Introduction
Lung cancer, the most frequently diagnosed cancer
and the leading cause of tumor death, was estimated to account for
nearly 1.6 million deaths and more than 1.8 million new cases
worldwide in 2012 (1), a sharp rise
from 2008 (2). Approximately 80% of
all lung cancer cases are non-small cell lung cancer (NSCLC), and
lung adenocarcinoma (LUAD) is a major pathologic subtype of NSCLC,
constituting approximately 40% of diagnosed lung cancers (3). The 5-year survival rate of LUAD has
not improved significantly over the past few decades despite great
research efforts. Therefore, our focus was improving the
early-stage diagnosis of LUAD, specifically identifying and
validating biomarkers for early diagnosis and prognosis.
MicroRNAs (miRNAs) are single-stranded non-coding
RNA molecules (~20 nucleotides long) that regulate gene expression
at the post-transcriptional level by transcriptional suppression
(4–6). miRNAs are frequently dysregulated in
various cancers, including LUAD (7), hepatocellular carcinoma (8), gastric (9) and esophageal cancer (10), and may play roles as both oncogenes
and antioncogenes (5,11). miRNAs is a major regulatory tool for
the epigenome and is also associated with phenotypic consequences
of diseases (12). Therefore,
identification of LUAD-related miRNAs may contribute to early
diagnosis and prognosis.
Studies of LUAD have reported aberrantly expressed
miRNAs that regulate initiation, development and metastasis
(13–15). However, small studies do not have
sufficient power to relate dysregulated miRNAs to clinical
features. Recently, Xie et al (16) analyzed 1341 NSCLCs in The Cancer
Genome Atlas (TCGA) (http://cancergenome.nih.gov/) and found that genetic
variants in regulatory regions of miRNAs might be related to lung
cancer risk and survival. However, there are few studies involving
both large-scale samples and microarray detection, and the
relationships between tumor-specific key miRNAs and clinical
features remain uncertain. The TCGA database is a large-scale
public data platform from which sequencing data on miRNA and
detailed clinical features of LUAD can be downloaded (17,18).
To improve the reliability and accuracy of the present study,
miRNAs in LUAD were identified using the TCGA dataset.
miRNA sequencing data from 463 LUAD samples and 42
adjacent non-tumor lung tissue samples were collected from the TCGA
database. This is the first study that has grouped tumor-specific
micRNA profiles from the TCGA with pivotal clinical features (TNM
stage and lymphatic metastases) in LUAD. Quantitational real-time
polymerase chain reaction (qRT-PCR) was used to verify the accuracy
of miRNA results in LUAD tumor tissues and adjacent non-tumor lung
tissues. The biological function of the LUAD-specific key miRNA
miR-30a-3p was further analyzed. The present study was
designed to help find potential biomarkers for diagnosis and
prognosis based on detailed tumor-specific miRNA expression
profiles in LUAD.
Materials and methods
Patients and samples
The LUAD and adjacent non-tumor tissue miRNA
sequencing data and related clinical information for 521 LUAD
patients were obtained from the TCGA Data Portal (as of March
2016). Patient exclusion criteria were as follows: i) first
histologic diagnosis was not LUAD; ii) presence of another
malignancy besides LUAD; iii) incomplete data for analysis; and iv)
overall survival more than five years. A total of 463 LUAD patients
were included in this study. miRNA expression profiles for normal
lung tissue samples were obtained from adjacent non-tumor lung
tissues (n=42). According to the staging system of the Union for
International Cancer Control (UICC), 359 cases were well and
moderately differentiated LUAD (stage I–II) and 104 were poorly
differentiated LUAD (stage III–IV). In addition, among these 463
LUAD patients were 170 patients with lymphatic metastasis and 295
with non-lymphatic metastasis. The present study meets the
publication guidelines of the TCGA.
In addition, 53 LUAD tissue specimens (tumor tissues
and paired adjacent non-cancerous tissues) that we collected from
Chinese Han population were obtained from the Nanjing Chest
Hospital Medical School of Southeast University. Tissues were
frozen with RNAlater (Ambion, Foster City, CA, USA) and stored at
−80°C immediately after surgical resection until further analysis.
Samples were collected with a detailed paper pathology report and a
quality assessment report verifying collection of tumor and/or
adjacent non-tumor lung tissues. Informed consent forms were
obtained from all the patients. The present study was approved by
the ethics committee of the Zhongda Hospital Southeast
University.
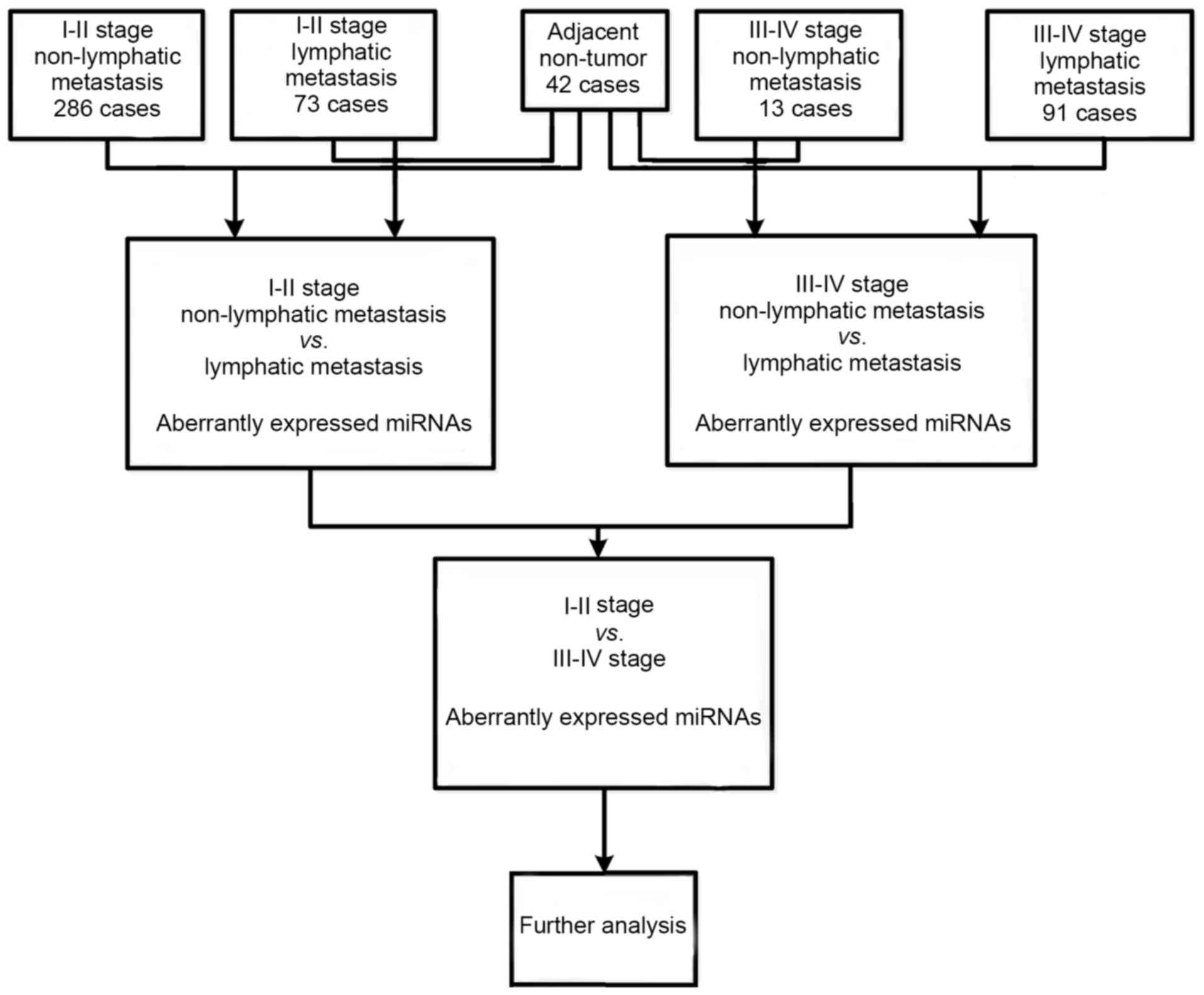
Identification of abnormally expressed
intersection miRNAs in LUAD
To identify miRNAs abnormally expressed in LUAD
compared with normal lung tissues, the ‘Level 3’ raw counts of
miRNA expression from the TCGA database were analyzed using
Illumina HiSeq Systems (Illumina, Inc., Hayward, CA, USA), 463 LUAD
samples and 42 normal controls were not further normalized, because
these data were already normalized by TCGA. Then, abnormally
expressed miRNAs were compared in Level 3, including LUAD tumor
tissues vs. adjacent non-tumor lung tissues, lymphatic vs.
non-lymph node metastasis in LUAD patients and stage I–II vs. stage
III–IV. Finally, intersection miRNAs were selected for further
analysis. A flow chart for bioinformatics analysis is presented in
Fig. 1.
The correlations between LUAD-specific
intersection miRNAs, clinical features and overall survival
According to the comparative analysis of LUAD miRNA
sequencing data in TCGA, LUAD-specific intersection miRNAs were
selected. The correlations between LUAD-specific intersection
miRNAs and clinical features, including race, sex, age, TNM stage,
lymphatic metastasis and patient outcome were further analyzed.
Subsequently, to correlate specific intersection miRNAs with
patient prognostic characteristics, the univariate Cox proportional
hazards regression model was used to analyze the association
between specific intersection miRNAs and LUAD patient survival. The
Kaplan-Meier and log-rank methods (Mantel-Haenszel test) were used
to test the equality of survival distributions in different groups
subjected to comparison (19).
Hazard ratios (HRs) for a 2-fold change in gene expression level
from univariate Cox regression analysis were used to identify
LUAD-specific intersection miRNAs associated with overall survival
(20). miRNAs defined as having a
protective signature showed HR<1 and those defined as high-risk
had HR for death >1.
Total RNA extraction and qRT-PCR
verification
We randomly selected 5 specific key miRNAs in the
above bioinformatics analysis and measured their actual expression
levels in 53 diagnosed LUAD patients tumor tissues and adjacent
non-tumor tissues by qRT-PCR. U6 was used as an internal normalized
reference to confirm reliability and validity. Reverse
transcription reactions using the ReverTra Ace® qPCR RT
kit (FSQ-101; Toyobo, Shanghai, China) was conducted in two steps
according to the manufacturer's protocol. First, the mixture
containing 1 µg of RNA samples was incubated in a 96-well plate for
5 min at 65°C and held at 4°C. Then, the 9 µl mixture, which
comprised 2 µl 5X RT buffer, 0.5 µl RT Enzyme Mix, 0.5 µl
miRNA-specific stem-loop RT primers (Guangzhou RiboBio, Co., Ltd.,
Guangzhou, China), and 6 µl ddH2O, was incubated in a
96-well plate at 37°C for 15 min, 98°C for 5 min and subsequently
held at 4°C.
Real-time PCR was performed to detect the expression
level of the candidate miRNAs with the StepOne Plus™PCR System
(Applied Biosystems, Foster City, CA, USA). QRT-PCR was then
performed using Thunderbird™ SYBR® qPCR Mix (QPS-201;
Toyobo) according to the manufacturer's protocol. The PCR reaction
components were 2 µl of cDNA, 10 µl of Thunderbird SYBR®
qPCR Mix, 0.6 µl (1 µl/pmol) PCR primers (ribobio), and 13.2 µl
RNase-free water. The reaction was performed at 95°C for 1 min,
followed by 45 cycles of 95°C for 15 sec, 60°C for 30 sec and 72°C
for 30 sec. A dissociation curve was analyzed from 60 to 95°C. The
Ct-value for each sample was calculated with the ∆∆Ct method
(21), and fold change results were
presented as 2−∆∆Ct, where ∆∆Ct = (CtmiRNAs -
CtU6)tumor - (CtmiRNAs -
CtU6)adjacent non-tumor tissues.
Functional enrichment analysis
DIANA-mirPath software (http://diana.cslab.ece.ntua.gr/pathways/) was applied
to perform online gene enrichment analysis of putative targets of
25 LUAD-specific key miRNAs, comparing each set of miRNA targets to
all known Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways.
Then LUAD-specific key miRNAs and signaling pathways were evaluated
using the negative logarithm of P-value (−lg P-value), in which the
value was proportional to relevance. Furthermore, interaction and
reaction networks between genes were analyzed according to the
pathway data available from the KEGG database (http://www.Genome.jp/kegg/pathway.html).
Cell culture and detection of the
expression of miR-30a-3p in lung cell lines
HBE cells and human LUAD cells (A549) were purchased
from the Chinese Academy of Sciences Type Culture collection Cell
Library Committee (Shanghai, China). HBE cells and A549 cells were
separately cultured in Dulbeccos modified Eagles medium (DMEM) and
1640 cell-culture medium (HyClone Laboratories, Inc., Logan, UT,
USA), supplemented with 10% fetal bovine serum (FBS; Gibco,
Carlsbad, CA, USA), and 100 U/ml penicillin and 100 mg/ml
streptomycin (HyClone Laboratories), in a humidified 5%
CO2 incubator at 37°C. The expression status of
miR-30a-3p was further confirmed by qRT-PCR before they were
used in experiments.
Transfection of miR-30a-3p mimic
Transfections were performed using riboFect™ CP
Transfection kit (Guangzhou RiboBio) according to the manufacturers
instructions. A549 cells were trypsinized during logarithmic growth
period and resuspended in normal growth medium at 1×105
cells/ml. Cells (2×105/well) were seeded in a 6-well
plate with 1640 cell-culture medium the day before transfection,
then transfected with 5 µl miR-30a-3p mimic (50 nM; Guangzhou
RiboBio)/mimic negative control (Guangzhou RiboBio). The
transfection complexes were dispensed into wells and incubated at
37°C for 24 h. After transfection, qRT-PCR was used to test the
expression of miR-30a-3p transfected cells and confirm
transfection efficiency.
Cell proliferation detection
The MTT [3-(4, 5-dimethyl)-2, 5-diphenyl-2H] assay
was used to assess cell proliferation. Five thousand A549 cells
were seeded into each well of a 96-well plate. Cells were
transfected in triplicate repeated wells with 50 nM
miR-30a-3p mimic and mimic negative control and then
cultured in complete culture medium for 24 h. The culture
supernatants were removed, and 200 µl MTT solution (0.5 mg/ml) was
added into each well and incubated for 4 h at 37°C. Then
supernatants were removed, and 150 µl dimethyl sulphoxide (DMSO)
solution was added into each well and vibrated to dissolve formazan
crystals. Optical density (OD) value was measured at 570 nm in a
microtiter plate reader.
Flow cytometric analysis
A549 cells (n=2×105) were seeded into
each well of a 6-well plate. Cells were transfected in sextuplicate
repeated wells with 50 nM miR-30a-3p mimic and mimic
negative control, and then were cultured in complete culture medium
for 48 h. Cells were harvested by trypsinization in a tube and were
washed twice in ice-cold phosphate-buffered saline (PBS). After
staining with fluorescein isothiocyanate (FITC)-Annexin V and
propidium iodide (PI) using the FITC Annexin V/PI apoptosis
detection kit (Becton-Dickinson, Franklin Lakes, NJ, USA) according
to the manufacturers protocol. The cells were analyzed for
apoptosis using a flow cytometer (FACScan; Becton-Dickinson).
Apoptosis assessment by Hoechst 33258
staining
A549 cells were placed in 24-well plates
(5×104/well) and transfected with 50 nM
miR-30a-3p mimic and mimic negative control, and then
cultured in complete culture medium for 48 h. After incubation,
cells were washed in PBS and incubated with the DNA dye Hoechst
33258 according to the manufacturer's protocol. The results were
visualized under a fluorescent microscope with excitation at a
wavelength of 350 nm and measured at 460 nm. Each test was carried
out at least three times.
Western blot assays
After homogenization of the transfected cells,
protein concentration was measured using the BCA protein assay
(Generay Biotech, Co., Ltd., Shanghai, China). Protein samples were
separated using 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis and transferred onto nitrocellulose membranes.
After membranes were blocked in 10% BSA for 2 h, immunoblotting was
conducted by incubation with the primary antibodies overnight at
4°C. In the present study, GAPDH (D16H11) XP® Rabbit mAb
(37 kDa) (22) and AKT3 (E1Z3W)
Rabbit mAb (60 kDa) (23)
antibodies were purchased from Cell Signaling (Danvers, MA, USA).
After incubation with HRP-conjugated goat anti-rabbit IgG (D110058;
Sangon Biotech Shanghai, Co., Ltd., Shanghai, China) cells were
incubated with secondary antibodies for 1 h at room temperature.
Immunoreactive bands were visualized using a chemiluminescent
substrate (Millipore, Darmstadt Germany) and analyzed with an
automatic chemical luminescence/fluorescence image analysis system
called the Gel Imaging System (Tanon Science & Technology, Co.,
Ltd, Shanghai, China).
Statistical analysis
The data are shown as mean ± SD and statistical
comparisons were made using the Students t-test. The significance
level was set as 0.05 as default to control the false discovery
rate (FDR). Values of P<0.05 were considered statistically
significant. Statistical analyses were performed using SPSS 19.0.
Operating characteristic curve (ROC) was used to determine the
specific key miRNAs as the sensitivity and specificity of detection
of LUAD.
Result
Identification of abnormally expressed
miRNAs in LUAD patients
In the present study, a total of 1030 miRNAs were
identified from TCGA ‘Level 3’ LUAD RNA-Sequencing data. Then 118
LUAD-associated abnormally expressed miRNAs were identified between
463 LUAD tumor tissue samples and 42 adjacent non-tumor tissues.
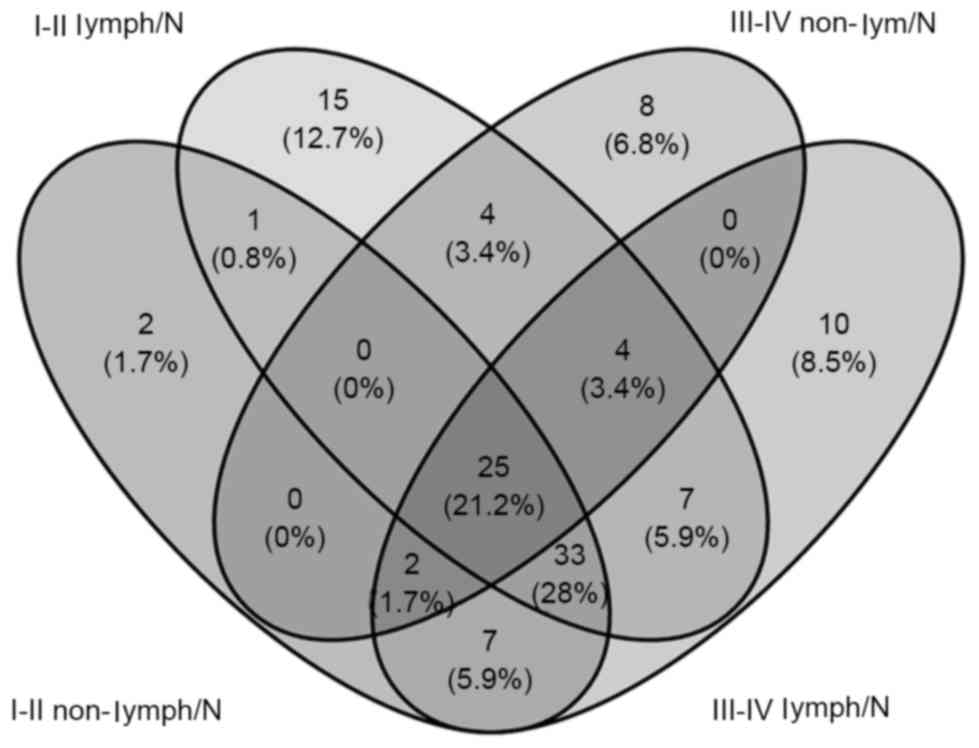
Furthermore, we compared these 118 miRNAs between tumor stage and
lymphatic metastasis. Seventy aberrantly expressed miRNAs were
selected from comparisons of stage I–II (non-lymphatic metastasis)
LUAD patient tissues and adjacent non-tumor lung tissues, 43 from
comparisons of stage I–II (lymphatic metastasis) with adjacent
non-tumor lung tissues, 89 from comparisons of stage III–IV
(non-lymphatic metastasis) and adjacent non-tumor tissues and 88
from comparisons of stage III–IV (lymphatic metastasis) and
adjacent non-tumor tissues. To further confirm data reliability, we
selected 25 aberrantly expressed miRNAs (12 downregulated and 13
upregulated) from the intersection of the above four groups
(Fig. 2 and Table I).
 | Table I.Abnormal expression of intersection
miRNAs in LUAD. |
Table I.
Abnormal expression of intersection
miRNAs in LUAD.
| miRNA | Accession | Regulation |
miRNA_Sequencing | Fold change | -log(p) | -log(FDR) |
|---|
|
hsa-miR-30a-3p | MIMAT0000088 | Down |
CUUUCAGUCGGAUGUUUGCAGC | −5.738±0.425 | 7.000 | 6.082 |
|
hsa-miR-139-5p | MIMAT0000250 | Down |
UCUACAGUGCACGUGUCUCCAGU | −4.356±0.219 | 3.615 | 1.930 |
|
hsa-miR-133a-3p | MIMAT0000427 | Down |
UUUGGUCCCCUUCAACCAGCUG | −7.1591±2.015 | 4.447 | 2.652 |
|
hsa-miR-378a-3p | MIMAT0000732 | Down |
ACUGGACUUGGAGUCAGAAGGC | −4.789±0.418 | 5.021 | 3.194 |
|
hsa-miR-451a | MIMAT0001631 | Down |
AAACCGUUACCAUUACUGAGUU | −6.3753±1.660 | 3.609 | 1.926 |
|
hsa-miR-486-5p | MIMAT0002177 | Down |
UCCUGUACUGAGCUGCCCCGAG | −19.740±3.61 | 7.000 | 5.397 |
|
hsa-miR-30c-2-3p | MIMAT0004550 | Down |
CUGGGAGAAGGCUGUUUACUCU | −11.1261±0.805 | 7.000 | 6.082 |
|
hsa-miR-139-3p | MIMAT0004552 | Down |
UGGAGACGCGGCCCUGUUGGAGU | −12.599±0.974 | 6.323 | 4.427 |
|
hsa-miR-221-5p | MIMAT0004568 | Down |
ACCUGGCAUACAAUGUAGAUUU | −4.1656±0.472 | 4.840 | 3.022 |
|
hsa-miR-338-5p | MIMAT0004701 | Down |
AACAAUAUCCUGGUGCUGAGUG | −4.8169±0.599 | 6.097 | 4.205 |
|
hsa-miR-378c | MIMAT0016847 | Down |
ACUGGACUUGGAGUCAGAAGAGUGG | −3.9291±0.199 | 5.854 | 3.975 |
|
hsa-miR-3614-5p | MIMAT0017992 | Down |
CCACUUGGAUCUGAAGGCUGCCC | −2.897±0.272 | 4.932 | 3.109 |
|
hsa-miR-33a-5p | MIMAT0000091 | Up |
GUGCAUUGUAGUUGCAUUGCA | 4.683±0.385 | 6.398 | 4.535 |
|
hsa-miR-96-5p | MIMAT0000095 | Up |
UUUGGCACUAGCACAUUUUUGCU | 5.375±0.271 | 7.000 | 5.449 |
|
hsa-miR-196a-5p | MIMAT0000226 | Up |
UAGGUAGUUUCAUGUUGUUGGG | 9.443±4.866 | 3.691 | 2.131 |
|
hsa-miR-182-5p | MIMAT0000259 | Up |
UUUGGCAAUGGUAGAACUCACACU | 6.415±0.526 | 4.862 | 3.131 |
|
hsa-miR-210-3p | MIMAT0000267 | Up |
CUGUGCGUGUGACAGCGGCUGA | 28.035±5.343 | 7.000 | 6.149 |
|
hsa-miR-142-3p | MIMAT0000434 | Up |
UGUAGUGUUUCCUACUUUAUGGA | 6.608±0.888 | 3.695 | 2.134 |
|
hsa-miR-9-5p | MIMAT0000441 | Up |
UCUUUGGUUAUCUAGCUGUAUGA | 20.480±3.673 | 6.347 | 4.482 |
|
hsa-miR-135b-5p | MIMAT0000758 | Up |
UAUGGCUUUUCAUUCCUAUGUGA | 4.980±0.548 | 3.699 | 2.138 |
|
hsa-miR-143-5p | MIMAT0004599 | Up |
GGUGCAGUGCUGCAUCUCUGGU | 3.143±0.604 | 7.000 | 5.449 |
|
hsa-miR-127-5p | MIMAT0004604 | Up |
CUGAAGCUCAGAGGGCUCUGAU | 3.283±0.403 | 3.918 | 2.321 |
|
hsa-miR-708-5p | MIMAT0004926 | Up |
AAGGAGCUUACAAUCUAGCUGGG | 5.228±0.396 | 6.372 | 4.506 |
|
hsa-miR-708-3p | MIMAT0004927 | Up |
CAACUAGACUGUGAGCUUCUAG | 5.993±0.250 | 4.842 | 3.113 |
|
hsa-miR-3607-3p | MIMAT0017985 | Up |
ACUGUAAACGCUUUCUGAUG | 9.643±4.630 | 5.577 | 3.760 |
The correlations between LUAD-specific
miRNAs and clinical features
The 25 LUAD-specific intersection miRNAs were
further analyzed according to their expression and clinical
features (race, sex, age, TNM stage, lymphatic metastasis and
patient outcome assessment at diagnosis in the TCGA database).
Fifteen specific miRNAs were significantly aberrantly expressed in
clinical feature comparisons (P<0.05; Table II).
 | Table II.The correlations between
LUAD-specific intersection miRNAs and clinical features. |
Table II.
The correlations between
LUAD-specific intersection miRNAs and clinical features.
| Comparisons | Downregulated | Upregulated |
|---|
| Race (White vs.
Asian) |
|
miR-96-5p |
| Gender (female vs.
male) | miR-133a-3p,
miR-139a-3p, miR-30a-3p, miR-30c-2-3p, | miR-127-5p,
miR-196a-5p |
| Age (≤60 vs. >
60 years) | miR-338-5p,
miR-378c |
|
| TNM stage (I–II vs.
III–IV) | miR-221-5p,
miR-378a-3p, miR-451a, miR-486-5p |
|
| Lymphatic
metastasis (no vs. yes) | miR-133a-3p,
miR-139-5p, miR-221-5p |
|
| Patient outcome
assessment (dead vs. alive) | miR-133a-3p,
miR-139-5p, miR-221-5p, miR-30a-3p,
miR-30c-2-3p, miR-378c, |
miR-33a-5p |
One miRNA (miR-96-5p) was aberrantly
expressed in race, 6 miRNAs (miR-133a-3p,
miR-139a-3p, miR-30a-3p, miR-30c-2-3p,
miR-127-5p and miR-196-5p) were aberrantly expressed
in sex, 2 miRNAs (miR-338-5p and miR-378c) were
aberrantly expressed in age, 4 miRNAs (miR-221-5p,
miR-378a-3p, miR-451a and miR-486-5p) were
aberrantly expressed in TNM stage, 3 miRNAs (miR-133a-3p,
miR-139-5p and miR-221-5p) were aberrantly expressed
in lymphatic metastasis and 6 miRNAs (miR-133a-3p,
miR-139-5p, miR-221-5p, miR-30a-3p,
miR-378c and miR-33a-5p) were aberrantly expressed in
patient outcome.
Survival analysis
A univariate Cox model was used to investigate the
relationship between the the 25 LUAD-specific intersection miRNAs
and overall survival. Three of these miRNAs were significantly
associated with overall survival status in 463 LUAD patients
(log-rank P<0.05): 2 (miR-378c and miR-221-5p)
negatively (P<0.05) and 1 (miR-142-3p) positively
(P<0.05) (Fig. 3).
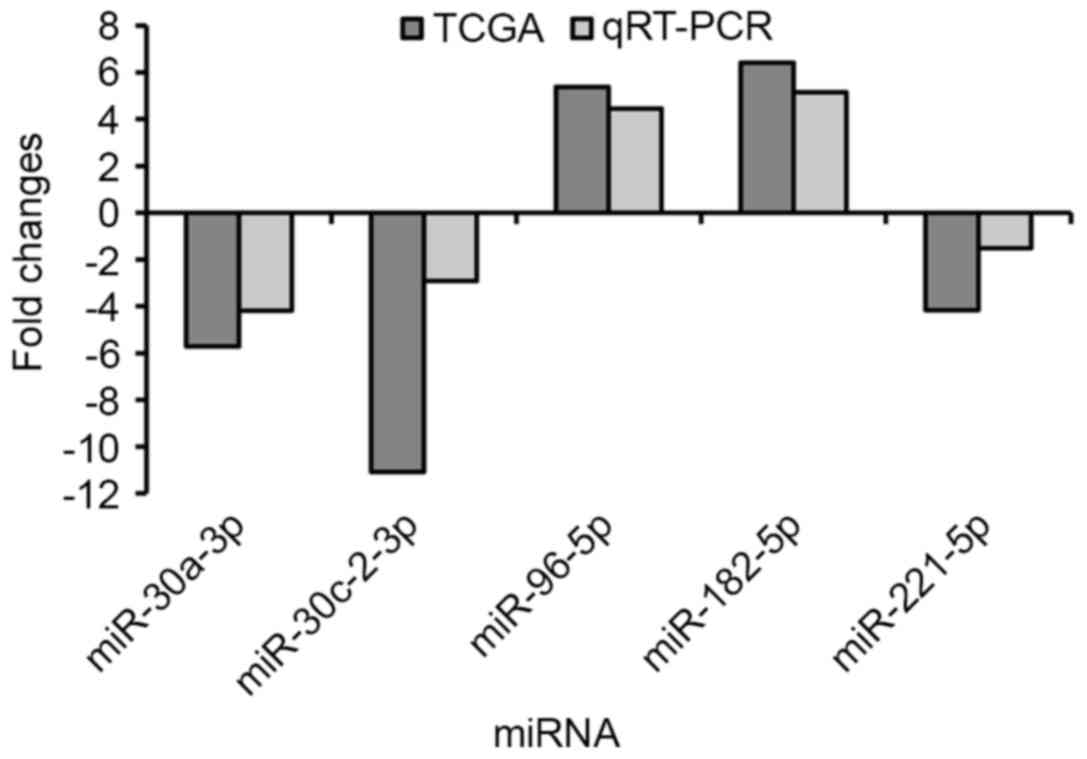
qRT-PCR verification
Finally, we randomly selected 5 specific key miRNAs
(miR-30a-3p, miR-30c-2-3p, miR-96-5p,
miR-182-5p and miR-221-5p) to validate the
reliability and validity of the results of the above miRNA
analysis. We applied the paired t-test to assess the differences
between the 53 newly diagnosed LUAD tumor tissues and the adjacent
non-tumor lung tissues. The results showed that miR-30a-3p,
miR-30c-2-3p and miR-221-5p were downregulated in
LUAD tumor tissues compared with adjacent non-tumor lung tissues,
while miR-96-5p and miR-182-5p were upregulated in
LUAD tumor tissues (Fig. 4). The
results from the qRT-PCR validation in 53 newly diagnosed LUAD
patients were consistent with the above bioinformatics results
(Table I).
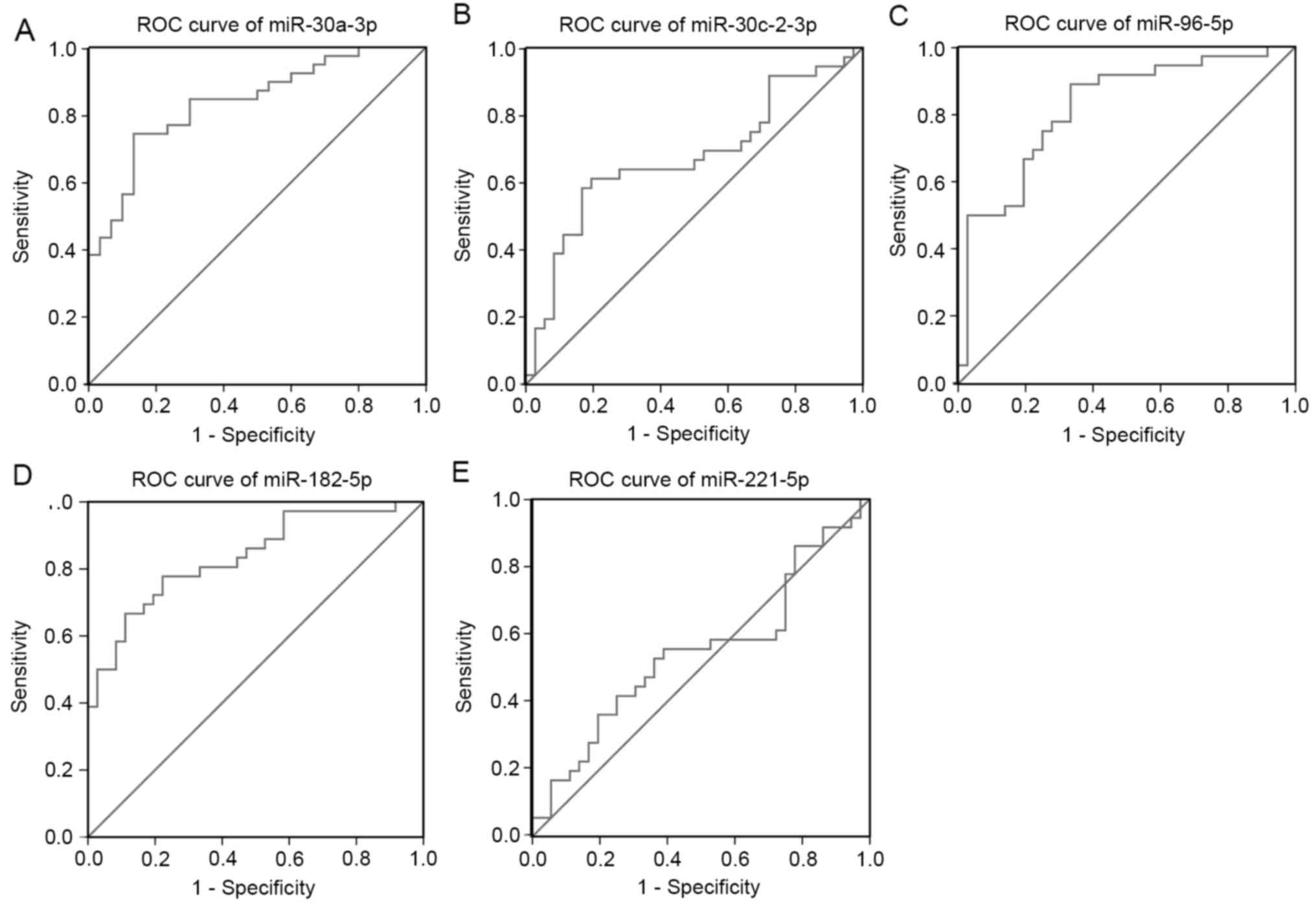
ROC curve analysis of specific key
miRNAs
ROC curve analysis demonstrated that the area under
curve (AUC)=0.837, 0.819 and 0.835 for miR-30a-3p,
miR-96-5p and miR-182-5p (P<0.01; Fig. 5A, C and D), which are the score both
higher the cut-off (0.7) and could be considerable biomarker for
early diagnosis of LUAD. ROC analysis measured an AUC=0.674 and
0.546 for miR-30c-2-3p and miR-221-5p (P<0.01;
Fig. 5B and E), which both are
scores very close to the cut-off (0.7) needed for a considerable
biomarker for early diagnosis of LUAD.
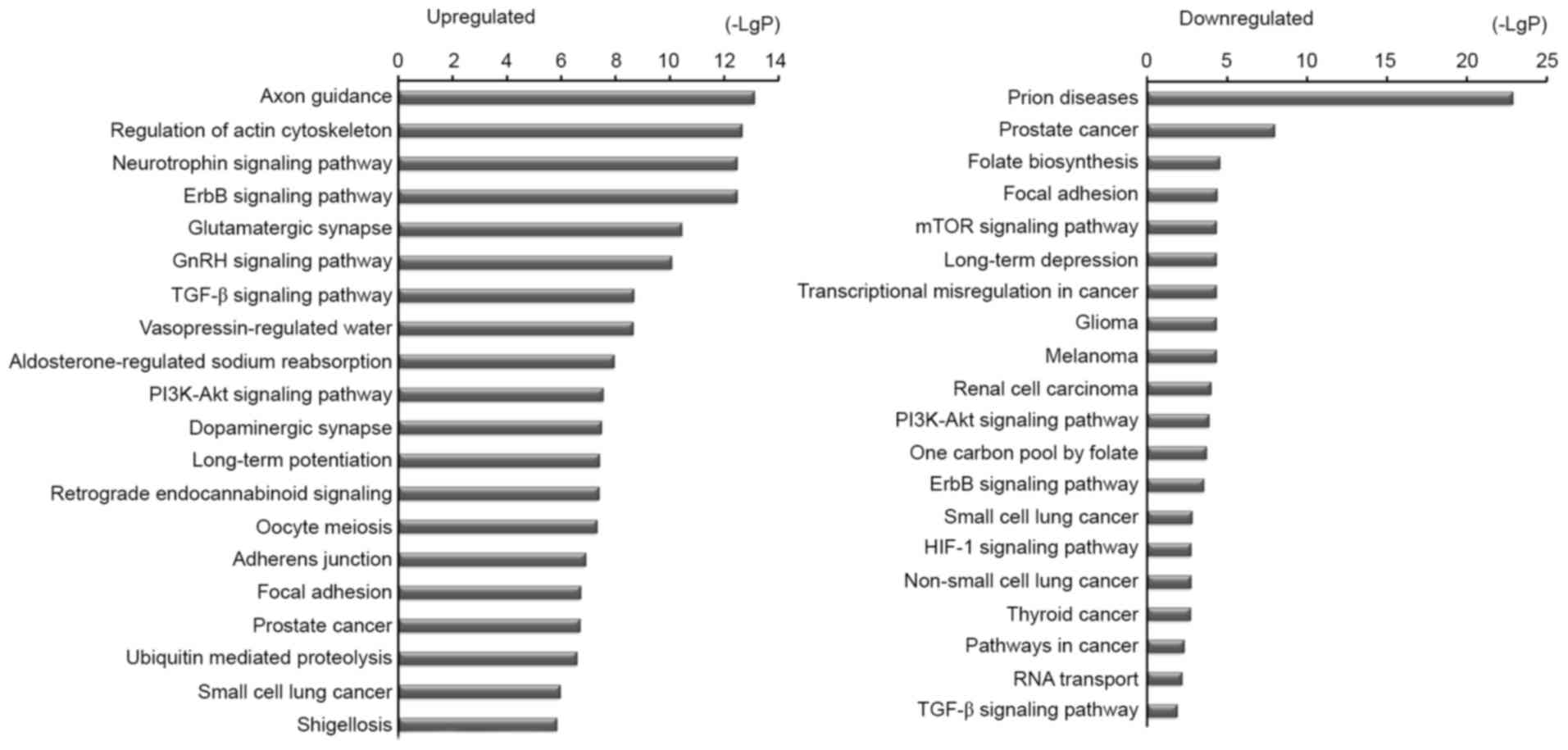
Functional enrichment analysis
To further confirm a possible link between miRNA and
cell function, DIANA-mirPath software was used to perform
differential expression analysis of miRNAs involved in signaling
pathways. The bio-informatics analysis indicated that
miR-30a-3p plays an important role in NSCLC (Fig. 6). It was also indicated that several
genes were regulated by miR-30a-3p to activate the process
of apoptosis in NSCLC (Fig. 7); for
example, AKT3 played an important activating role.
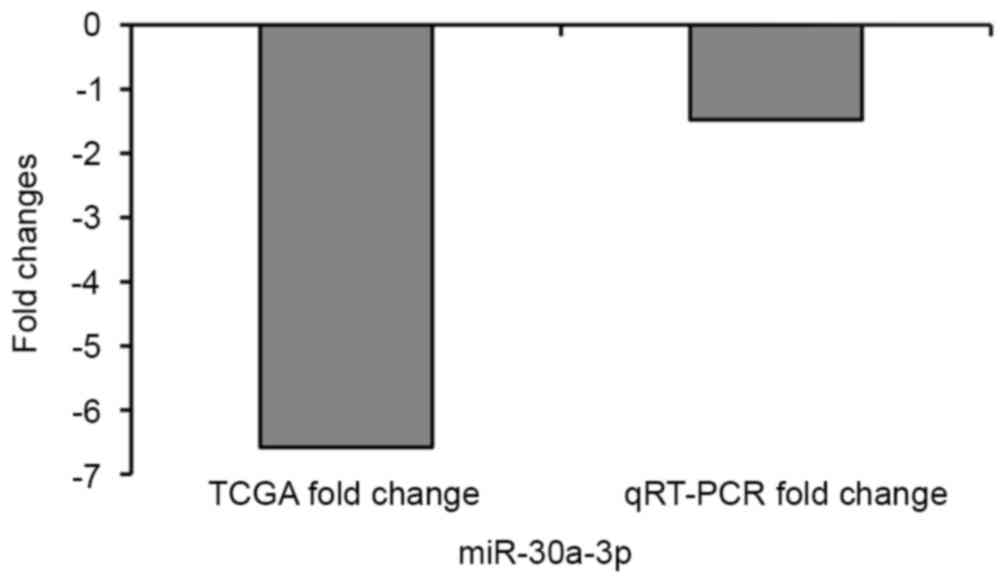
qRT-PCR verification of miR-30a-3p
expression in lung cell lines
miR-30a-3p was detected in analysis of
miRNAs. QRT-PCR was used to detect the expression of
miR-30a-3p expression in HBE cells and A549 cells. The
results showed that the qRT-PCR findings were consistent with the
TCGA findings (Fig. 8).
Influence of miR-30a-3p mimic on
morphology and proliferation of A549 cells
miR-30a-3p mimic was transfected into A549
cells to upregulate the expression of miR-30a-3p. qPCR was
performed to determine the efficiency of transfection with the
miR-30a-3p mimic. After transfection of the
miR-30a-3p mimic, the expression of miR-30a-3p
increased significantly (1852.04-fold), compared with negative
control (Table III). Light
microscopy revealed no significant difference in morphology of
miR-30a-3p transfected A549 cells, nor was there any
difference in cell density compared with the negative control
group. Furthermore, the MTT assay was used to assess the effect of
upregulated miR-30a-3p on the viability of A549 cells. The
MTT assay showed that after upregulation of miR-30a-3p, the
proliferation of A549 cells was significantly inhibited (P=0.000;
Fig. 9A).
 | Table III.The influence of the expression of
miR-30a-3p after miR-30a-3p mimic transfection. |
Table III.
The influence of the expression of
miR-30a-3p after miR-30a-3p mimic transfection.
|
| ΔCT (mean ±
SD) | ΔΔCT) (mean ±
SD |
2−ΔΔCT |
|---|
| miR-30a-3p
mimic group | −0.859±0.233 | −10.855±0.228 | 1852.040 |
| Negative
control | 9.548±1.106 |
|
|
Examination of miR-30a-3p-induced
apoptosis
PI and Annexin V were used to dye A549 cells, and
the apoptosis rate was assessed by flow cytometry. The total
apoptosis rate of cells in the miR-30a-3p mimic group
(17.100%) was significantly higher than among cells in the negative
control (9.400%) (P=0.022; Fig.
9B). Furthermore, nuclear staining with Hoechst 33258 and
electron microscopic examination demonstrated that in comparison
with negative control cells, typical apoptotic morphological
changes (such as cell shrinkage, condensation of cytoplasm and
chromatin compaction), were more frequent in miR-30a-3p
mimic group cells (Fig. 9C).
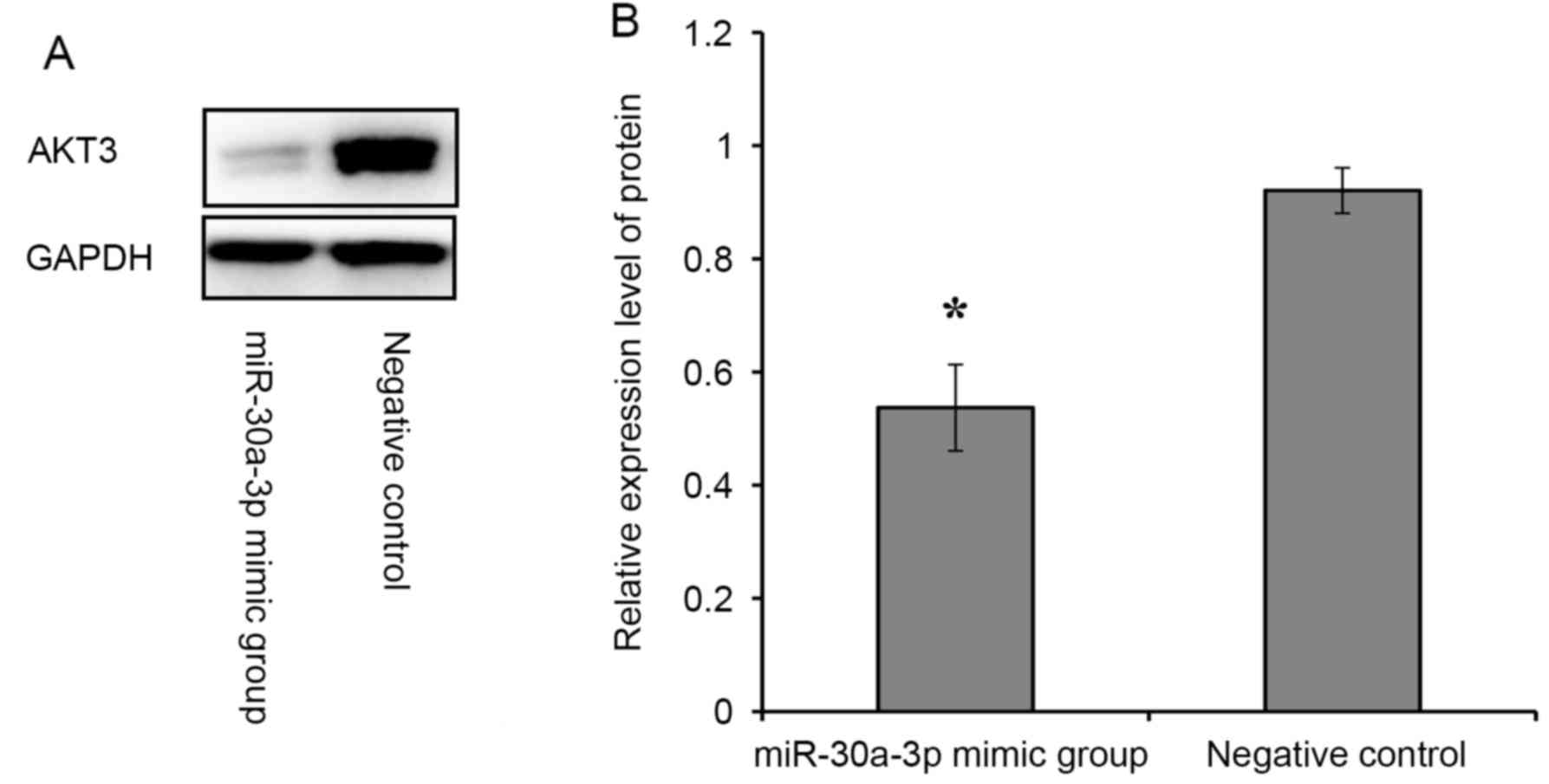
Western blot assays
Western blot was applied to measure the effect of
expression of the AKT3 gene on protein levels. The
expression of the AKT3 gene significantly decreased
(P=0.002; Fig. 10) protein levels
in the miR-30a-3p mimic group compared with the negative
control. This implies that miR-30a-3p might promote A549
cell apoptosis through negative regulation of the AKT3
gene.
Discussion
LUAD is the most frequent subtype of lung cancer,
with high global incidence and mortality (24). Because of immense heterogeneity in
multiple parameters (molecular, pathology, surgery and radiology)
among LUAD patients, the development of individualized therapy and
accurate prognosis have remained a huge challenge (25). Although major improvements in
diagnosis, medical treatment and surgical technology of LUAD have
been made over the past few decades, the 5-year disease-free
survival rate remains <10% (26). In the past decade, targeted therapy
has been improved, and many target genes, including PIK3CA,
PTEN and KRAS, have been identified as having a
potentially powerful clinical impact (27,28).
However, because of the large numbers of genes and the low
prevalence of mutations, miRNAs may be more effective than gene
expression profiles in classifying various human cancers (29). miRNAs are small, conserved,
non-coding regulatory RNAs that play important roles in initiation
and development of carcinoma, and they can interact with the miRNA
binding sites on the 3 untranslated region (3UTR) and coding
sequence of the target gene to regulate normal gene expression
(30). Moreover, there is
increasing evidence that miRNAs are closely related to initiation
and development of LUAD (31,32).
Therefore, identification of tumor-related miRNAs and the
regulatory mechanisms involved in LUAD has received increasing
attention.
In the present study, we identified aberrantly
expressed miRNAs in LUAD from the TCGA database. Based on miRNA
sequencing profiles in TCGA, we investigated the distribution of
LUAD miRNAs in reference to different clinical features and overall
survival. We further randomly selected five specific key miRNAs
(miR-30a-3p, miR-30c-2-3p, miR-96-5p,
miR-182-5p and miR-221-5p) from among 25 specific key
miRNAs and measured their expression in 53 LUAD tissue specimens
with qRT-PCR. Then, DIANA-miRPath software was used to analyze
these 25 specific key miRNAs. miR-30a-3p, known to be
involved in the NSCLC pathway and relevant in prognosis of LUAD,
was selected for cell functional studies and target gene
validation.
Among the 25 specific key miRNAs identified in our
bioinformatics analysis, several have been reported to be
dysregulated in cancers, including miR-30a-3p (33) miR-139-5p (34), miR-133a-3p (35) and miR-221-5p (36). miR-486-5p was previously
reported to be downregulated in NSCLC tissues and cell lines, and
was shown to target CDK4 to regulate cell proliferation,
apoptosis and cell cycle progression. Moreover, the upstream
promoter of miR-486-5p was shown to be strongly regulated by
methylation in NSCLC (37). Xiao
et al (38) investigated
miR-142-3p in NSCLC, and their results suggested that
miR-142-3p may be a tumor suppressor through downregulating
HMGB1 in NSCLC. In addition, miR-378a-3p,
miR-451a, miR-30c-2-3p, miR-210-3p and
miR-708-5p were also reported to be dysregulated in lung
cancer and might be related to initiation and development of lung
cancer. Eighteen of these 25 specific key miRNAs were not reported
in lung cancer.
With respect to the associations between the 25
LUAD-specific key miRNAs and clinical features, including race,
sex, age, TNM stage, lymphatic metastasis and patient outcome, we
found 15 of these 25 LUAD-specific key miRNAs to be related to
clinical features, including miR-133a-3p,
miR-139a-3p, miR-30a-3p and miR-30c-2-3p. All
15 were reported to be associated with human cancer. For example,
Chen et al (39) found
miR-451a to be downregulated in LUAD and was associated with more
advanced pathological stage, larger tumor diameter, and lymphatic
metastasis. Only two miRNAs (miR-451a and miR-142-3p)
were associated with LUAD clinical features. When associations
between 25 LUAD-specific key miRNAs and patient survival were
analyzed, three miRNAs were found to be related to LUAD overall
survival.
Five of the 25 LUAD-specific key miRNAs
(miR-30a-3p, miR-30c-2-3p, miR-96-5p,
miR-182-5p and miR-221-5p) were randomly selected to
verify expression of LUAD-specific key miRNAs and the accuracy of
bioinformatics analysis using qRT-PCR. The expression data from
TCGA and verification results from 53 diagnosed LUAD patients were
in 100% agreement, suggesting the credibility of our bioinformatics
analysis.
Based on the correlations between LUAD-specific key
miRNAs, clinical features from the TCGA, and functional enrichment
analysis, miR-30a-3p was found to be strongly related to
patient prognosis. Furthermore, it was shown that miR-30a-3p
might directly target AKT3 to regulate cell apoptosis. It
has been reported that inhibiting miR-30a-3p expression can
increase HIF2α levels in human clear cell renal cell
carcinoma cells and promote cell proliferation, angiogenesis and
tumor growth (40). Ma et al
(33) detected the expression of
miR-30a-3p in colorectal cancer using qRT-PCR compared with
adjacent non-tumor tissues and demonstrated that it was descreased.
Rodríguez et al (41) found
that higher expression of miR-30a-3p was one benefit of
treatment for breast cancer. In combination, the above findings
suggest that miR-30a-3p might play an important role in
tumor progression and prognosis formation in multiple cancers.
Finally, we analyzed the cell function of miR-30a-3p in A549
cells and found that miR-30a-3p might regulate cell
apoptosis through targeting AKT3. It was also found that
transfection with AKT3 siRNA in T98G cells could induce
apoptosis (42).
In summary, we identified LUAD-specific key miRNAs
from hundreds of candidate miRNAs identified from large-scale
samples in the TCGA database, and identified aberrant expression
profiles of cancer-specific key miRNAs under different clinical
features. Moreover, we also analyzed the association among
aberrently expressed miRNAs, clinical features and overall
survival. QRT-PCR verification was further used to confirm the
reliability and validity of expression of LUAD-specific key miRNAs
and bioinformatics analysis. ROC was used to determine the specific
key miRNAs as the sensitivity and specificity of detection of LUAD.
miR-30a-3p was selected to assess cell function, and the
results confirmed the credibility of functional enrichment
analysis. The aberrantly expressed key miRNAs identified in LUAD
may provide clues as to sensitive biomarkers in LUAD. Our studies
provide novel insight into finding potential biomarkers for
diagnosis and prognosis of LUAD.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (nos. 81472939, 81502783 and
81673132), the Qing Lan Project, the 333 project of Jiangsu
province, the Liu Da Ren Cai Gao Feng Project of Jiangsu Province,
and the Fundamental Research Funds for the central universities and
Innovative Research Project for postgraduates in Colleges of
Jiangsu province.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meng W, Ye Z, Cui R, Perry J,
Dedousi-Huebner V, Huebner A, Wang Y, Li B, Volinia S, Nakanishi H,
et al: MicroRNA-31 predicts the presence of lymph node metastases
and survival in patients with lung adenocarcinoma. Clin Cancer Res.
19:5423–5433. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li CY, Liang GY, Yao WZ, Sui J, Shen X,
Zhang YQ, Peng H, Hong WW, Ye YC, Zhang ZY, et al: Identification
and functional characterization of microRNAs reveal a potential
role in gastric cancer progression. Clin Transl Oncol. 19:162–172.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Babu KR and Muckenthaler MU: miR-20a
regulates expression of the iron exporter ferroportin in lung
cancer. J Mol Med (Berl). 94:347–359. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu W, Dang S, Feng Q, Liang J, Wang Y and
Fan N: MicroRNA-542-3p inhibits the growth of hepatocellular
carcinoma cells by targeting FZD7/Wnt signaling pathway. Biochem
Biophys Res Commun. 482:100–105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ren C, Chen H, Han C, Fu D, Wang D and
Shen M: High expression of miR-16 and miR-451 predicating better
prognosis in patients with gastric cancer. J Cancer Res Clin Oncol.
142:2489–2496. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang X, Li M, Wang Z, Han S, Tang X, Ge Y,
Zhou L, Zhou C, Yuan Q and Yang M: Silencing of long noncoding RNA
MALAT1 by miR-101 and miR-217 inhibits proliferation, migration,
and invasion of esophageal squamous cell carcinoma cells. J Biol
Chem. 290:3925–3935. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kent OA and Mendell JT: A small piece in
the cancer puzzle: microRNAs as tumor suppressors and oncogenes.
Oncogene. 25:6188–6196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Borel C and Antonarakis SE: Functional
genetic variation of human miRNAs and phenotypic consequences. Mamm
Genome. 19:503–509. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Takamizawa J, Konishi H, Yanagisawa K,
Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y,
et al: Reduced expression of the let-7 microRNAs in human lung
cancers in association with shortened postoperative survival.
Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Becker-Santos DD, Thu KL, English JC,
Pikor LA, Martinez VD, Zhang M, Vucic EA, Luk MT, Carraro A,
Korbelik J, et al: Developmental transcription factor NFIB is a
putative target of oncofetal miRNAs and is associated with tumour
aggressiveness in lung adenocarcinoma. J Pathol. 240:161–172. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mou X and Liu S: MiR-485 inhibits
metastasis and EMT of lung adenocarcinoma by targeting Flot2.
Biochem Biophys Res Commun. 477:521–526. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xie K, Wang C, Qin N, Yang J, Zhu M, Dai
J, Jin G, Shen H, Ma H and Hu Z: Genetic variants in regulatory
regions of microRNAs are associated with lung cancer risk.
Oncotarget. 7:47966–47974. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sui J, Li YH, Zhang YQ, Li CY, Shen X, Yao
WZ, Peng H, Hong WW, Yin LH, Pu YP, et al: Integrated analysis of
long non-coding RNA-associated ceRNA network reveals potential
lncRNA biomarkers in human lung adenocarcinoma. Int J Oncol.
49:2023–2036. 2016.PubMed/NCBI
|
|
18
|
Li CY, Liang GY, Yao WZ, Sui J, Shen X,
Zhang YQ, Peng H, Hong WW, Ye YC, Zhang ZY, et al: Integrated
analysis of long non-coding RNA competing interactions reveals the
potential role in progression of human gastric cancer. Int J Oncol.
48:1965–1976. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xie J and Liu C: Adjusted Kaplan-Meier
estimator and log-rank test with inverse probability of treatment
weighting for survival data. Stat Med. 24:3089–3110. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Branders S and Dupont P: A balanced hazard
ratio for risk group evaluation from survival data. Stat Med.
34:2528–2543. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sana J, Faltejskova P, Svoboda M and Slaby
O: Novel classes of non-coding RNAs and cancer. J Transl Med.
10:1032012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang DY, Wu YN, Huang JQ, Wang W, Xu M,
Jia JP, Han G, Mao BB and Bi WZ: Hippo/YAP signaling pathway is
involved in osteosarcoma chemoresistance. Chin J Cancer. 35:472016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mo X, Cao Q, Liang H, Liu J, Li H and Liu
F: MicroRNA-610 suppresses the proliferation of human glioblastoma
cells by repressing CCND2 and AKT3. Mol Med Rep. 13:1961–1966.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kerr KM: Pulmonary adenocarcinomas:
Classification and reporting. Histopathology. 54:12–27. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yoshizawa A, Motoi N, Riely GJ, Sima CS,
Gerald WL, Kris MG, Park BJ, Rusch VW, Travis WD, et al: Impact of
proposed IASLC/ATS/ERS classification of lung adenocarcinoma:
prognostic subgroups and implications for further revision of
staging based on analysis of 514 stage I cases. Mod pathol.
24:653–664. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ortea I, Rodríguez-Ariza A, Chicano-Gálvez
E, Vacas Arenas MS and Jurado Gámez B: Discovery of potential
protein biomarkers of lung adenocarcinoma in bronchoalveolar lavage
fluid by SWATH MS data-independent acquisition and targeted data
extraction. J Proteomics. 138:106–114. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
De Luca A, Maiello MR, DAlessio A,
Pergameno M and Normanno N: The RAS/RAF/MEK/ERK and the PI3K/AKT
signalling pathways: Role in cancer pathogenesis and implications
for therapeutic approaches. Expert Opin Ther Targets. 16 Suppl
2:S17–S27. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cardarella S and Johnson BE: The impact of
genomic changes on treatment of lung cancer. Am J Respir Crit Care
Med. 188:770–775. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cai Y, Yu X, Hu S and Yu J: A brief review
on the mechanisms of miRNA regulation. Genomics Proteomics
Bioinformatics. 7:147–154. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shi X, Xu Y, Zhang C, Feng L, Sun Z, Han
J, Su F, Zhang Y, Li C and Li X: Subpathway-LNCE: Identify
dysfunctional subpathways competitively regulated by lncRNAs
through integrating lncRNA-mRNA expression profile and pathway
topologies. Oncotarget. 7:69857–69870. 2016.PubMed/NCBI
|
|
32
|
Wu C, Xu B, Zhou Y, Ji M, Zhang D, Jiang J
and Wu C: Correlation between serum IL-1β and miR-144-3p as well as
their prognostic values in LUAD and LUSC patients. Oncotarget.
7:85876–85887. 2016.PubMed/NCBI
|
|
33
|
Ma Y, Zhang P, Yang J, Liu Z, Yang Z and
Qin H: Candidate microRNA biomarkers in human colorectal cancer:
Systematic review profiling studies and experimental validation.
Int J Cancer. 130:2077–2087. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pantaleo MA, Ravegnini G, Astolfi A,
Simeon V, Nannini M, Saponara M, Urbini M, Gatto L, Indio V,
Sammarini G, et al: Integrating miRNA and gene expression profiling
analysis revealed regulatory networks in gastrointestinal stromal
tumors. Epigenomics. 8:1347–1366. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wei Y, He R, Wu Y, Gan B, Wu P, Qiu X, Lan
A, Chen G, Wang Q, Lin X, et al: Comprehensive investigation of
aberrant microRNA profiling in bladder cancer tissues. Tumour Biol.
37:12555–12569. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pichler M, Stiegelbauer V,
Vychytilova-Faltejskova P, Ivan C, Ling H, Winter E, Zhang X,
Goblirsch M, Wulf-Goldenberg A, Ohtsuka M, et al: Genome-wide
microRNA analysis identifies miR-188-3p as novel prognostic marker
and molecular factor involved in colorectal carcinogenesis. Clin
Cancer Res. 23:1323–1333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shao Y, Shen YQ, Li YL, Liang C, Zhang BJ,
Lu SD, He YY, Wang P, Sun QL, Jin YX, et al: Direct repression of
the oncogene CDK4 by the tumor suppressor miR-486-5p in non-small
cell lung cancer. Oncotarget. 7:34011–34021. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xiao P and Liu WL: MiR-142-3p functions as
a potential tumor suppressor directly targeting HMGB1 in
non-small-cell lung carcinoma. Int J Clin Exp Pathol.
8:10800–10807. 2015.PubMed/NCBI
|
|
39
|
Chen Q, Hu H, Jiao D, Yan J, Xu W, Tang X,
Chen J and Wang J: miR-126-3p and miR-451a correlate with
clinicopathological features of lung adenocarcinoma: The underlying
molecular mechanisms. Oncol Rep. 36:909–917. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mathew LK, Lee SS, Skuli N, Rao S, Keith
B, Nathanson KL, Lal P and Simon MC: Restricted expression of
miR-30c-2-3p and miR-30a-3p in clear cell renal cell carcinomas
enhances HIF2α activity. Cancer Discov. 4:53–60. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Rodríguez-González FG, Sieuwerts AM, Smid
M, Look MP, Meijer-van Gelder ME, de Weerd V, Sleijfer S, Martens
JW and Foekens JA: MicroRNA-30c expression level is an independent
predictor of clinical benefit of endocrine therapy in advanced
estrogen receptor positive breast cancer. Breast Cancer Res Treat.
127:43–51. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shin K, Kim KH, Yoon MS, Suh DS, Lee JY,
Kim A and Eo W: Expression of interactive genes associated with
apoptosis and their prognostic value for ovarian serous
adenocarcinoma. Adv Clin Exp Med. 25:513–521. 2016. View Article : Google Scholar : PubMed/NCBI
|