role in mesenchymal phenotypes
in glioblastoma. Previous research has demonstrated that recurrent
glioblastoma exhibits a mesenchymal subtype and the levels of MMP-2
are directly correlated with mesenchymal transition (31). TGF-β-dependent signaling is one of
the critical signaling pathways in the process of EMT (32). FoxM1 was found to regulate TGF-β
signaling by interacting with Smad3 in glioblastoma and MEF cells
(34,35). Overexpression of FoxM1 promoted EMT
and metastasis of HCC by targeting SNAI1, which plays a critical
role in FoxM1-mediated EMT (33).
The high FoxM1 expression and low E-cadherin expression in gastric
cancer tissues play a critical role in the development and
progression of gastric cancer (34). WNT/β-catenin and transforming growth
factor (TGF-β)/SMAD signaling pathways which act downstream of
FOXM1 targets, drive cancer progression by inducing EMT (35). FoxM1 overexpression is responsible
for the acquisition of EMT and the CSC phenotype, which is in part
regulated by miR-200b (36). These
results indicate that miR-216b may play a vital role in tumour cell
migration and the process of EMT.
Based on the results of bioinformatic analysis, we
detected and verified that FoxM1 is a direct target gene of
miR-216b in human glioma cells. In the present study, we found that
overexpression of miR-216b reduced glioma cell proliferation and
invasion. Thus, we conclude that miR-216b may have a role in the
treatment of human glioma. Additionally forced expression of FoxM1
lacking the 3′UTR impeded the inhibition of proliferation and
invasion by miR-216b overexpression, which indicates that FoxM1 is
a potential target of miR-216b in human glioma. These data
implicate that miR-216b has a potential role in the treatment of
human glioma.
In conclusion, the present study demonstrated that
miR-216b is downregulated in glioma and overexpression of miR-216b
suppressed the proliferation, migration and invasion abilities by
directly targeting FoxM1. These results reveal that re-expression
of miR-216b may be a promising therapeutic target for glioma.
References
|
1
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
Classification of Tumors of the Central Nervous System: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Geiger GA, Fu W and Kao GD:
Temozolomide-mediated radiosensitization of human glioma cells in a
zebrafish embryonic system. Cancer Res. 68:3396–3404. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zheng Y, Lin L and Zheng Z: TGF-alpha
induces upregulation and nuclear translocation of Hes1 in glioma
cell. Cell Biochem Funct. 26:692–700. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zheng H, Ying H, Wiedemeyer R, Yan H,
Quayle SN, Ivanova EV, Paik JH, Zhang H, Xiao Y, Perry SR, et al:
PLAGL2 regulates Wnt signaling to impede differentiation in neural
stem cells and gliomas. Cancer Cell. 17:497–509. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
de Moor CH, Meijer H and Lissenden S:
Mechanisms of translational control by the 3′ UTR in development
and differentiation. Semin Cell Dev Biol. 16:49–58. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Berezikov E, Guryev V, van de Belt J,
Wienholds E, Plasterk RH and Cuppen E: Phylogenetic shadowing and
computational identification of human microRNA genes. Cell.
120:21–24. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pichler M and Calin GA: MicroRNAs in
cancer: From developmental genes in worms to their clinical
application in patients. Br J Cancer. 113:569–573. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin Z, Zhao J, Wang X, Zhu X and Gong L:
Overexpression of microRNA-497 suppresses cell proliferation and
induces apoptosis through targeting paired box 2 in human ovarian
cancer. Oncol Rep. 36:2101–2107. 2016.PubMed/NCBI
|
|
10
|
Quintavalle C, Garofalo M, Zanca C, Romano
G, Iaboni M, De del Basso Caro M, Martinez-Montero JC, Incoronato
M, Nuovo G, Croce CM, et al: miR-221/222 overexpession in human
glioblastoma increases invasiveness by targeting the protein
phosphate PTPμ. Oncogene. 31:858–868. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xie YK, Huo SF, Zhang G, Zhang F, Lian ZP,
Tang XL and Jin C: CDA-2 induces cell differentiation through
suppressing Twist/SLUG signaling via miR-124 in glioma. J
Neurooncol. 110:179–186. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen L, Li H, Han L, Zhang K, Wang G, Wang
Y, Liu Y, Zheng Y, Jiang T, Pu P, et al: Expression and function of
miR-27b in human glioma. Oncol Rep. 26:1617–1621. 2011.PubMed/NCBI
|
|
13
|
Zheng WW, Zhou J, Zhang CH and Liu XS:
MicroRNA-216b is downregulated in hepatocellular carcinoma and
inhibits HepG2 cell growth by targeting Forkhead box protein M1.
Eur Rev Med Pharmacol Sci. 20:2541–2550. 2016.PubMed/NCBI
|
|
14
|
Korver W, Schilham MW, Moerer P, van den
Hoff MJ, Dam K, Lamers WH, Medema RH and Clevers H: Uncoupling of S
phase and mitosis in cardiomyocytes and hepatocytes lacking the
winged-helix transcription factor Trident. Curr Biol. 8:1327–1330.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu M, Dai B, Kang SH, Ban K, Huang FJ,
Lang FF, Aldape KD, Xie TX, Pelloski CE, Xie K, et al: FoxM1B is
overexpressed in human glioblastomas and critically regulates the
tumorigenicity of glioma cells. Cancer Res. 66:3593–3602. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Myatt SS and Lam EW: The emerging roles of
forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 7:847–859.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Scholzen T and Gerdes J: The Ki-67
protein: From the known and the unknown. J Cell Physiol.
182:311–322. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mook OR, Frederiks WM and Van Noorden CJ:
The role of gelatinases in colorectal cancer progression and
metastasis. Biochim Biophys Acta. 1705:69–89. 2004.PubMed/NCBI
|
|
19
|
Kim SY, Lee YH and Bae YS: MiR-186,
miR-216b, miR-337-3p, and miR-760 cooperatively induce cellular
senescence by targeting α subunit of protein kinase CKII in human
colorectal cancer cells. Biochem Biophys Res Commun. 429:173–179.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Deng M, Tang H, Zhou Y, Zhou M, Xiong W,
Zheng Y, Ye Q, Zeng X, Liao Q, Guo X, et al: miR-216b suppresses
tumor growth and invasion by targeting KRAS in nasopharyngeal
carcinoma. J Cell Sci. 124:2997–3005. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zheng L, Zhang X, Yang F, Zhu J, Zhou P,
Yu F, Hou L, Xiao L, He Q and Wang B: Regulation of the P2X7R by
microRNA-216b in human breast cancer. Biochem Biophys Res Commun.
452:197–204. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu FY, Zhou SJ, Deng YL, Zhang ZY, Zhang
EL, Wu ZB, Huang ZY and Chen XP: MiR-216b is involved in
pathogenesis and progression of hepatocellular carcinoma through
HBx-miR-216b-IGF2BP2 signaling pathway. Cell Death Dis.
6:e16702015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Egeli U, Tezcan G, Cecener G, Tunca B,
Sevinc E Demirdogen, Kaya E, Ak S, Dundar HZ, Sarkut P, Ugras N, et
al: miR-216b targets FGFR1 and confers sensitivity to radiotherapy
in pancreatic ductal adenocarcinoma patients without EGFR or KRAS
mutation. Pancreas. 45:1294–1302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Deng M, Liu JF, Gu YX, Zheng GP and He ZM:
miR-216b suppresses cell proliferation and invasion by targeting
PKCα in nasopharyngeal carcinoma cells. Zhonghua Zhong Liu Za Zhi.
35:645–650. 2013.(In Chinese). PubMed/NCBI
|
|
25
|
Zhang N, Wei P, Gong A, Chiu WT, Lee HT,
Colman H, Huang H, Xue J, Liu M, Wang Y, et al: FoxM1 promotes
β-catenin nuclear localization and controls Wnt target-gene
expression and glioma tumorigenesis. Cancer Cell. 20:427–442. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Koo CY, Muir KW and Lam EW: FOXM1: From
cancer initiation to progression and treatment. Biochim Biophys
Acta. 1819:28–37. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu GY, Shi BZ and Li Y: FoxM1 regulates
Sirt1 expression in glioma cells. Eur Rev Med Pharmacol Sci.
18:205–211. 2014.PubMed/NCBI
|
|
28
|
Raychaudhuri P and Park HJ: FoxM1: A
master regulator of tumor metastasis. Cancer Res. 71:4329–4333.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang Z, Zhang G and Kong C: FOXM1
participates in PLK1-regulated cell cycle progression in renal cell
cancer cells. Oncol Lett. 11:2685–2691. 2016.PubMed/NCBI
|
|
30
|
Luo X, Yao J, Nie P, Yang Z, Feng H, Chen
P, Shi X and Zou Z: FOXM1 promotes invasion and migration of
colorectal cancer cells partially dependent on HSPA5
transactivation. Oncotarget. 7:26480–26495. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mahabir R, Tanino M, Elmansuri A, Wang L,
Kimura T, Itoh T, Ohba Y, Nishihara H, Shirato H, Tsuda M, et al:
Sustained elevation of Snail promotes glial-mesenchymal transition
after irradiation in malignant glioma. Neuro Oncol. 16:671–685.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xue J, Lin X, Chiu WT, Chen YH, Yu G, Liu
M, Feng XH, Sawaya R, Medema RH, Hung MC, et al: Sustained
activation of SMAD3/SMAD4 by FOXM1 promotes TGF-β-dependent cancer
metastasis. J Clin Invest. 124:564–579. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Meng FD, Wei JC, Qu K, Wang ZX, Wu QF, Tai
MH, Liu HC, Zhang RY and Liu C: FoxM1 overexpression promotes
epithelial-mesenchymal transition and metastasis of hepatocellular
carcinoma. World J Gastroenterol. 21:196–213. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang J, Chen XY, Huang KJ, Wu WD, Jiang
T, Cao J, Zhou LS, Qiu ZJ and Huang C: Expression of FoxM1 and the
EMT-associated protein E-cadherin in gastric cancer and its
clinical significance. Oncol Lett. 12:2445–2450. 2016.PubMed/NCBI
|
|
35
|
Chiu WT, Huang YF, Tsai HY, Chen CC, Chang
CH, Huang SC, Hsu KF and Chou CY: FOXM1 confers to
epithelial-mesenchymal transition, stemness and chemoresistance in
epithelial ovarian carcinoma cells. Oncotarget. 6:2349–2365. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bao B, Wang Z, Ali S, Kong D, Banerjee S,
Ahmad A, Li Y, Azmi AS, Miele L and Sarkar FH: Over-expression of
FoxM1 leads to epithelial-mesenchymal transition and cancer stem
cell phenotype in pancreatic cancer cells. J Cell Biochem.
112:2296–2306. 2011. View Article : Google Scholar : PubMed/NCBI
|