Introduction
Esophageal cancer is a highly lethal malignancy. As
the main type of esophageal cancer, esophageal squamous cell
carcinoma (ESCC) accounts for ~90% of all esophageal cancer cases
(1,2). However, patients with ESCC are usually
diagnosed at advanced stages (3).
Recently, advances in the application of combined chemotherapy and
radiotherapy, alone or as an adjunct regimen to surgery, have
improved the prognosis of ESCC patients (4). Cumulative evidence suggests that a
number of oncogenic and tumor-suppressive genes are associated with
the initiation and progression of ESCC (3). Yet, the molecular mechanisms
underlying the deregulation of the cellular phenotype in ESCC have
not been fully clarified. Elucidation of the genetic alterations
and underlying molecular pathways involved in ESCC may facilitate
the identification of novel targets, as well as improve disease
diagnosis and therapy.
MicroRNAs (miRNAs), a kind of endogenous RNA gene
products consisting of 18–25 nucleotides, have been identified as
important regulators of human malignancies (5). Recently, using detection techniques
such as RNA sequencing and microarray, there are various findings
that have illustrated the expression profile of miRNAs in ESCC.
Moreover, research concerning miRNAs and their target genes and the
molecular mechanisms involved in the carcinogenesis of ESCC
suggests the enormous therapeutic-clinical potential of miRNAs
(6). miR-25 is one of the oncogenes
has been reported to be upregulated in several cancers such as
hepatocellular carcinoma (7), lung
(8), prostate (9) and gastric cancer (10). In addition, plasma miRNA profiles
have revealed miR-25 as a novel diagnostic and monitoring biomarker
in ESCC (11). Desmocollin 2
(DSC2), a desmosomal cadherin protein which promotes cell
aggressiveness by redistributing adherens junctions and activating
β-catenin signaling in ESCC, has been identified as a downstream
target of miR-25 (12). However,
the expression profile of miR-25 in ESCC tissues, its specific role
and target in ESCC, have been scarcely studied. It has been
previously reported that the F-box and WD repeat domain-containing
7 (FBXW7) protein is a target of miR-25 in multiple human
malignancies (13–15), and it has been confirmed that FBXW7,
a tumor-suppressor, is frequently inactivated or deleted in ESCC
(16). However, whether miR-25
targets FBXW7 in ESCC, thus implicated in cancer development,
remains unknown.
In the present study, we clarified the clinical
significance of miR-25 expression in 75 pairs of tumor tissues from
ESCC patients. miR-25 expression in tissues was detected using
quantitative reverse transcription-polymerase chain reaction
(qRT-PCR). In addition, we investigated the potential relationship
between miR-25 levels and clinicopathological factors and overall
survival (OS) of ESCC. We also aimed to ascertain whether miR-25
influences the malignant behaviors of ESCC cells by in vitro
experiments. Finally, we determined whether miR-25 targets FBXW7
protein in ESCC.
Materials and methods
Clinical sample collection
We retrospectively enrolled 75 patients (aged 34–82
years; mean, 60.85±10.79 years) diagnosed with primary ESCC who
underwent surgical treatment, without preoperative local or
systemic treatment, at the Affiliated Hospital of Jiangsu
University, between May 2007 and May 2010. All resected ESCC
tissues and corresponding non-cancerous tissues were immediately
frozen and stored in liquid nitrogen until use. Medical records
were used to ascertain the patient medical histories, including
age, sex, recurrence and pathological information such as tumor
size, differentiation, lymph node metastasis, vascular and neural
invasion, and tumor-node-metastasis (TNM) stage. The
characteristics of all patients are listed in Table I. The present study was approved by
the Clinical Research Ethics Committee of the Affiliated Hospital
of Jiangsu University, and informed consent was obtained from all
patients and controls.
 | Table I.The relationship between miR-25
expression and clinicopathological parameters. |
Table I.
The relationship between miR-25
expression and clinicopathological parameters.
| Characteristics | Total (n=75) | Low expression | High expression | P-value |
|---|
| Age (years) |
| 62.73±9.70 | 60.85±10.79 | 0.219 |
| Sex |
|
|
| 0.817 |
| Male | 60 | 30 | 30 |
|
|
Female | 15 | 7 | 8 |
|
|
Differentiation |
|
|
| 0.176 |
|
Well | 18 | 6 | 12 |
|
|
Moderate, poor | 57 | 31 | 26 |
|
| Tumor size
(cm) |
|
|
| 0.491 |
|
≤4.0 | 39 | 21 | 18 |
|
|
>4.0 | 36 | 16 | 20 |
|
| Depth of tumor
invasion |
|
|
| 0.032a |
| T1 | 18 | 13 | 5 |
|
|
T2/3 | 57 | 24 | 33 |
|
| Lymph node
metastasis |
|
|
| 0.338 |
|
Absent | 26 | 15 | 11 |
|
|
Present | 49 | 22 | 27 |
|
| Vascular
invasion |
|
|
| 0.211 |
|
Absent | 36 | 20 | 16 |
|
|
Present | 39 | 17 | 22 |
|
| Neural
invasion |
|
|
| 0.357 |
|
Absent | 34 | 19 | 15 |
|
|
Present | 41 | 18 | 23 |
|
| TNM stage |
|
|
| 0.036a |
|
I+II | 33 | 20 | 13 |
|
|
III | 42 | 1 | 25 |
|
Quantitative real-time polymerase
chain reaction (qRT-PCR)
Total RNA was extracted from tissues and cells using
TRIzol® reagent (Invitrogen, Carlsbad, CA, USA). The
reverse transcription (RT) reaction was carried out using the
TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems,
Foster City, CA, USA). RT reaction was processed at 16°C for 30
min, 42°C for 30 min, and 85°C for 5 min. Gene expression levels
were quantified using the ABI 7900 Real-Time PCR system (Applied
Biosystems) at 95°C for 10 min, followed by 40 cycles of 95°C for
15 sec, and 60°C for 60 sec. The relative levels of miR-25
expression were calculated from the relevant signals by
normalization with the signal of RNU6B expression. PCR reactions of
each sample were conducted in triplicate. The 2−ΔΔCt
method was used to quantify the relative levels of gene
expression.
Cell lines and transfection
The human esophageal epithelial cell line (SHEE) and
malignant esophageal carcinoma cell lines (KYSE150 and KYSE450)
were purchased from the Cell Bank of Central South University. All
cells were cultured in RPMI-1640 medium (Invitrogen) supplemented
with 10% FBS (Gibco, Carlsbad, CA, USA), 100 U/ml penicillin and
100 mg/ml streptomycin (Invitrogen). Cells were incubated at 37°C
and supplemented with 5% CO2 in a humidified
chamber.
Reagents and cell transfection
Lipofectamine 3000 transfection reagent (Invitrogen)
was used. The miR-25 and FBXW7 inhibitor and negative control were
purchased from Invitrogen (Shanghai, China). Cells were seeded in
96- or 6-well plates 24 h before the experiment. For luciferase
reporter assay, the 3-untranslated region (3′UTR) of FBXW7 which
contains putative binding sites of miR-25 was amplified by PCR and
subcloned into the psiCHECK-2 vector within XhoI and
NotI restriction sites (Promega, Madison, WI, USA) as
previously described (13).
Cell migration and invasion
assays
Cell migration and invasion assays were carried out
using Transwell assay. In brief, transfected cells were harvested,
suspended (4×104/well) in 500 µl serum-free medium and
then loaded onto the upper compartment of the chamber, and 500 µl
of complete medium was added to the lower compartment. For the
invasion assay, inserts containing 8-µm pore filters were coated
with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). After a
24-h incubation, cells were fixed with methanol at 4°C for 0.5 h.
Non-invaded cells were removed from the upper surface of the filter
carefully with a cotton swab. Invaded cells on the lower side of
the filter were stained using Giemsa for 2 h at room temperature
and counted from 10 random fields under a microscope at a
magnification of ×200.
Luciferase reporter assay
For luciferase reporter assay, KYSE150 and KYSE450
cells were grown in 24-well plates and co-transfected with the
miR-25 inhibitor or control (NC-inhibitor) and the full length
3′UTR of FBXW7. After transfection for 48 h, the cells were
collected and the relative luciferase activity was measured using
the Dual-Luciferase Reporter Assay System (Promega). The experiment
was independently repeated three times.
Statistical analysis
Each experiment was performed in triplicate. All
statistical analyses were performed using SPSS 20.0 statistical
software (SPSS, Inc., Chicago, IL, USA). Differences between groups
were evaluated by the Student's t-test for continuous variables and
χ2 test for categorical variables. For OS mortality,
only patients who specifically died from ESCC, and not as the
result of a different disease, were included. The associations
between miR-25 expression and prognosis of patients were analyzed
using the Kaplan-Meier method, and differences in survival were
estimated using the log-rank test. Prognostic factors were examined
by univariate and multivariate analyses (Cox proportional hazards
regression model). P-value <0.05 was considered statistically
significant.
Results
Overexpression of miR-25 in human ESCC
tissues
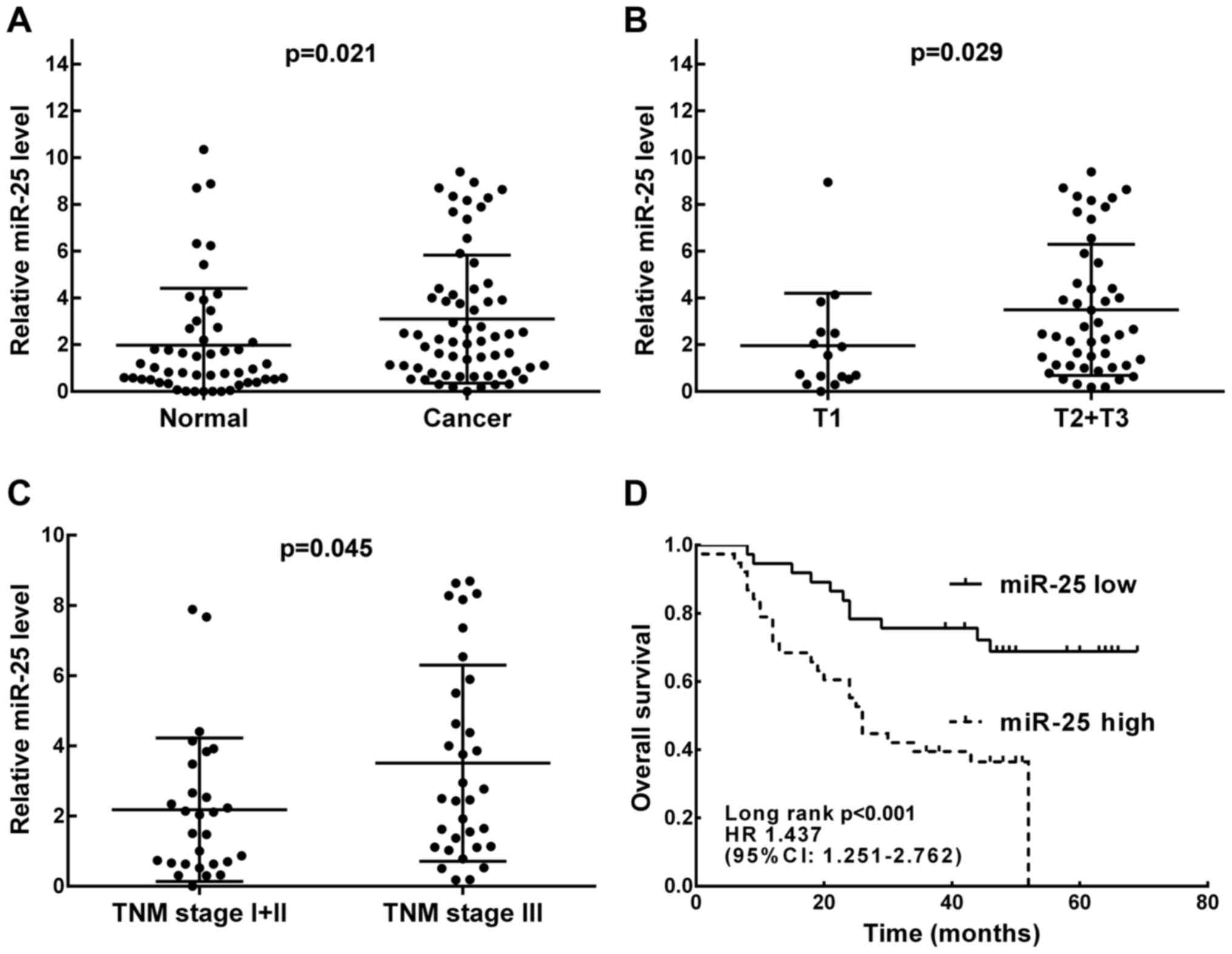
We detected the expression of miR-25 in tumors and
corresponding adjacent normal tissues using qRT-PCR assay. As shown
in Fig. 1A, the expression of
miR-25 in ESCC tissues was upregulated in comparison with that in
the adjacent normal tissues. Moreover, miR-25 expression levels
were further analyzed in cancer tissue samples with different tumor
depth and TNM stage. As shown in Fig.
1B and C, the expression of miR-25 was obviously higher in
patients with deeper tumor invasion (T2-3, P=0.029) and higher TNM
stage (stage III–IV, P=0.045).
Correlations between miR-25 expression
and clinicopathologic features of ESCC patients
To further investigate the correlation between
miR-25 expression and clinicopathologic features, using the median
value of miR-25 expression level in all ESCC tissues as the cut-off
value, all 75 patients were divided into a low-miR-25 expression
group (n=38) and a high-miR-25 expression group (n=37). As shown in
Table I, high miR-25 expression was
closely related to depth of tumor invasion (P=0.032) and TNM stage
(P=0.036). However, no relationship was found between miR-25 and
other clinicopathologic features including age, sex, tumor size,
lymph node metastasis, vascular and neural invasion, or tumor
differentiation (all P>0.05).
Correlation of miR-25 expression and
survival of ESCC patients
To further investigate the correlation of miR-25
expression with the survival of ESCC patients, Kaplan-Meier curve
with long-rank analysis were performed. As shown in Fig. 1D, the OS rates in the high and low
miR-25 expression group were 35.1 and 68.4%, respectively
(P<0.001). Univariate analysis of OS revealed that depth of
tumor invasion (P=0.016), lymph node metastasis (P=0.003), neural
invasion (P=0.024), TNM stage (P=0.001) and miR-25 expression
(P=0.002) were all factors associated with prognosis. Further
multivariate analysis results showed that in addition to TNM stage
(HR, 3.530; 95% CI, 1.213–5.270; P=0.004), miR-25 expression was
also an independent predictor of OS (HR, 2.528; 95% CI,
1.513–8.241; P=0.013) for ESCC patients (Table II).
 | Table II.Univariate and multivariate analyses
of different prognostic factors for OS in 75 patients with
ESCC. |
Table II.
Univariate and multivariate analyses
of different prognostic factors for OS in 75 patients with
ESCC.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Prognostic
factors | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (years) |
|
|
|
|
|
|
|
>60/≤60 | 0.987 | 0.958–1.017 | 0.383 |
|
|
|
| Sex |
|
|
|
|
|
|
|
Male/female | 1.133 | 0.516–2.488 | 0.756 |
|
|
|
| Tumor size |
|
|
|
|
|
|
|
≤4/>4 cm | 1.374 | 1.712–2.649 | 0.343 |
|
|
|
| Depth of tumor
invasion |
|
|
|
|
|
|
|
T1/T2+T3 | 4.304 | 1.318–14.056 | 0.016a |
|
|
|
|
Differentiation |
|
|
|
|
|
|
|
Poor/well + moderate | 1.404 | 0.690–2.856 | 0.349 |
|
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
|
Presence/absence | 3.749 | 1.554–9.042 | 0.003a |
|
|
|
| Vascular
invasion |
|
|
|
|
|
|
|
Presence/absence | 1.595 | 0.815 −3.121 | 0.173 |
|
|
|
| Neural
invasion |
|
|
|
|
|
|
|
Presence/absence | 2.270 | 1.112–4.635 | 0.024a |
|
|
|
| TNM stage |
|
|
|
|
|
|
|
I+II/III | 4.232 | 1.841–9.731 | 0.001a | 3.530 | 1.213–5.270 | 0.004a |
| miR-25
expression |
|
|
|
|
|
|
|
High/low | 3.158 | 1.539–6.480 | 0.002a | 2.528 | 1.513–8.241 | 0.013a |
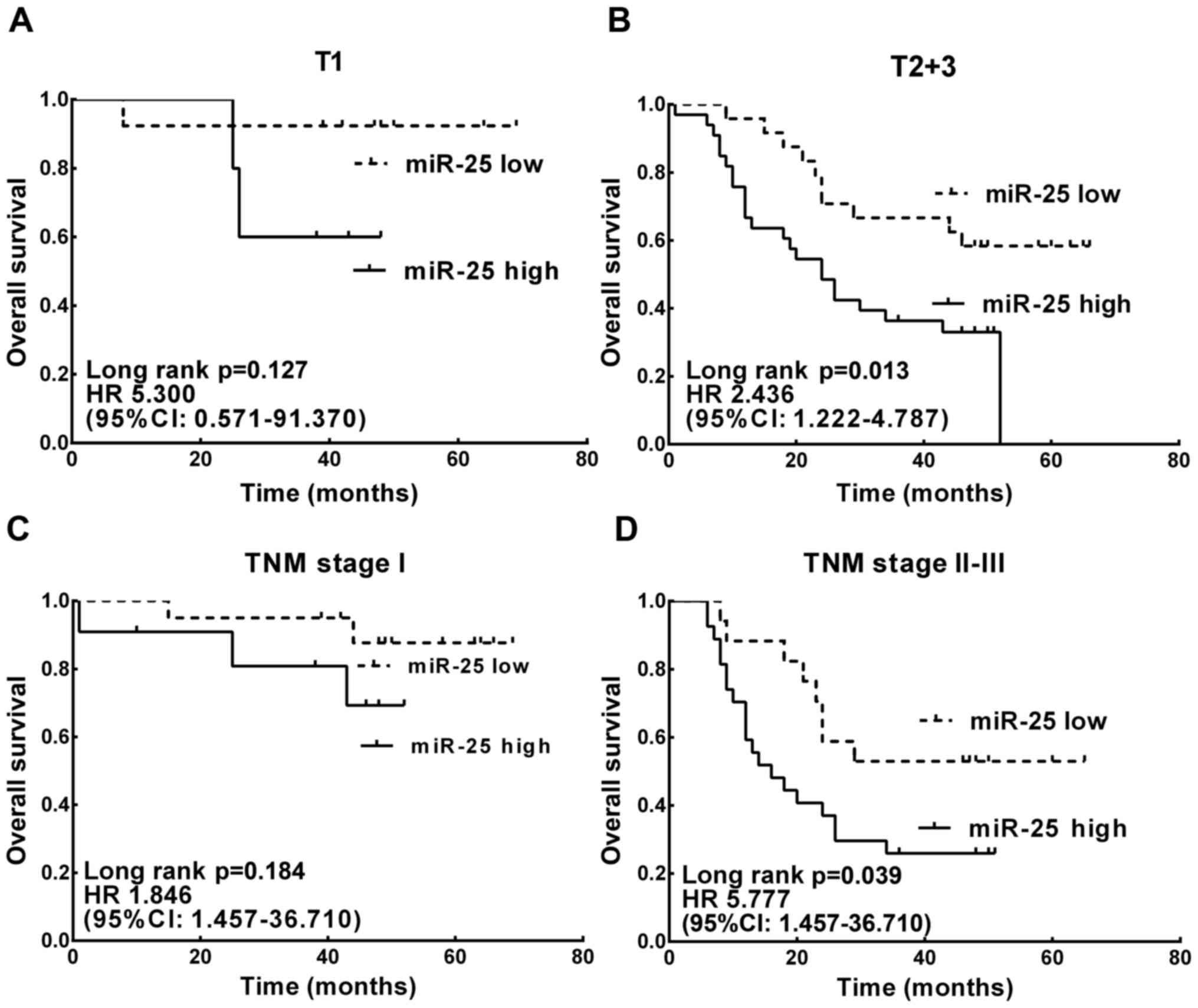
The prognostic value of miR-25 expression in patient
subgroups classified by depth of tumor invasion and stage was also
evaluated. High miR-25 expression was found significantly
correlated with worse survival in patients with deeper tumor depth
(T2-3) (HR, 2.436; 95% CI, 1.222–4.787; P=0.013; Fig. 2B) and advanced TNM stage (stage
III–IV; HR, 5.777; 95% CI, 1.457–36.710; P=0.017; Fig. 2D).
Knockdown of miR-25 suppresses ESCC
cell migration and invasion
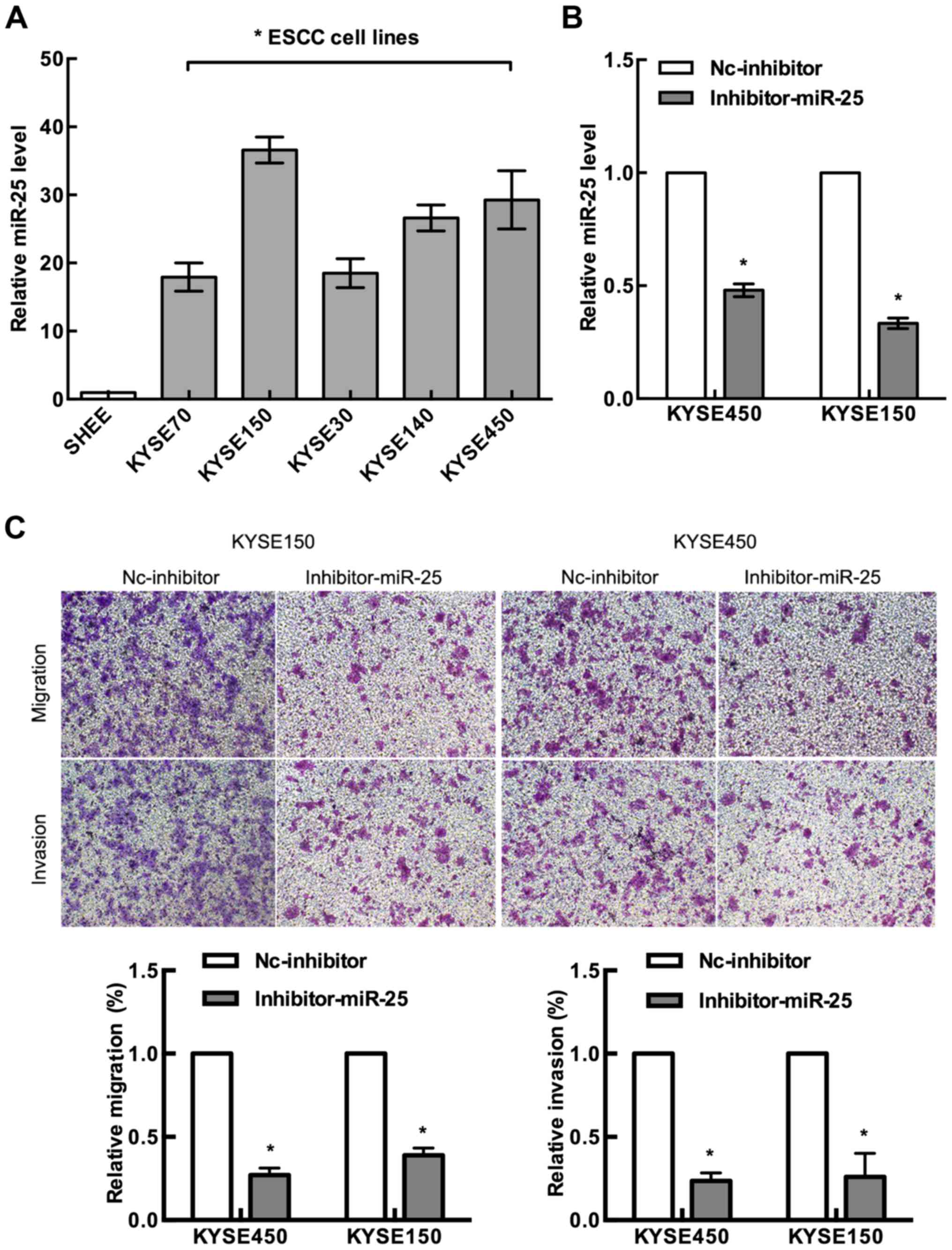
As shown in Fig. 3A,
expression levels of miR-25 were investigated in six human ESCC
cell lines and human esophageal epithelial cell line (SHEE), and
miR-25 was found to be significantly upregulated in ESCC cells as
compared with that in the normal human esophageal epithelial cell
line (P<0.05). To explore the functions of miR-25 in ESCC cell
lines, we knocked down miR-25 by transfecting human ESCC cell lines
KYSE150 and KYSE450 with the miR-25 inhibitor (Fig. 3B).
To investigate whether the decreased expression of
miR-25 affects the migration and invasion of ESCC cells, we
examined the migration rate of the miR-25 inhibitor-transfected
KYSE150 and KYSE450 cells by Transwell assay. Our results revealed
that both cell lines had a marked decrease in cell migration
compared with their control counterparts (Fig. 3C). We also examined the ability of
the miR-25-inhibitor-transfected cells to invade the Matrigel
matrix using the Boyden chamber assay. We found that knockdown of
miR-25 significantly reduced both KYSE150 and KYSE450 cell invasion
compared with the control group (Fig.
3C). These data suggest that miR-25 upregulation may be an
important event in the metastasis of ESCC cells.
Knockdown of miR-25 upregulates the
expression of FBXW7 in ESCC cells
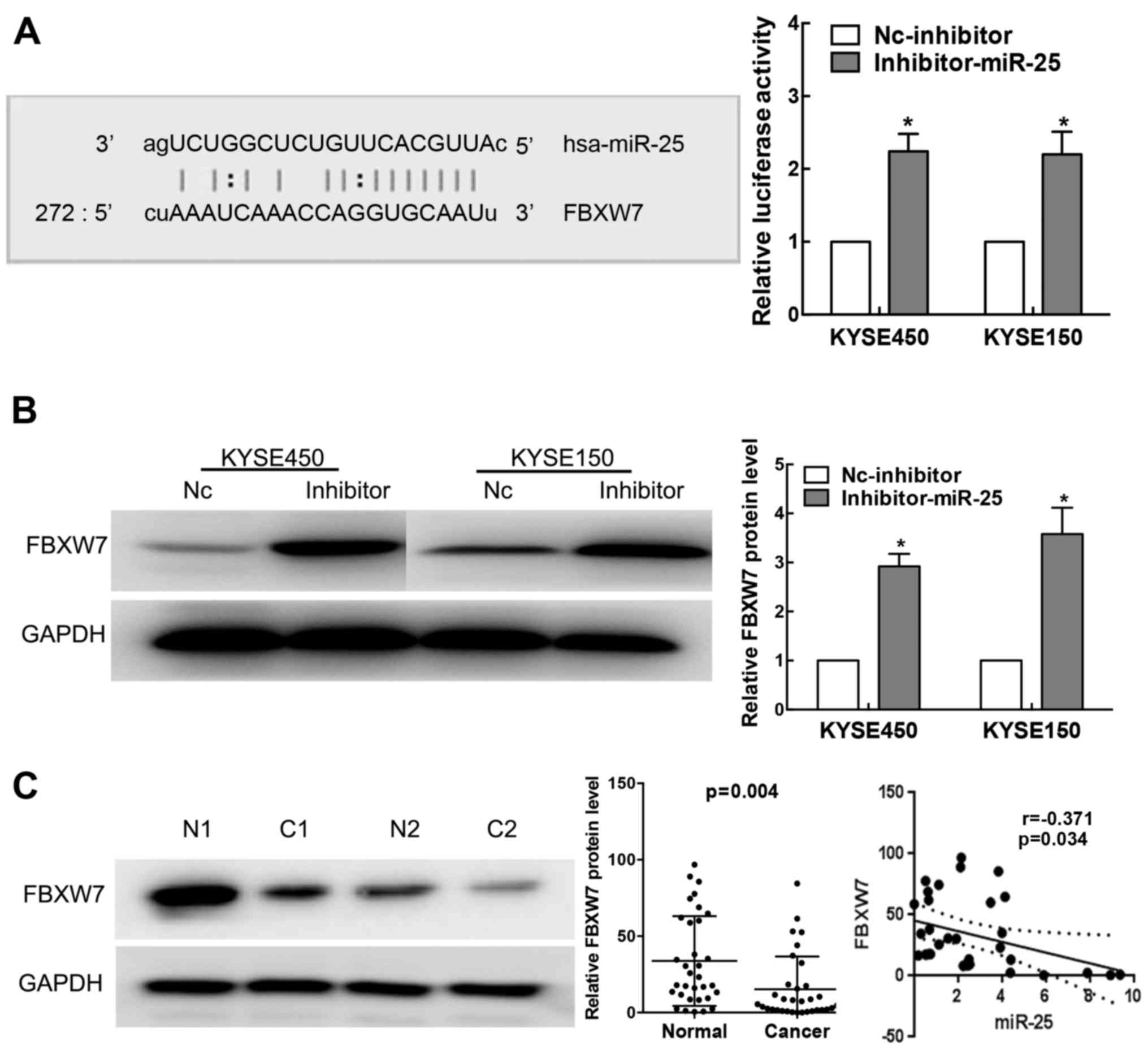
The TargetScan database identified the putative
target site of miR-25 in the 3′UTR of FBXW7 (Fig. 4A left). To investigate whether
miR-25 is implicated in human ESCC by targeting FBXW7, we cloned
the full length 3′UTR of FBXW7 into luciferase reporter vectors and
conducted luciferase activity assay. Our results showed that
luciferase activity was significantly increased by the transfection
of miR-25 inhibitor in KYSE150 and KYSE450 cells (Fig. 4A right; P<0.01).
We also detected the expression level of FBXW7 in
human ESCC cells transfected with the miR-25 inhibitor or negative
control using western blot analysis. Our results showed that miR-25
knockdown led to marked increases in FBXW7 protein expression in
the KYSE150 and KYSE450 cells (Fig.
4B). We further analyzed the expression of FBXW7 protein in
ESCC patient tissues by western blotting. As shown in Fig. 4C, FBXW7 was significantly decreased
in our ESCC tissues when compared with that in the normal tissues
(P=0.004). Moreover, by Pearson correlation analysis, we found that
the expression level of miR-25 was negatively correlated with FBXW7
(r=-0.371; P=0.034) protein levels (Fig. 4C).
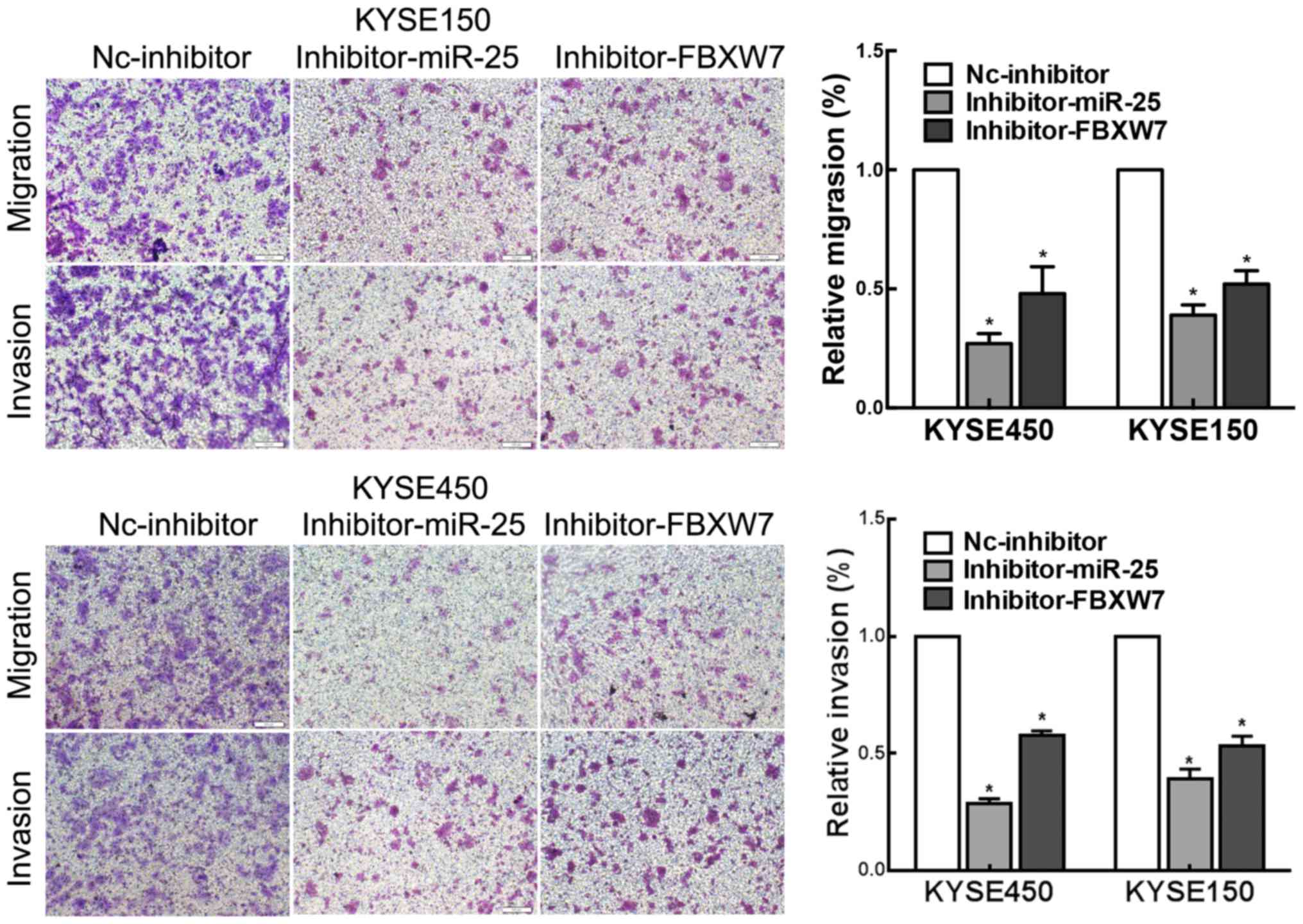
FBXW7-knockdown partially rescues the
effect of downregulated miR-25 in ESCC cells
Finally, we conducted in vitro experiments to
investigate whether FBXW7 functions in a miR-25-mediated manner in
ESCC. Transwell assays showed that FBXW7-knockdown partially
rescued the effects of the downregulation of miR-25 on ESCC cell
migration and invasion compared with these parameters observed in
the controls (Fig. 5). Our results
indicated that miR-25 modulates ESCC cell migration and invasion in
a FBXW7-mediated manner.
Discussion
Esophageal squamous cell carcinoma (ESCC) is one of
the most common malignant tumors, with an overall 5-year survival
rate of only ~10–20% in the Chinese population (17). In recent years, a large number of
miRNAs have been detected to be dysregulated in ESCC and play
important roles in ESCC (6,18,19).
The present study demonstrated that the expression
of miR-25 was significantly overexpressed in human ESCC tissues. It
also showed for the first time that miR-25 was obviously increased
in patients with deeper tumor invasion (T2-3) and higher TNM stage
(stage III and IV), which indicates that miR-25 may act as an
oncogene in ESCC. In vitro functional experiments
demonstrated that miR-25 was associated with ESCC cell migration
and invasion. Further experiments identified that FBXW7 is a direct
target gene of miR-25. In addition, the expression level of miR-25
was negatively correlated with FBXW7 protein levels, indicated that
FBXW7 may act as a downstream regulator of miR-25 in ESCC. In
brief, the present study showed that miR-25 may play an important
role in the carcinogenesis and development of human ESCC.
miR-25, a member of the miR-25-93-106 family, is
located at human 7q22.1. It has been reported to be overexpressed
in a variety of human malignancies, such as hepatocellular
carcinoma (7), lung (8), prostate (9) and gastric cancer (10), and is therefore considered to be an
oncogene. Komatsu et al (11) investigated miR-25 expression in ESCC
tissues and patient plasma and found that the relative expression
of miR-25 was significantly higher in ESCC tissues and cell lines
than in normal tissues and fibroblasts. In addition, the level of
plasma miR-25 in ESCC patients was significantly reduced in
postoperative samples than in preoperative samples and was
significantly increased during tumor recurrences. Meanwhile, serum
miR-25 was found to be an independent risk factor for OS in ESCC
(20). All these results indicate
that miR-25 is a valuable biomarker for reflecting the tumor
dynamics of ESCC. However, to date, the molecular mechanism
underlying the oncogenic functions of miR-25 in ESCC remain
elusive.
As the substrate recognition component of an
evolutionarily conserved SCF (complex of SKP1, CUL1 and F-box
protein)-type ubiquitin ligase complex, FBXW7 coordinates the
ubiquitin-dependent proteolysis of several critical cellular
regulators, thereby controlling essential processes, such as cell
cycle, differentiation and apoptosis (21). Accumulating research indicates that
FBXW7 is a vital regulator that contributes to human tumorigenesis,
as FBXW7 regulates the stability of multiple oncoprotein
substrates, including cyclin E, c-Myc, Notch, c-Jun, mammalian
target of rapamycin and MCL1, thus contributing to the
carcinogenesis and development of cancer (22–26).
In addition, FBXW7 is a p53-dependent tumor suppressor and its
activation by p53 results in ubiquitination-mediated suppression of
several oncoproteins (27). In the
present study, FBXW7 protein expression was significantly reduced
in ESCC tissues, and thus FBXW7 may be a tumor suppressor in
ESCC.
FBXW7 has been shown to be a target of
cancer-related miRNAs in ESCC. Kurashige et al found that
miR-223 is an miRNA significantly upregulated in ESCC tissues, and
its overexpression confers a poor prognosis. Moreover, miR-223 may
downregulate cell cycle regulators c-Myc and c-Jun proteins by
targeted FBXW7 (28). Meanwhile,
previous studies also illustrated that miR-25 exhibits its
oncogenic role in different cancers via targeting different
proteins, such as LATS2 in gastric cancer (29), and CDC42 and BTG2 in NSCLC (9,30,31).
It is well known that one miRNA can exhibit its function in a
specific tumor by target multiple genes, and one protein-coding
gene can be targeted by multiple miRNAs in a specific tumor; it is
not the one miRNA-one mRNA relationship (32). Although whether or not miR-25
regulates FBXW7 expression in ESCC has not been reported, several
studies have identified FBXW7 as a target gene of miR-25. miR-25
can promote cell proliferation by repressing FBXW7 expression in
non-small cell lung and gastric cancer, and prostatic small cell
neuroendocrine carcinoma (13–15).
In the present study, by luciferase reporter analysis, miR-25 was
able to bind to the 3′UTR of FBXW7 in ESCC cells. Moreover,
knockdown of miR-25 upregulated FBXW7 protein expression. In
addition, our western blot analysis results showed a significant
increase in FBXW7 protein in the ESCC KYSE150 and KYSE450 cells
transfected with the miR-25 inhibitor. These results provide solid
evidence that one miRNA can be involved in specific malignancies by
targeting different genes.
In conclusion, our results showed that the
expression level of miR-25 was significantly upregulated in ESCC
tissues, and high miR-25 expression was associated with tumor depth
and TNM stage. In addition, upregulation of miR-25 was an
unfavorable prognostic factor for OS of ESCC. These findings
suggest that miR-25 may be a novel prognostic indicator in ESCC,
particularly for patients with deeper tumor invasion and advanced
TNM stage, and a potential target for diagnosis and gene therapy.
Furthermore, miR-25 downregulated the expression of ESCC-related
tumor-suppressor FBXW7, and knockdown of miR-25 inhibited ESCC
migration and invasion. Our results showed that understanding the
complex regulation of miR-25 with regard to its target gene
expression in ESCC may be valuable for exploring potential
therapeutic methods for ESCC, and gene therapy targeting miR-25
should be further investigated as a potential alternative strategy
for ESCC therapy.
Acknowledgements
The present study was funded by the Postgraduate
Research and Innovation Project of Jiangsu Province (grant no.
CXZZ12_0708).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mariette C, Piessen G and Triboulet JP:
Therapeutic strategies in oesophageal carcinoma: Role of surgery
and other modalities. Lancet Oncol. 8:545–553. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hammond SM: An overview of microRNAs. Adv
Drug Deliv Rev. 87:3–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chu Y, Zhu H, Lv L, Zhou Y and Huo J:
MiRNA s in oesophageal squamous cancer. Neth J Med. 71:69–75.
2013.PubMed/NCBI
|
|
7
|
Wang C, Wang X, Su Z, Fei H, Liu X and Pan
Q: MiR-25 promotes hepatocellular carcinoma cell growth, migration
and invasion by inhibiting RhoGDI1. Oncotarget. 6:36231–36244.
2015.PubMed/NCBI
|
|
8
|
Wu T, Chen W, Kong D, Li X, Lu H, Liu S,
Wang J, Du L, Kong Q, Huang X, et al: miR-25 targets the modulator
of apoptosis 1 gene in lung cancer. Carcinogenesis. 36:925–935.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zoni E, van der Horst G, van de Merbel AF,
Chen L, Rane JK, Pelger RC, Collins AT, Visakorpi T, Snaar-Jagalska
BE, Maitland NJ, et al: miR-25 modulates invasiveness and
dissemination of human prostate cancer cells via regulation of
αv- and α6-integrin expression. Cancer Res.
75:2326–2336. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li BS, Zuo QF, Zhao YL, Xiao B, Zhuang Y,
Mao XH, Wu C, Yang SM, Zeng H, Zou QM, et al: MicroRNA-25 promotes
gastric cancer migration, invasion and proliferation by directly
targeting transducer of ERBB2, 1 and correlates with poor survival.
Oncogene. 34:2556–2565. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Komatsu S, Ichikawa D, Hirajima S,
Kawaguchi T, Miyamae M, Okajima W, Ohashi T, Arita T, Konishi H,
Shiozaki A, et al: Plasma microRNA profiles: Identification of
miR-25 as a novel diagnostic and monitoring biomarker in
oesophageal squamous cell carcinoma. Br J Cancer. 111:1614–1624.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fang WK, Liao LD, Li LY, Xie YM, Xu XE,
Zhao WJ, Wu JY, Zhu MX, Wu ZY, Du ZP, et al: Down-regulated
desmocollin-2 promotes cell aggressiveness through redistributing
adherens junctions and activating beta-catenin signalling in
oesophageal squamous cell carcinoma. J Pathol. 231:257–270. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xiang J, Hang JB, Che JM and Li HC: MiR-25
is up-regulated in non-small cell lung cancer and promotes cell
proliferation and motility by targeting FBXW7. Int J Clin Exp
Pathol. 8:9147–9153. 2015.PubMed/NCBI
|
|
14
|
Lu D, Davis MP, Abreu-Goodger C, Wang W,
Campos LS, Siede J, Vigorito E, Skarnes WC, Dunham I, Enright AJ,
et al: MiR-25 regulates Wwp2 and Fbxw7 and promotes reprogramming
of mouse fibroblast cells to iPSCs. PLoS One. 7:e409382012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gong J, Cui Z, Li L, Ma Q, Wang Q, Gao Y
and Sun H: MicroRNA-25 promotes gastric cancer proliferation,
invasion, and migration by directly targeting F-box and WD-40
domain protein 7, FBXW7. Tumour Biol. 36:7831–7840. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang L, Ye X, Liu Y, Wei W and Wang Z:
Aberrant regulation of FBW7 in cancer. Oncotarget. 5:2000–2015.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao QY and Fang JY: Early esophageal
cancer screening in China. Best Pract Res Clin Gastroenterol.
29:885–893. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kong KL, Kwong DL, Chan TH, Law SY, Chen
L, Li Y, Qin YR and Guan XY: MicroRNA-375 inhibits tumour growth
and metastasis in oesophageal squamous cell carcinoma through
repressing insulin-like growth factor 1 receptor. Gut. 61:33–42.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ren LH, Chen WX, Li S, He XY, Zhang ZM, Li
M, Cao RS, Hao B, Zhang HJ, Qiu HQ, et al: MicroRNA-183 promotes
proliferation and invasion in oesophageal squamous cell carcinoma
by targeting programmed cell death 4. Br J Cancer. 111:2003–2013.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu C, Wang C, Guan X, Liu Y, Li D, Zhou X,
Zhang Y, Chen X, Wang J, Zen K, et al: Diagnostic and prognostic
implications of a serum miRNA panel in oesophageal squamous cell
carcinoma. PLoS One. 9:e922922014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cao J, Ge MH and Ling ZQ: Fbxw7 tumor
suppressor: A vital regulator contributes to human tumorigenesis.
Medicine. 95:e24962016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nakayama KI and Nakayama K: Ubiquitin
ligases: Cell-cycle control and cancer. Nat Rev Cancer. 6:369–381.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mao JH, Kim IJ, Wu D, Climent J, Kang HC,
DelRosario R and Balmain A: FBXW7 targets mTOR for degradation and
cooperates with PTEN in tumor suppression. Science. 321:1499–1502.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Welcker M and Clurman BE: FBW7 ubiquitin
ligase: A tumour suppressor at the crossroads of cell division,
growth and differentiation. Nat Rev Cancer. 8:83–93. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Inuzuka H, Shaik S, Onoyama I, Gao D,
Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, et al:
SCFFBW7 regulates cellular apoptosis by targeting MCL1
for ubiquitylation and destruction. Nature. 471:104–109. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wertz IE, Kusam S, Lam C, Okamoto T,
Sandoval W, Anderson DJ, Helgason E, Ernst JA, Eby M, Liu J, et al:
Sensitivity to antitubulin chemotherapeutics is regulated by MCL1
and FBW7. Nature. 471:110–114. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Perez-Losada J, Mao JH and Balmain A:
Control of genomic instability and epithelial tumor development by
the p53-Fbxw7/Cdc4 pathway. Cancer Res. 65:6488–6492. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kurashige J, Watanabe M, Iwatsuki M,
Kinoshita K, Saito S, Hiyoshi Y, Kamohara H, Baba Y, Mimori K and
Baba H: Overexpression of microRNA-223 regulates the ubiquitin
ligase FBXW7 in oesophageal squamous cell carcinoma. Br J Cancer.
106:182–188. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang M, Wang X, Li W and Cui Y: miR-107
and miR-25 simultaneously target LATS2 and regulate proliferation
and invasion of gastric adenocarcinoma (GAC) cells. Biochem Biophys
Res Commun. 460:806–812. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang T, Chen T, Li Y, Gao L, Zhang S, Wang
T and Chen M: Downregulation of miR-25 modulates non-small cell
lung cancer cells by targeting CDC42. Tumour Biol. 36:1903–1911.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He Z, Liu Y, Xiao B and Qian X: miR-25
modulates NSCLC cell radio-sensitivity through directly inhibiting
BTG2 expression. Biochem Biophys Res Commun. 457:235–241. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Peter ME: Targeting of mRNAs by multiple
miRNAs: The next step. Oncogene. 29:2161–2164. 2010. View Article : Google Scholar : PubMed/NCBI
|