Introduction
Hepatocellular carcinoma (HCC) is one of the six
malignancies in the world and with a mortality rate among patients
(1,2). Although advances in medicine and
surgical procedures have improved the treatment of HCC, the
prognosis of patients with HCC is poor with a low long-term
survival rate due to a high recurrence rate and postoperative
metastasis (3–5). The accurate molecular mechanism that
contributes to the occurrence and development of HCC is unclear.
Thus, it is necessary to find new individualized molecules which
are involved in HCC and clarify their roles, to further develop new
therapeutics for HCC treatment.
FGF18 is a member of the fibroblast growth factor
(FGF) family (6). The FGF family
includes 22 members, which possess mitogenic and cell survival
activities, and are involved in a variety of biological processes,
such as embryonic development, cell growth, morphogenesis, tissue
repair and angiogenesis (7). FGF18
is an essential mitogen in embryonic limb development and is
required for differentiation during osteogenesis and chondrogenesis
(8–10). Additionally, as a pleiotropic growth
factor, FGF18 stimulates proliferation in a number of tissues, most
notably the liver (11) and small
intestine (8,12). Recently, FGF18 was reported to be
highly expressed in colon cancer and ovarian cancer. Increased
FGF18 expression, which plays a key role in controlling tumor
growth and invasion, was associated with tumor progression and poor
overall survival in patients (13–15).
FGF18 can also affect both the tumor and the connective tissue
cells of the tumor microenvironment (16,17).
Overexpression of FGF18 promoted tumor progression by modulating
migration, invasion and tumorigenicity of cancer cells arguably
through different signaling pathways (13,15).
These findings demonstrated that FGF18 is important for a subset of
ovarian cancers and may serve as a therapeutic target. However,
there is still no study concerning the regulation mechanism of
FGF18 in HCC.
The occurrence of cancers undergoes multiple
complicated stages that are caused by many factors. Thus, there is
a lack of effective treatments. The strategy of gene-direct
inhibition may be an effective method in cancer therapy. MicroRNAs
(miRNAs) are a class of small RNAs that can regulate gene
expression at the mRNA level and function in various cellular
processes and biological activities. They can be used to regulate
the development of cancer, through cell apoptosis, invasion and
proliferation (18). Numerous
miRNAs in HCC have been widely investigated, and have also been
identified as diagnostic and prognostic markers as well as
therapeutic candidates for HCC. This strategy may provide accurate
clinical decisions for HCC diagnosis and treatment (19,20).
In the present study, we demonstrated that the
expression of FGF18, which was investigated in HCC, was regulated
by miR-139. In addition, the effects of FGF18 suppression by
miR-139 on the biological processes of HCC cells were also
revealed.
Materials and methods
Clinical specimens, cell culture and
transfection
Human hepatocellular carcinoma (HCC) and matched
adjacent non-tumor tissues were collected from 15 patients at the
Department of Oncology, The First People's Hospital of Nantong. The
present study was approved by the Institute Research Ethics
Committee of the First People's Hospital of Nantong. Human HCC cell
lines HepG2, Huh7 and normal control liver cell line LO2, as well
as human embryonic kidney 293 (HEK293) cells were all obtained from
Biomics Biotechnologies Co., Ltd. (Nantong, China), cultured in
Dulbecco's modified Eagle's medium (DMEM) and supplemented with 10%
fetal bovine serum (FBS) (Thermo Fisher Scientific, Inc., Waltham,
MA, USA) at 37°C in a humidified incubator with 5%
CO2.
miR-139 mimics were used for upregulation of
endogenous miR-139, and a miR-139 sequence-scrambled RNA was used
as a negative control miRNA (NC_miR). Lipofectamine®
2000 transfection reagent (Thermo Fisher Scientific, Inc.) was used
for transfection of miRNAs into cells according to the
manufacturer's instructions. miR-139 mimics and NC_miR were
obtained from Biomics Biotechnologies Co., Ltd. (Nantong,
China).
Dual-luciferase reporter assay
The 3 untranslated region (3′UTR) region
complementary DNA sequence of human FGF18 mRNA (NCBI Reference
Sequence: NM_003862.2) was amplified by polymerase chain reaction
(PCR) and constructed into the pGL3-vector (Promega, Madison, WI,
USA) as a dual-luciferase miRNA target expression vector (wild-type
FGF18 3′UTR, wtFGF18-3′UTR) to evaluate miR-139 activity in cells.
The FGF18 3′UTR mutant vector was also constructed as the negative
control (mutant FGF18 3′UTR, mFGF18-3′UTR). Briefly, HEK293 cells
were seeded into a 24-well plate and grown for 24 h. Then
wtFGF18-3′UTR or mFGF18-3′UTR was co-tansfected with miR-139 mimics
or NC_miR using Lipofectamine® 2000 transfection reagent
(Thermo Fisher Scientific, Inc.); pRL-TK (Promega) was
co-transfected as an internal control. After transfection for 48 h,
cells were collected and lysed, firefly and Renilla
luciferase activities in cell lysates were detected using the DLR
assay system (Promega) according to the manufacturer's
instructions.
Real-time quantitative PCR
(RT-qPCR)
After cells were treated with miRNAs as
aforementioned, the total RNA of cells was extracted using
TRIzol® reagent (Thermo Fisher Scientific, Inc.) and
small RNA enriched with miRNAs was isolated using
mirPremier® microRNA isolation kit (Sigma-Aldrich, St.
Louis MO, USA) according to the manufacturers instructions.
Detection of miRNA expression levels in tissues or cells was
performed by stem-loop RT-qPCR (21). U6 small RNA or GAPDH was used as an
internal control for miRNA or mRNA detection, respectively. RT-qPCR
reactions were carried out using the SYBR-Green One-Step RT-qPCR
kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The relative miRNA or mRNA expression
levels were evaluated by 2−ΔΔCt method (22). The primer sequences were FGF18
forward, 5′-ACCTTCGGTAGTCAAGTC-3′ and reverse,
5′-CGTGTAGTTGTTCTCCAG-3′; GAPDH forward, 5′-GAGTCCACTGGCGTCTTC-3′
and reverse, 5′-GATGATCTTGAGGCTGTTGTC-3′.
Western blot analysis
Cells were plated in a 6-well plate, after treatment
for 48 h as aforementioned. The cells were collected and lysed in a
cell RIPA lysis and extraction buffer (Thermo Fisher Scientific,
Inc.) on ice. Centrifugation to collect the proteins, then
separation by polyacrylamide gel electrophoresis (PAGE) and
transfer to a polyvinylidene difluoride (PVDF) membrane (Millipore,
Billerica, MA, USA)followed. Subsequently, the membrane was
incubated with a primary antibody rabbit anti-human FGF18 (1:1,000
dilution) or a mouse anti-human β-actin antibody (1:5,000 dilution)
(all from Abcam, Cambridge, MA, USA) as an internal control. After
being washed with Tris-buffered saline and Tween-200 (TBST) the
membrane was incubated with a horseradish peroxidase-conjugated
secondary antibody (1:5,000 dilution) (Abcam) for 1.5 h at room
temperature, and then washed in TBST. The specific proteins were
detected with ECL substrate (Thermo Fisher Scientific, Inc.).
Cell proliferation assay
The proliferation abilities of cells were assessed
using Cell Counting Kit-8 (CCK-8) (Sigma-Aldrich). Briefly,
5×103 cells/well were seeded on a 96-well plate before
treatment and grown to ~70% confluence for 24 h. Post-treatment at
0, 24, 48, 72 and 96 h, the cells were washed with
phosphate-buffered saline (PBS) three times, and replaced with 100
µl/well of PBS, and then 10 µl of FCCK working solution was added
to each well. After incubation at 37°C in an incubator for 30 min,
the fluorescence intensity of each well was assessed at 535 nm
using a fluorescence microplate reader (BioTek, Winooski, VT,
USA).
Cell apoptosis assay
Cell apoptosis was determined by flow cytometric
(FCM) analysis using Annexin V-FITC/propidium iodide (PI) double
staining (Sigma-Aldrich). The cells were seeded on a 6-well plate
for 24 h of growth and treated as aforementioned for 48 h. Briefly,
1×105 cells/well were harvested and washed with PBS, and
then re-suspended in Annexin-binding buffer, followed by incubation
with FITC-conjugated Annexin V and propidium iodide (PI) for 15 min
at room temperature. Finally, the cells were analyzed by FCM (BD
Biosciences, Franklin Lakes, NJ, USA).
Cell migration assay
A cell wound scratch assay was used to determine the
migration abilities of HCC cells. Briefly, 1×104
cells/well were plated on a 12-well plate for 24 h of growth and
treated as aforementioned. Cell wounds were made to confluent
monolayer cells with a pipette tip post 48-h treatment and the
cells were washed with DMEM twice. Cell migration was monitored at
0 and 48 h after the scratch.
Cell invasion assay
The invasion abilities of cells were detected by
Transwell assay. Cells/well (2×105) were seeded into a
24-well plate for 24 h and treated for 48 h, then suspended in DMEM
at a density of 1×106 cells/ml. Transwell chambers
(Corning, NY, USA) were incubated with DMEM for 1 h before
treatments; the upper and lower chamber was separated by an 8-µm
pore polycarbonate membrane which was coated with 50 µl of 0.5
mg/ml Matrigel (BD Biosciences). A cell suspension of 100 µl was
added into each upper chamber with 600 µl DMEM containing 10% FBS
or conditioned medium which was the cell supernatant with treatment
as aforementioned for 48 h. The cells on the top surface of the
membrane were carefully removed using cotton swabs post-treatment
for 24 h. The cells on the Transwell chambers were fixed in 10%
formaldehyde for 30 sec, and then stained with 0.5% crystal violet
solution. After being washed with PBS, the cells on the top surface
of the membrane were carefully removed again, and the cells on the
bottom surface of the membrane were observed and counted under a
microscope.
Tube formation assay
An in vitro angiogenesis model was used to
evaluate the inhibitory effects of miRNAs on angiogenesis. Briefly,
a 24-well plate was coated with 100 µl/well Matrigel (BD
Biosciences) and incubated at 37°C for 30 min. Human umbilical vein
endothelial cells (HUVECs) at a concentration of 1×105
cells/well were re-suspended in conditioned medium (which was the
supernatant of the cells treated with miRNAs for 48 h), then seeded
on a Matrigel coated 24-well plate and cultured in a 37°C/5%
CO2 incubator for 24 h. The number of branching points
was counted under a microscope.
Statistical analysis
All the experiments were performed independently
three times. The data are shown as the mean values ± standard
deviation (SD). Statistical analyses were performed using SPSS 19.0
software (SPSS, Inc., Chicago, IL, USA), and the results were
analyzed using one way ANOVA followed by post hoc test to assess
statistical significance. All P-values were based on a two-side
statistical analysis and P<0.05 was considered to indicate
statistical significance.
Results
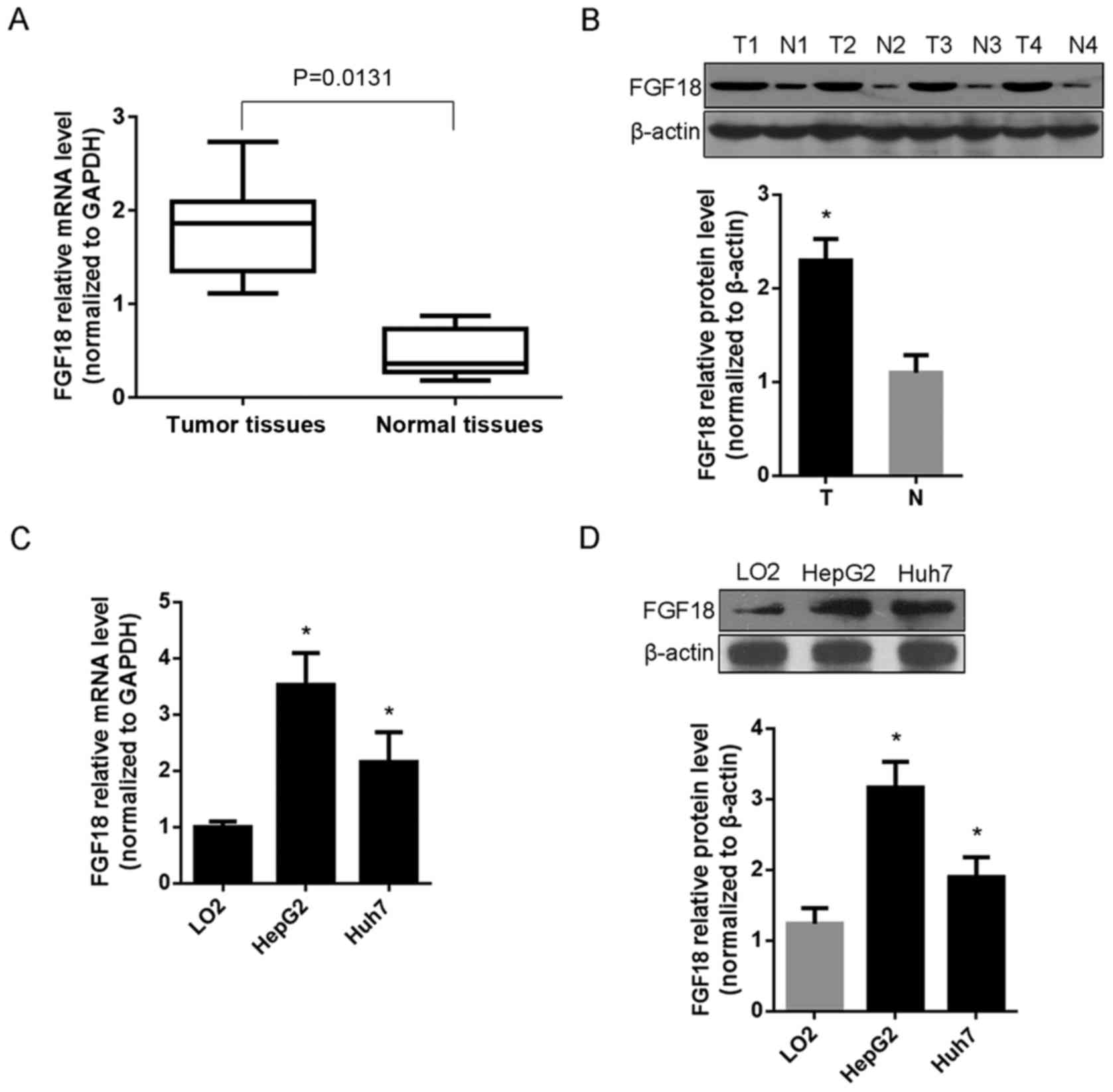
FGF18 expression in human HCC patients
and cells
The expression of FGF18 was detected by RT-qPCR and
western blot assay in 15 cases of HCC tissues and matched adjacent
normal tissues, as well as in HCC cell lines HepG2 and Huh7.
Compared with normal tissues, the results revealed that FGF18 mRNA
and protein levels were highly expressed in HCC patients (P=0.0131)
(Fig. 1A and B). Moreover, the mRNA
and protein levels of FGF18 were also highly expressed in HCC cell
lines HepG2 and Huh7, compared with normal liver cell LO2
(P<0.05) (Fig. 1C and D).
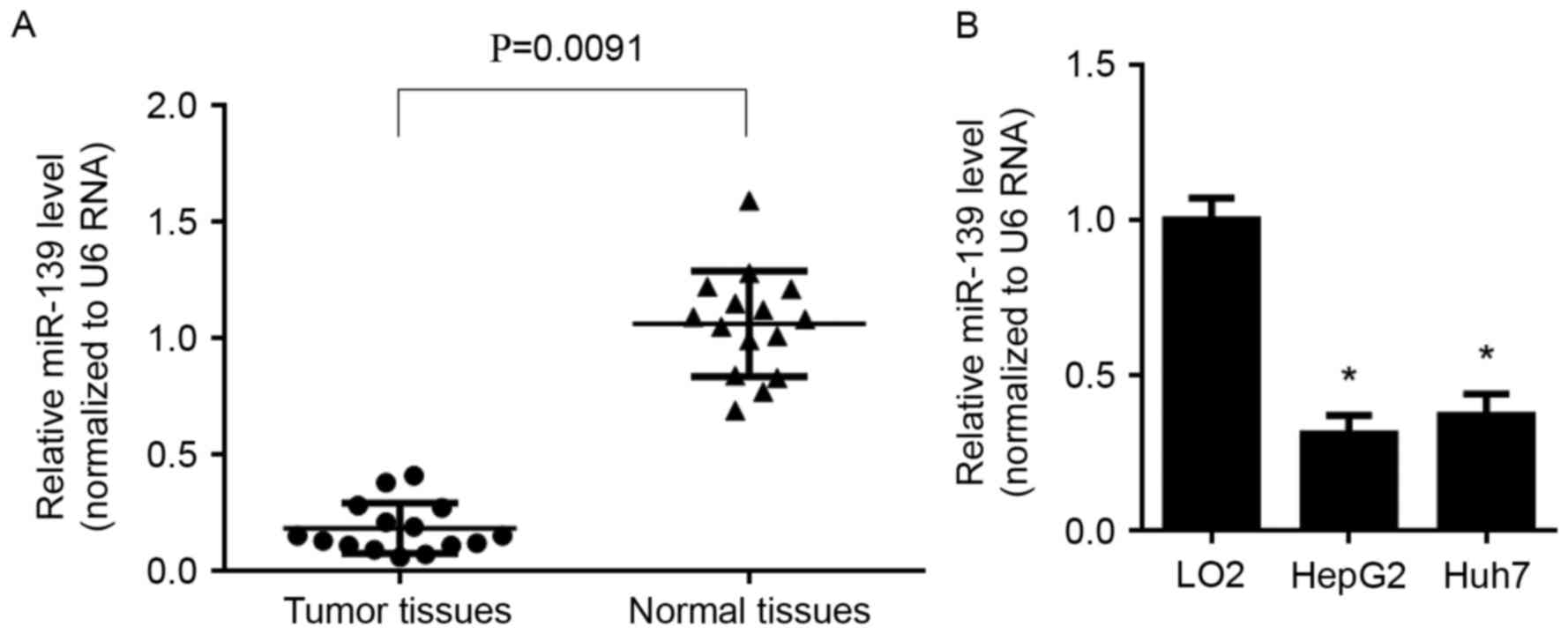
Expression profile of miR-139 in human
HCC patients and cells
The expression of miR-139 was detected by RT-qPCR
both in HCC tissues and cell lines. The results revealed that,
miR-139 was significantly decreased in HCC tissues (P=0.0091)
(Fig. 2A), compared with normal
tissues, it was also lowly expressed in both HepG2 and Huh7 cells,
compared with that in LO2 cells (P<0.05) (Fig. 2B).
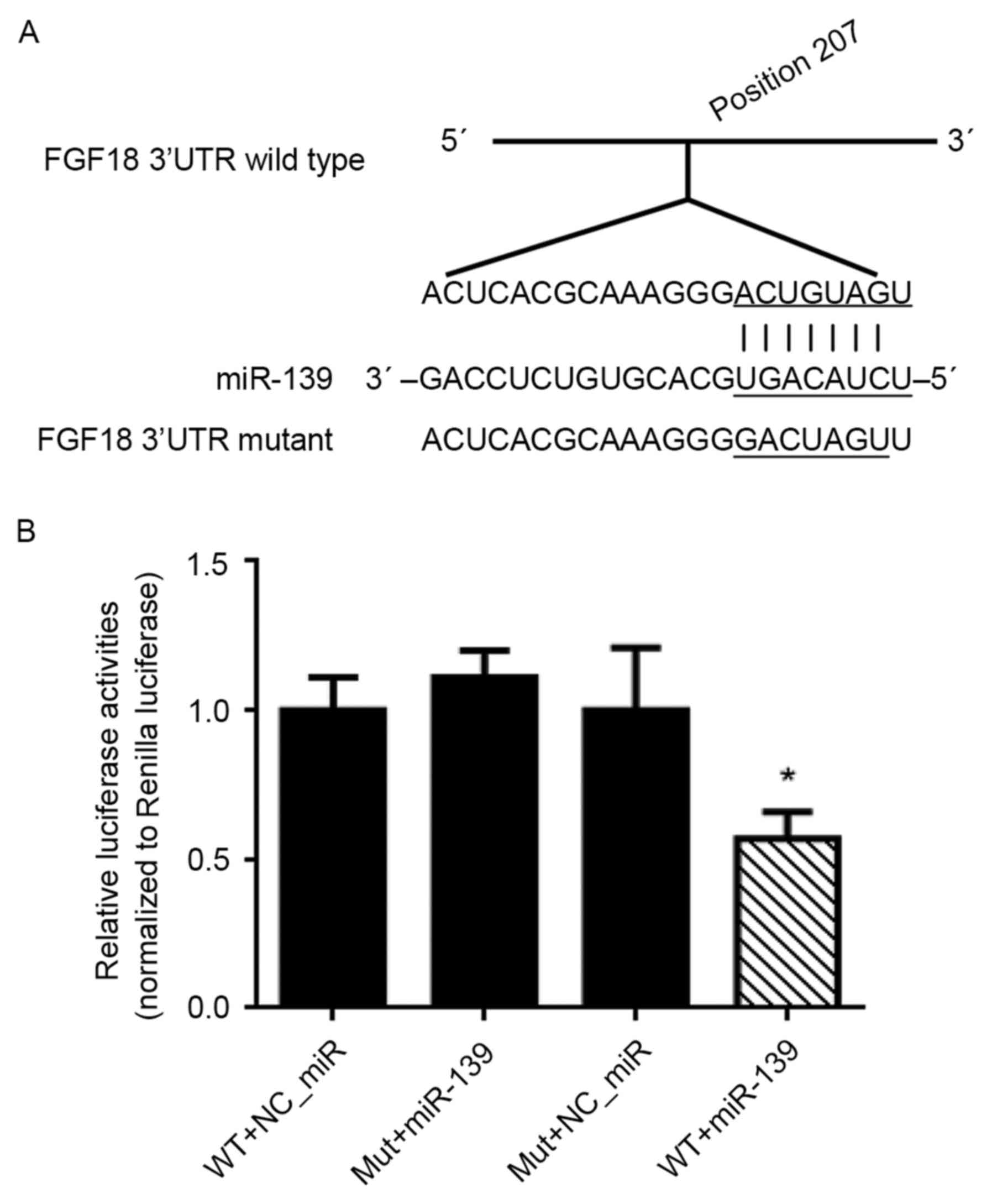
miR-139 targeting the 3′UTR of FGF18
mRNA
The target site of miR-139 with the position 207 of
the FGF18 mRNA sequence was predicated by web online software
(http://www.targetscan.org), and then the
dual-luciferase reporter (DLR) assay was used to observe the site
binding and inhibition effects. The reporter vectors of the
wild-type wtFGF18-3′UTR and mutant-type mFGF18-3′UTR were
constructed (Fig. 3A). The result
of the DLR assay revealed that co-tansfection of HEK293 cells with
miR-139 mimics and wtFGF18-3′UTR, resulted in a marked decrease of
luciferase activities, compared with NC_miR transfected with
wtFGF18-3′UTR or mFGF18-3′UTR, and miR-139 transfected with
mFGF18-3′UTR (Fig. 3B).
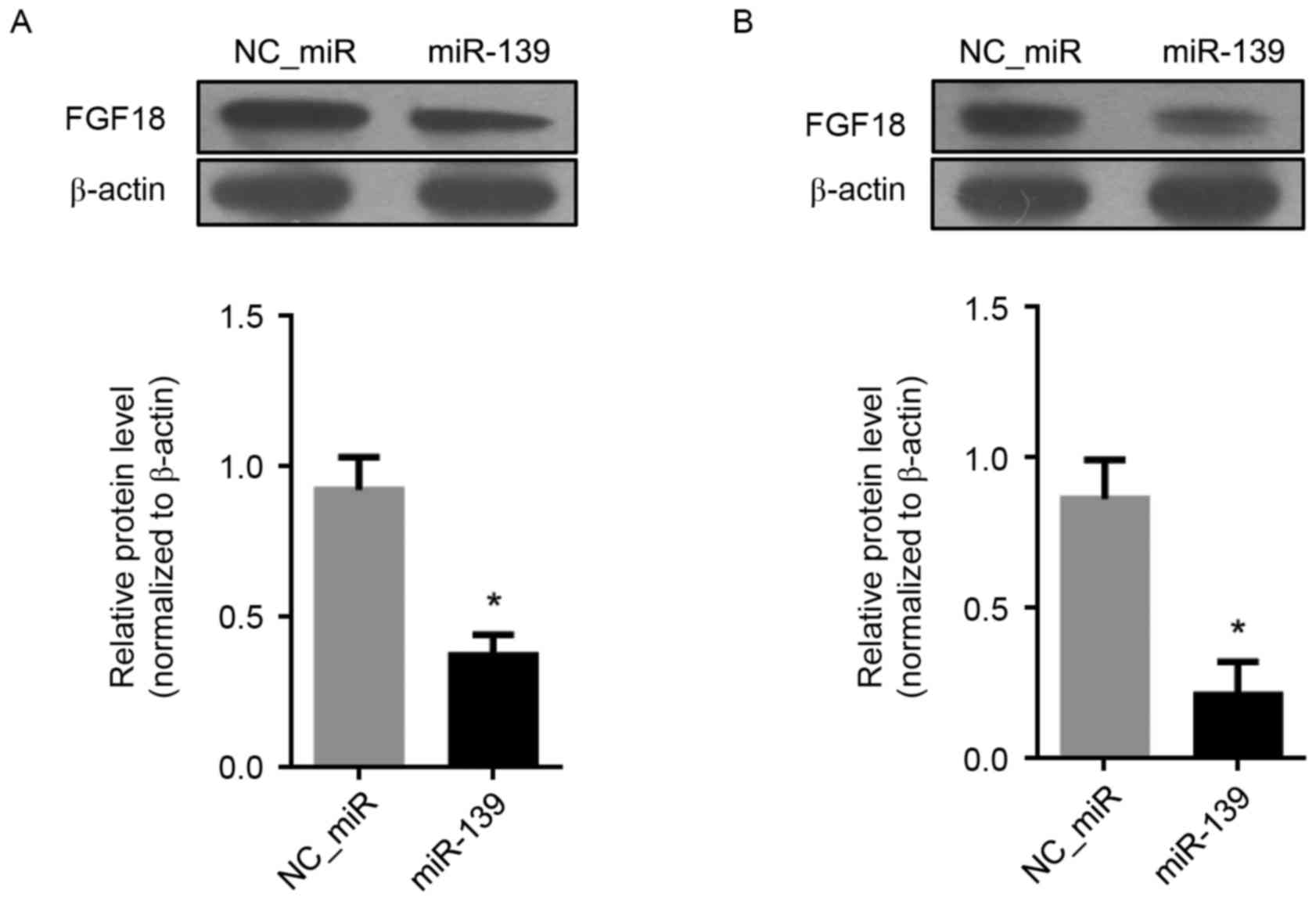
Inhibition of FGF18 by miR-139 in HCC
cells
The inhibitory effect of miR-139 on the expression
of FGF18 was detected by western blot assay. Compared with
NC_miR-treated cells, the results revealed that the FGF18 protein
levels were suppressed by miR-139 in both HCC cells HepG2 and Huh7
(P<0.05) (Fig. 4A and B).
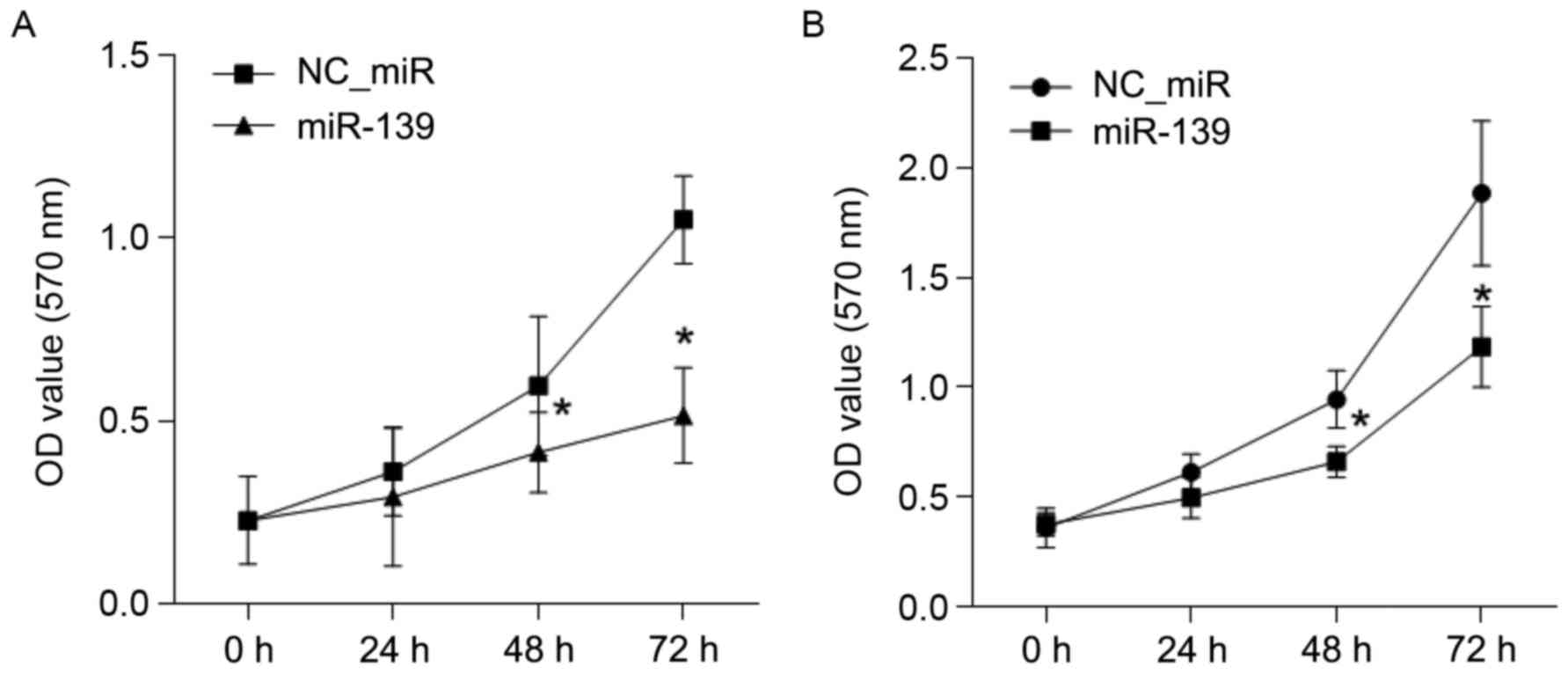
The growth effect of FGF18 on HCC
cells is inhibited by miR-139
To investigate the inhibition effect of miR-139 on
the growth of HCC cells, a CCK8 assay was performed. The results
revealed that the growth of HCC cell lines, HepG2 and Huh7, were
both inhibited by miR-139 at 48 and 72 h, compared with the
NC_miR-treated cells (P<0.05) (Fig.
5).
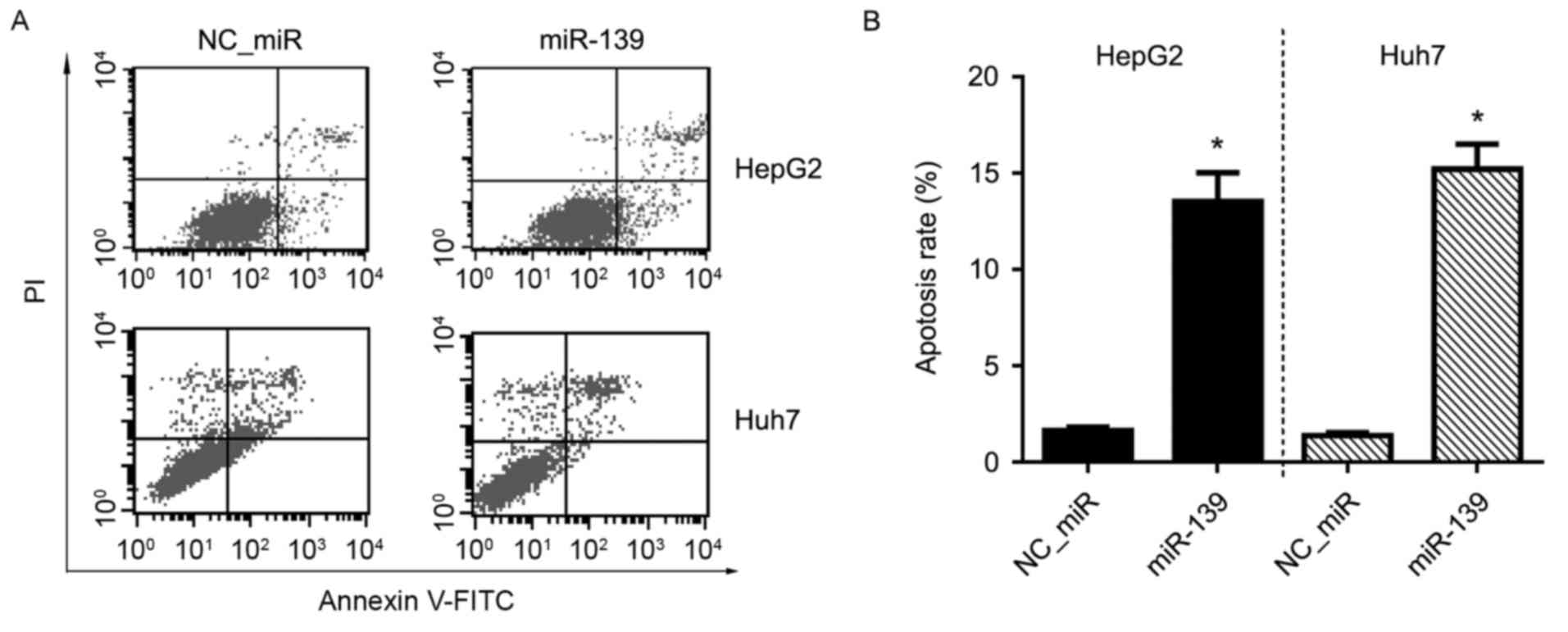
Apoptosis effect of FGF18 on HCC cells
is inhibited by miR-139
The inhibitory effect of miR-139 on the apoptosis of
HCC cells was determined by flow cytometric (FCM) analysis after
Annexin V-FITC/PI double staining. The results revealed that the
apoptosis of HCC cell lines, HepG2 and Huh7, were both inhibited by
miR-139 when compared with the NC_miR-treated cells (P<0.05)
(Fig. 6).
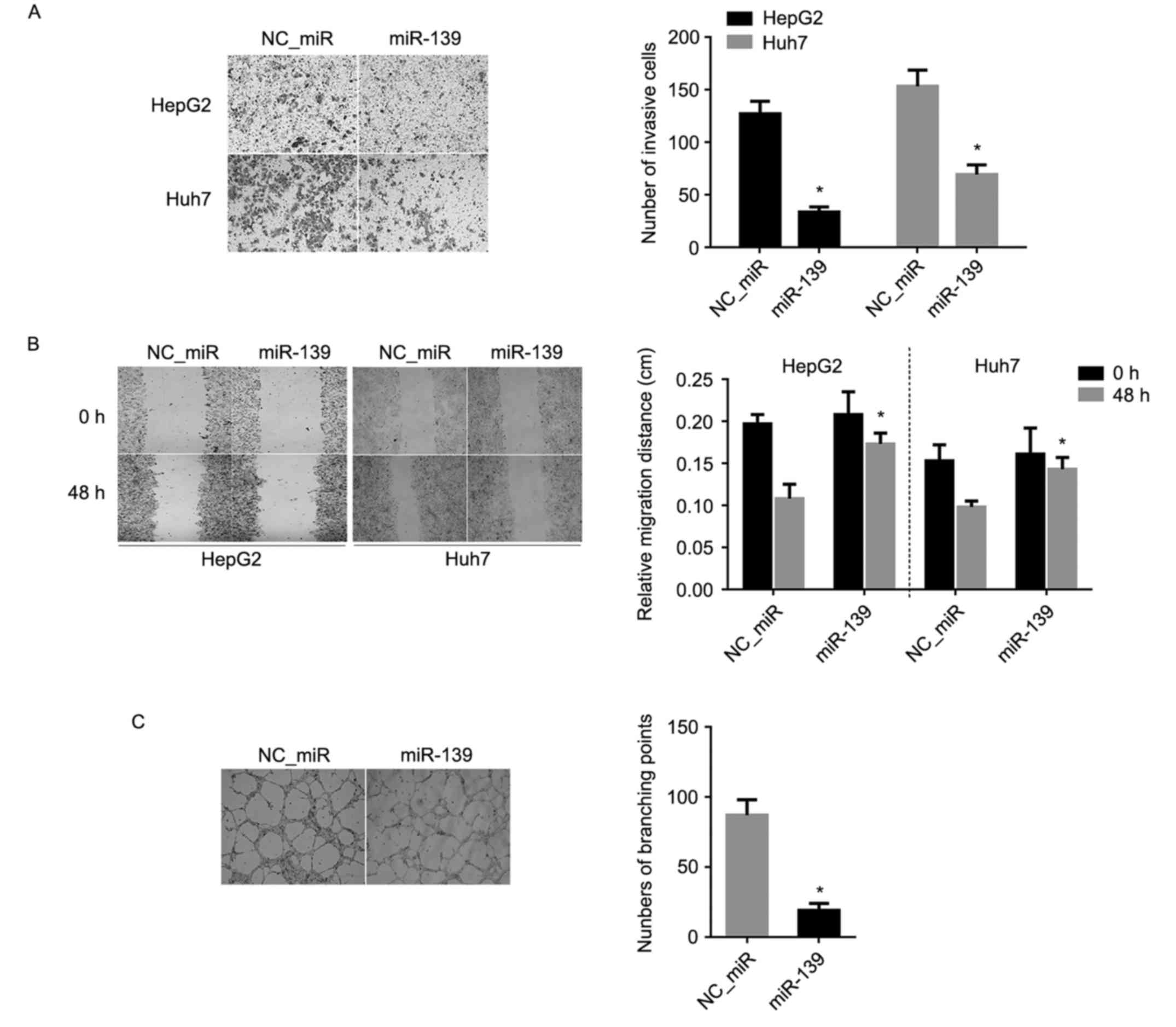
Invasion and migration effect of FGF18
on HCC cells is inhibited by miR-139
To observe the inhibitory effect of invasion and
migration abilities, Transwell and cell wound scratch assays were
separately used. The result of the Transwell assay revealed that
the invasion abilities of HCC cell lines, HepG2 and Huh7, were
decreased significantly after miR-139 treatment, compared with the
NC_miR-treated cells (P<0.05) (Fig.
7A). Similarly, the result of the cell wound scratch assay
revealed that the migratory abilities of HCC cell lines, HepG2 and
Huh7, were also markedly decreased post-miR-139 treatment, compared
with the NC_miR-treated cells (P<0.05) (Fig. 7B).
Tube formation effect of FGF18 on HCC
cells is inhibited by miR-139
Tube formation was evaluated in a HUVEC angiogenesis
model. HUVECs stimulated by the conditioned medium derived from HCC
cells were transfected with miR-139 mimics and NC_miR. HUVECs
treated with the conditioned medium of miR-139 mimics were
significantly inhibited to form extensive and enclosed tube
networks and when compared with the untreated cells, the number of
branching points was less than the NC_miR-treated cells (P<0.05)
(Fig. 7C).
Discussion
High expression of fibroblast growth factor 18
(FGF18) is related to the development and prognosis in several
types of cancer, and has a relationship with the cell
proliferation, invasion and angiogenesis (13–15).
However, its expression and functions in HCC was still unclear. In
the present study, high expression of FGF18 on the mRNA and protein
level was demonstrated in HCC tissues and cell lines, such as HepG2
and Huh7 (Fig. 1).
miRNAs are a class of small RNAs 19–23 nt in length
(23), that have been reported as
novel biomarkers in many carcinomas and that play essential roles
in carcinogenesis (24). Evidence
has suggested that miRNAs can act as tumor suppressors or promoters
in various carcinomas (25). Recent
studies indicated that miR-139 was reported as a tumor suppressor
in many types of cancer, such as breast cancer (26), acute myeloid leukemia (27), bladder (28), colorectal (29), cervical (30) and lung cancer (31). miR-139 was lowly expressed in these
cancers, while highly expressed in normal tissues or cells. miR-139
was found to induce cell apoptosis, suppress the tumor growth and
decrease tumor metastasis and drug sensitivity by inhibiting the
EMT signaling pathway (30,32). Overexpression of miR-139-5p resulted
in the inhibition of uterine leiomyoma cell growth (34). It suppressed glioma cell
proliferation and regulated the cell cycle (34). In the present study, we also found
that miR-139 was lowly expressed in HCC tumor tissues (Fig. 2A) and cell lines (both in HepG2 and
Huh7) (Fig. 2B).
Furthermore, the relationship between FGF18 and
miR-139 was investigated and predication of software revealed that
miR-139 can bind to the mRNA of FGF18 (Fig. 3A). The region of the binding site
was synthesized and cloned into a dual-luciferase reporter vector
and was tansfected with miR-139 mimics into HEK293 cells. The
results revealed that FGF18 was directly inhibited by miR-139
(Fig. 3B), suggesting that FGF18
was the target of miR-139.
Additionally, to explore whether FGF18 can be
regulated by miR-139 in HCC cells, HCC cell lines, HepG2 and Huh7
were used. In both cell lines, a high expression of miR-139
(transfection of miR-139 mimics) resulting in a decrease of the
protein level of FGF18 (Fig. 4),
indicated that FGF18 can be suppressed by miR-139 in HCC cells. The
biological functions of miR-139 influencing HCC cells were also
demonstrated. After HepG2 and Huh7 cells were treated with miR-139,
cell proliferation (Fig. 5),
apoptosis (Fig. 6), invasion and
migration were all inhibited, and the ability of angiogenesis
induction was also decreased (Fig.
7).
In brief, the results of the present study reveal
that FGF18, and miR-139 may be biomarkers or novel prognostic
indicators in HCC, and inhibition of FGF18, particularly by miR-139
may be a potential approach for HCC treatment.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang C, Zhang F, Fan H, Peng L, Zhang R,
Liu S and Guo Z: Sequence polymorphisms of mitochondrial D-loop and
hepatocellular carcinoma outcome. Biochem Biophys Res Commun.
406:493–496. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shindoh J, Hasegawa K, Inoue Y, Ishizawa
T, Nagata R, Aoki T, Sakamoto Y, Sugawara Y, Makuuchi M and Kokudo
N: Risk factors of post-operative recurrence and adequate surgical
approach to improve long-term outcomes of hepatocellular carcinoma.
HPB Oxf. 15:31–39. 2013. View Article : Google Scholar
|
|
6
|
Hu MC, Qiu WR, Wang YP, Hill D, Ring BD,
Scully S, Bolon B, DeRose M, Luethy R, Simonet WS, et al: FGF-18, a
novel member of the fibroblast growth factor family, stimulates
hepatic and intestinal proliferation. Mol Cell Biol. 18:6063–6074.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cinque L, Forrester A, Bartolomeo R,
Svelto M, Venditti R, Montefusco S, Polishchuk E, Nusco E, Rossi A,
Medina DL, et al: FGF signalling regulates bone growth through
autophagy. Nature. 528:272–275. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu Z, Lavine KJ, Hung IH and Ornitz DM:
FGF18 is required for early chondrocyte proliferation, hypertrophy
and vascular invasion of the growth plate. Dev Biol. 302:80–91.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hung IH, Schoenwolf GC, Lewandoski M and
Ornitz DM: A combined series of Fgf9 and Fgf18 mutant
alleles identifies unique and redundant roles in skeletal
development. Dev Biol. 411:72–84. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dahlberg LE, Aydemir A, Muurahainen N,
Gühring H, Edebo Fredberg H, Krarup-Jensen N, Ladel CH and Jurvelin
JS: A first-in-human, double-blind, randomised, placebo-controlled,
dose ascending study of intra-articular rhFGF18 (sprifermin) in
patients with advanced knee osteoarthritis. Clin Exp Rheumatol.
34:445–450. 2016.PubMed/NCBI
|
|
11
|
Itoh N, Nakayama Y and Konishi M: Roles of
FGFs as paracrine or endocrine signals in liver development,
health, and disease. Front Cell Dev Biol. 4:302016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Davidson D, Blanc A, Filion D, Wang H,
Plut P, Pfeffer G, Buschmann MD and Henderson JE: Fibroblast growth
factor (FGF) 18 signals through FGF receptor 3 to promote
chondrogenesis. J Biol Chem. 280:20509–20515. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Koneczny I, Schulenburg A, Hudec X,
Knöfler M, Holzmann K, Piazza G, Reynolds R, Valent P and Marian B:
Autocrine fibroblast growth factor 18 signaling mediates
Wnt-dependent stimulation of CD44-positive human colorectal adenoma
cells. Mol Carcinog. 54:789–799. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
El-Gendi S, Abdelzaher E, Mostafa MF and
Sheasha GA: FGF18 as a potential biomarker in serous and mucinous
ovarian tumors. Tumour Biol. 37:3173–3183. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wei W, Mok SC, Oliva E, Kim SH, Mohapatra
G and Birrer MJ: FGF18 as a prognostic and therapeutic biomarker in
ovarian cancer. J Clin Invest. 123:4435–4448. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sonvilla G, Allerstorfer S, Stättner S,
Karner J, Klimpfinger M, Fischer H, Grasl-Kraupp B, Holzmann K,
Berger W, Wrba F, et al: FGF18 in colorectal tumour cells:
Autocrine and paracrine effects. Carcinogenesis. 29:15–24. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Flannery CA, Fleming AG, Choe GH, Naqvi H,
Zhang M, Sharma A and Taylor HS: Endometrial cancer-associated
FGF18 expression is reduced by bazedoxifene in human endometrial
stromal cells in vitro and in murine endometrium. Endocrinology.
157:3699–3708. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Forner A: Hepatocellular carcinoma
surveillance with miRNAs. Lancet Oncol. 16:743–745. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hayes CN and Chayama K: MicroRNAs as
biomarkers for liver disease and hepatocellular carcinoma. Int J
Mol Sci. 17:2802016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shen S, Lin Y, Yuan X, Shen L, Chen J,
Chen L, Qin L and Shen B: Biomarker microRNAs for diagnosis,
prognosis and treatment of hepatocellular carcinoma: A functional
survey and comparison. Sci Rep. 6:383112016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee
DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Soifer HS, Rossi JJ and Saetrom P:
MicroRNAs in disease and potential therapeutic applications. Mol
Ther. 15:2070–2079. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Baker M: RNA interference: MicroRNAs as
biomarkers. Nature. 464:12272010. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cui L, Zhang X, Ye G, Zheng T, Song H,
Deng H, Xiao B, Xia T, Yu X, Le Y, et al: Gastric juice MicroRNAs
as potential biomarkers for the screening of gastric cancer.
Cancer. 119:1618–1626. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li HC, Chen YF, Feng W, Cai H, Mei Y,
Jiang YM, Chen T, Xu K and Feng DX: Loss of the Opa interacting
protein 5 inhibits breast cancer proliferation through
miR-139-5p/NOTCH1 pathway. Gene. 603:1–8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Krowiorz K, Ruschmann J, Lai C, Ngom M,
Maetzig T, Martins V, Scheffold A, Schneider E, Pochert N, Miller
C, et al: MiR-139-5p is a potent tumor suppressor in adult acute
myeloid leukemia. Blood Cancer J. 6:e5082016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yonemori M, Seki N, Yoshino H, Matsushita
R, Miyamoto K, Nakagawa M and Enokida H: Dual tumor-suppressors
miR-139-5p and miR-139-3p targeting matrix
metalloprotease 11 in bladder cancer. Cancer Sci.
107:1233–1242. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu H, Yin Y, Hu Y, Feng Y, Bian Z, Yao S,
Li M, You Q and Huang Z: miR-139-5p sensitizes colorectal cancer
cells to 5-fluorouracil by targeting NOTCH-1. Pathol Res Pract.
212:643–649. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang P, Xi J and Liu S: MiR-139-3p
induces cell apoptosis and inhibits metastasis of cervical cancer
by targeting NOB1. Biomed Pharmacother. 83:850–856. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sun C, Sang M, Li S, Sun X, Yang C, Xi Y,
Wang L, Zhang F, Bi Y, Fu Y, et al: Hsa-miR-139-5p inhibits
proliferation and causes apoptosis associated with down-regulation
of c-Met. Oncotarget. 6:39756–39792. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Q, Liang X, Wang Y, Meng X, Xu Y, Cai
S, Wang Z, Liu J and Cai G: miR-139-5p inhibits the
epithelial-mesenchymal transition and enhances the chemotherapeutic
sensitivity of colorectal cancer cells by downregulating BCL2. Sci
Rep. 6:271572016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen H, Xu H, Meng YG, Zhang Y, Chen JY
and Wei XN: miR-139-5p regulates proliferation, apoptosis, and cell
cycle of uterine leiomyoma cells by targeting TPD52. Onco Targets
Ther. 9:6151–6160. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dai S, Wang X, Li X and Cao Y:
MicroRNA-139-5p acts as a tumor suppressor by targeting ELTD1 and
regulating cell cycle in glioblastoma multiforme. Biochem Biophys
Res Commun. 467:204–210. 2015. View Article : Google Scholar : PubMed/NCBI
|