Introduction
Huntington-interacting protein 1 (HIP1) was
originally characterized using the yeast two-hybrid system in 1997
(1). HIP1 and its only known
mammalian homolog HIP1-related (HIP1r) are involved in
neurodegeneration based on the finding that HIP1 interacts with the
Huntington protein, which is mutated in Huntington's disease, and
acts as a component of the endocytic machinery, binding to
clathrin, AP2 and actin (2–4).
The overexpression of HIP1 is correlated with brain
(5), colon (6), and breast cancers (7). Additionally, HIP1 overexpression in
glioblastoma and oligodendroglioma results in a prolonged half-life
of growth factor receptors, such as EGFR and PDGF-βR (5). EGFR and its downstream pathways are
important for tumor cell invasion and proliferation. The
degradation of EGFR is partially achieved by the internalization of
activated EGFR and its degradation in lysosomes (8,9).
The stabilization of EGFR levels by HIP1 in cancer
cells suggests that HIP1 is related to endocytosis and EGFR
degradation, but this relationship has not been definitively
established. In this study, the effect of HIP1 on the first step of
EGFR endocytosis and the mechanisms by which HIP1 mediates cancer
cell proliferation in an EGFR-dependent manner were examined.
Materials and methods
Materials
EGF and Alexa Fluor 647-EGF were purchased from
Sigma-Aldrich (St. Louis, MO, USA). Human PKR siRNA (SR303767) and
control siRNA (SR30004) were purchased from OriGene (Rockville, MD,
USA). The siRNAs against HIP1 were purchased from Ambion
(Foster City, CA, USA). Antibodies against clathrin and EGFR were
purchased from Abcam (Cambridge, UK). Tetramethylrhodamine
isothiocyanate-labeled affinity purified goat anti-mouse IgG and
goat anti-rabbit IgG (H+L) were purchased from KPL (Guildford, UK).
Electrochemiluminescence kit was bought for Boshide (Wuhan, China).
All other chemicals were purchased from Sigma-Aldrich.
Cell culture
Human hepatocarcinoma cells lines 7402, 7703, 7721,
Hep3B, HepG2, H460, SPCA1, SKOV-3, HeLa, and MCF-7 and glioma cells
U87, U251, C33a, PC-3, NCI-H1299, NCI-H446, and K562 were cultured
in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10%
fetal bovine serum, 2 mM L-glutamine, and 1X antibiotic-antimycotic
solution (15240-096; Invitrogen, Carlsbad, CA, USA) at 37°C in a
humidified atmosphere with 5% CO2.
siRNA and transfection
The sequences of siRNAs targeting human HIP1
were as follows: siRNA1, taattgagcgactatacagag; siRNA2,
acagcgatatagcaagctaaa; siRNA3, accgcttcatggag cagttta. These siRNAs
were designed using siRNA Target Finder developed by Ambion, Inc.
Cells were transfected with siRNA using Lipofectamine 2000
(11668-019; Invitrogen) according to the manufacturer's protocol.
Transfection was confirmed by an immunoblot analysis.
Western blot assay
Cells were collected, pelleted, and lysed in
ice-cold lysis buffer (25 mmol/l Tris-HCl, 1 mmol/l edetic acid,
150 mmol/l NaCl, 50 mmol/l NaF, 1% Triton-100, 1 mmol/l PMSF, 1
mg/l leupeptin, 1 µmol/l aprotinin, pH 7.6). Cell lysates were
centrifuged at 4°C and 12,000 g/min, rotated with polyclonal rabbit
anti-human antibody for 2 h, followed by precipitation with Protein
A Sepharose at 4°C overnight. Beads were washed five times with
cold wash buffer (20 mmol/l Tris, pH 7.8, 150 mmol/l NaCl, 1 mmol/l
EDTA, 0.1% Triton X-100, 100 µM PMSF and 1 mmol/l Na3VO4) and the
bound protein was eluted with Laemmli sample buffer and separated
by sodium dodecyl sulfate polyacrylamide gel electrophoresis
(SDS-PAGE). After SDS-PAGE, the protein was transferred to a
nitrocellulose membrane using a semidry transfer apparatus
(Bio-Rad, Hercules, CA, USA), blocked with 3% bovine serum albumin
in TBST [10 mmol/l Tris (pH 8.0), 150 mmol/l NaCl, 0.1% Tween-20],
incubated at room temperature for 1 h with rabbit anti-human HIP1,
EGFR, and GAPDH antibodies, rotated for 1 h at room temperature of
25°C, and washed with TBST three times. The protein on the
nitrocellulose membrane was then detected by
electrochemiluminescence using a kit with a goat anti-rabbit
antibody according to the manufacturer's instructions. Based on the
western blotting results, HeLa cells exhibited high coexpression of
HIP1 and EGFR and accordingly were used in subsequent
experiments.
EGF-EGFR internalization assay
HeLa cells were plated in 6-well dishes and grown to
~50% confluency. After cells were serum-starved for 2 h at 37°C,
they were incubated with serum-free DMEM containing Alexa Fluor
647-EGF at a final concentration of 1.5 ng/ml or 100 ng/ml for 1 h
at 4°C. Cells were then incubated at 37°C for 1 h to allow
internalization. EGFR internalization was stopped by placing the
cells at 4°C. The cells were then washed three times for 10 min
with PBS, fixed, and permeabilized with 4% paraformaldehyde and
0.1% Triton X-100 in PBS for 10 min. They were then washed with PBS
again at room temperature and visualized under a Zeiss LSM confocal
microscope (Oberkochen, Germany).
Colocalization assay
EGFR internalization experiments were carried out as
described previously. The cells were blocked with 5% normal goat
serum in PBS for 30 min, washed three times for 5 min each with
PBS, and then incubated with mouse anti-clathrin monoclonal
antibody (Abcam) diluted 1:50 in PBS for 3 h at room temperature.
After another three washes for 10 min each with PBS,
tetramethylrhodamine isothiocyanate-labeled affinity-purified goat
anti-mouse IgG and goat anti-rabbit IgG (H+L; KPL) diluted 1:50 in
PBS were added and incubated for 30 min. Following a final rinse
(3×10 min) with PBS, the cells were visualized under a Zeiss LSM
confocal microscope.
Image analysis
Images were collected using a Zeiss LSM510-Meta
laser scanning confocal microscope with a 63× water immersion
objective. Colocalization was calculated using ImageJ (NIH,
Bethesda, MD, USA) with the JACoP plug-in to estimate Manders
coefficients with automated thresholding. Statistical analyses were
implemented in GraphPad Prism (GraphPad, La Jolla, CA, USA).
Statistical analysis
Based on the image analysis data, differences among
treatment groups were examined by analysis of variance (ANOVA).
Data are represented as means ± SEM of three experiments. P<0.05
was considered statistically significant. When significant
differences were detected, specific post-hoc comparisons between
treatment groups were performed using Student-Newman-Keuls
tests.
Results
Expression of HIP1 and EGFR in tumor
cell lines
Using 17 cell lines, western blotting was performed
to identify cells with high expression levels of both HIP1 and
EGFR. As shown in Fig. 1, the 7703,
7721, SKOV-3, U251, and HeLa cell lines exhibited higher expression
levels of HIP1 than those of other cell lines. As shown in Fig. 2, the Hep3B, HeLa, PC-3, NCI-H1299,
and NCI-H466 cell lines had higher expression levels of EGFR than
those of other cell lines. Combining these results, the HeLa cell
line had high expression levels of both HIP1 and EGFR.
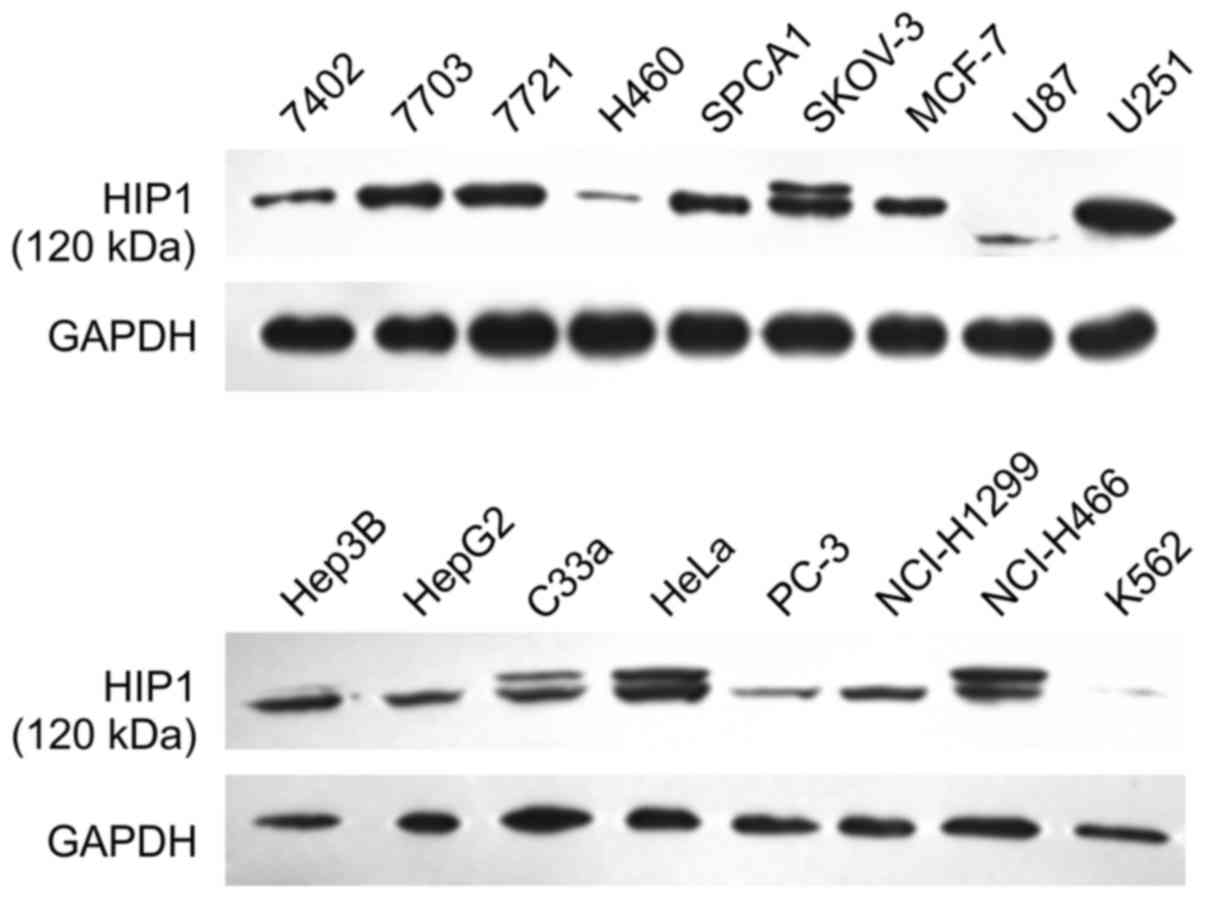 | Figure 1.HIP1 expression in seventeen cell
lines measured by western blotting. The cell lines were 7402, 7703,
7721, Hep3B, HepG2, H460, SPCA1, SKOV-3, HeLa, MCF-7, glioma cells
U87, U251, C33a, PC-3, NCI-H1299, NCI-H446, and K562 cells. 7703,
7721, SKOV-3, U251, and HeLa cells exhibited higher expression
levels of HIP1 than those of other cell lines. |
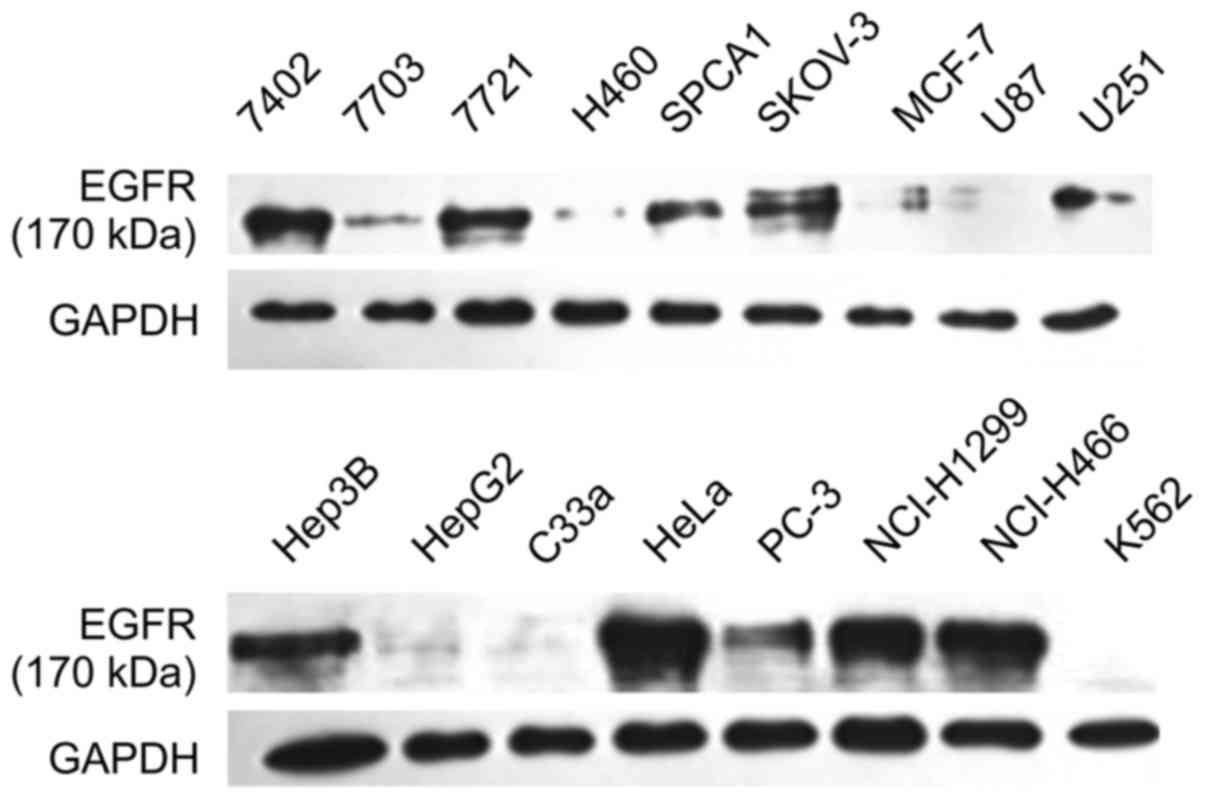 | Figure 2.EGFR expression in seventeen cell
lines measured by western blotting. The cell lines were 7402, 7703,
7721, Hep3B, HepG2, H460, SPCA1, SKOV-3, HeLa, MCF-7, glioma cells
U87, U251, C33a, PC-3, NCI-H1299, NCI-H446, and K562 cells. Hep3B,
HeLa, PC-3, NCI-H1299, and NCI-H466 cells exhibited higher
expression levels of EGFR than those of other cell lines. Combined
with the results presented in Fig.
1, HeLa cells had high expression of both HIP1 and EGFR. |
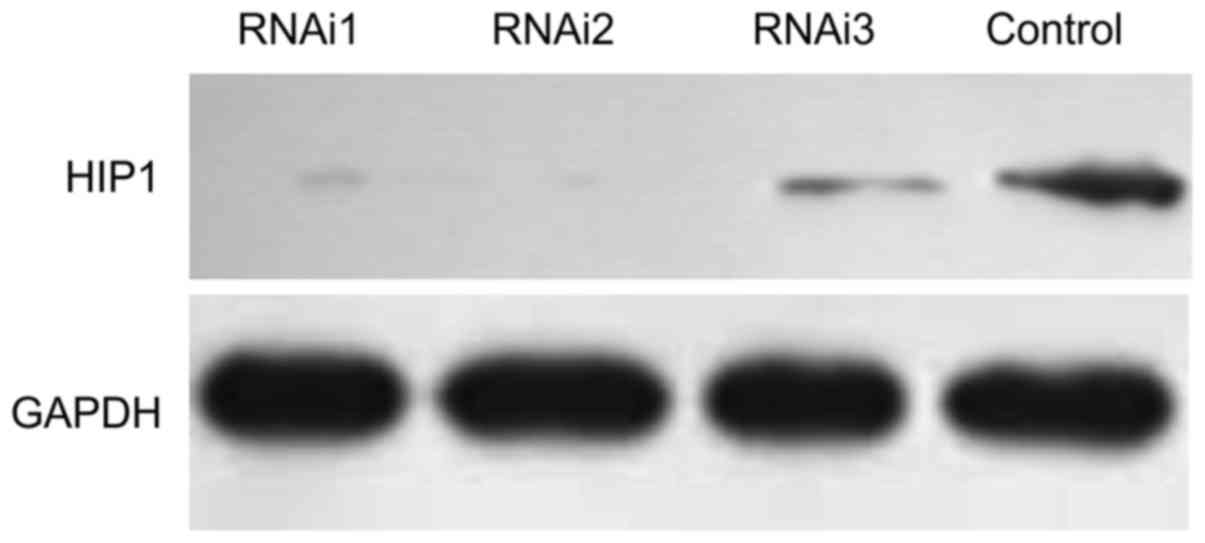
Knockdown of HIP1 in HeLa cells
Based on western blot assays, in HeLa cells, the
expression of HIP1 was significantly blocked by siRNA1
(taattgagcgactatacagag) and siRNA2 (acagcgatatagcaagctaaa), both of
which were more efficient compared with siRNA3
(accgcttcatggagcagttta) (Fig. 3).
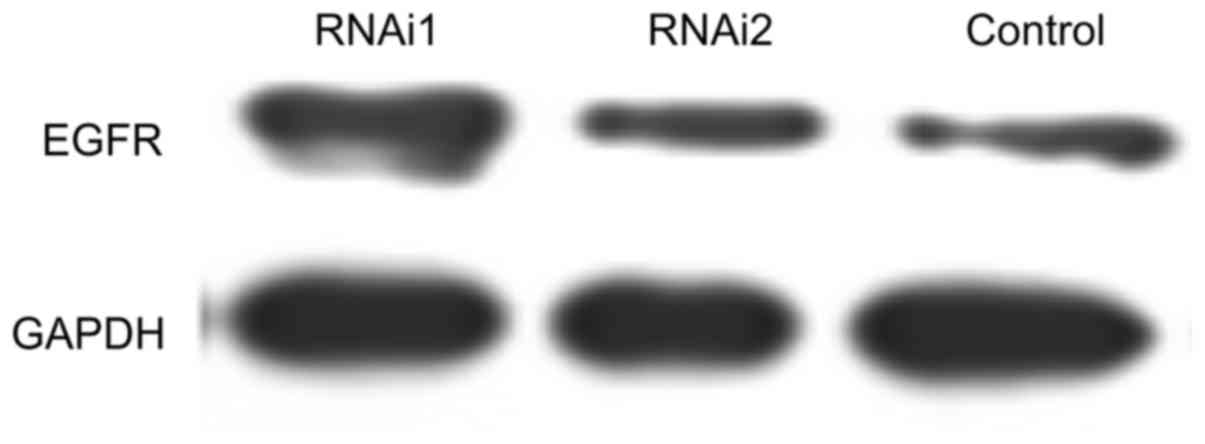
Neither siRNA1 nor siRNA2 influenced the expression levels of EGFR
and GAPDH in HeLa cells (Fig.
4).
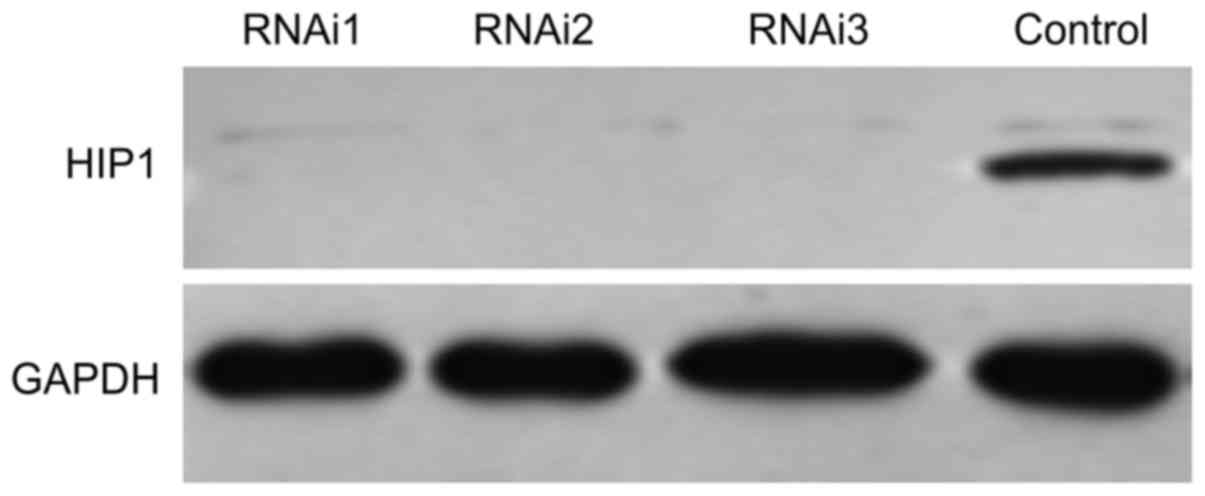
NCI-H1299 cells exhibited high expression of EGFR
and low expression of HIP1 (Figs. 1
and 2). Accordingly, NCI-H1299 was
chosen as a control to examine the effects of HIP1 siRNA. As
shown in Fig. 5, the expression of
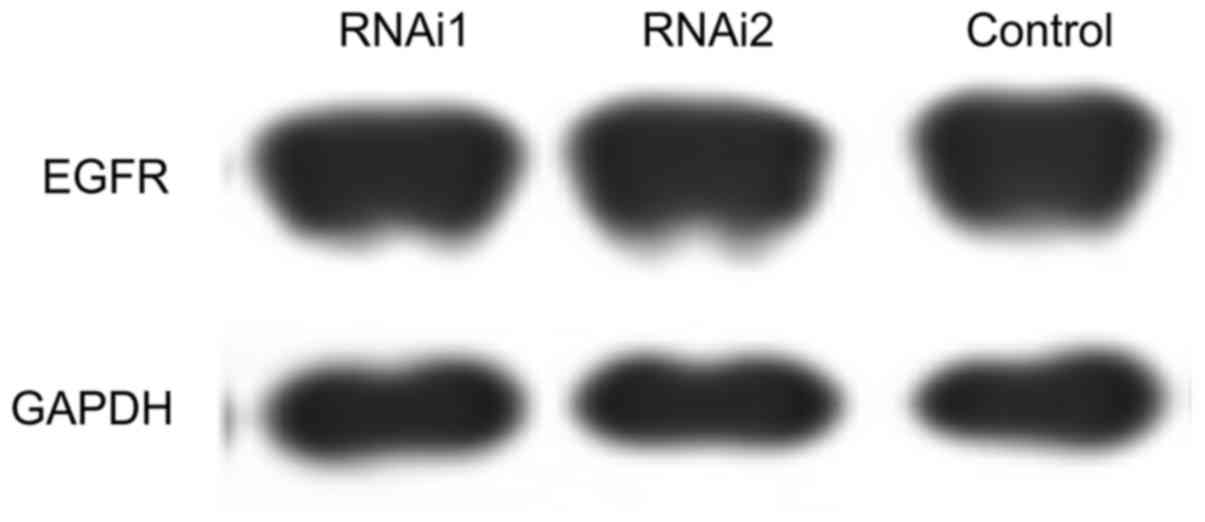
HIP1 was significantly blocked by siRNA1, siRNA2 and siRNA3. The
three siRNAs had no effect on the expression levels of EGFR and
GAPDH in NCI-H1299 cells (Fig.
6).
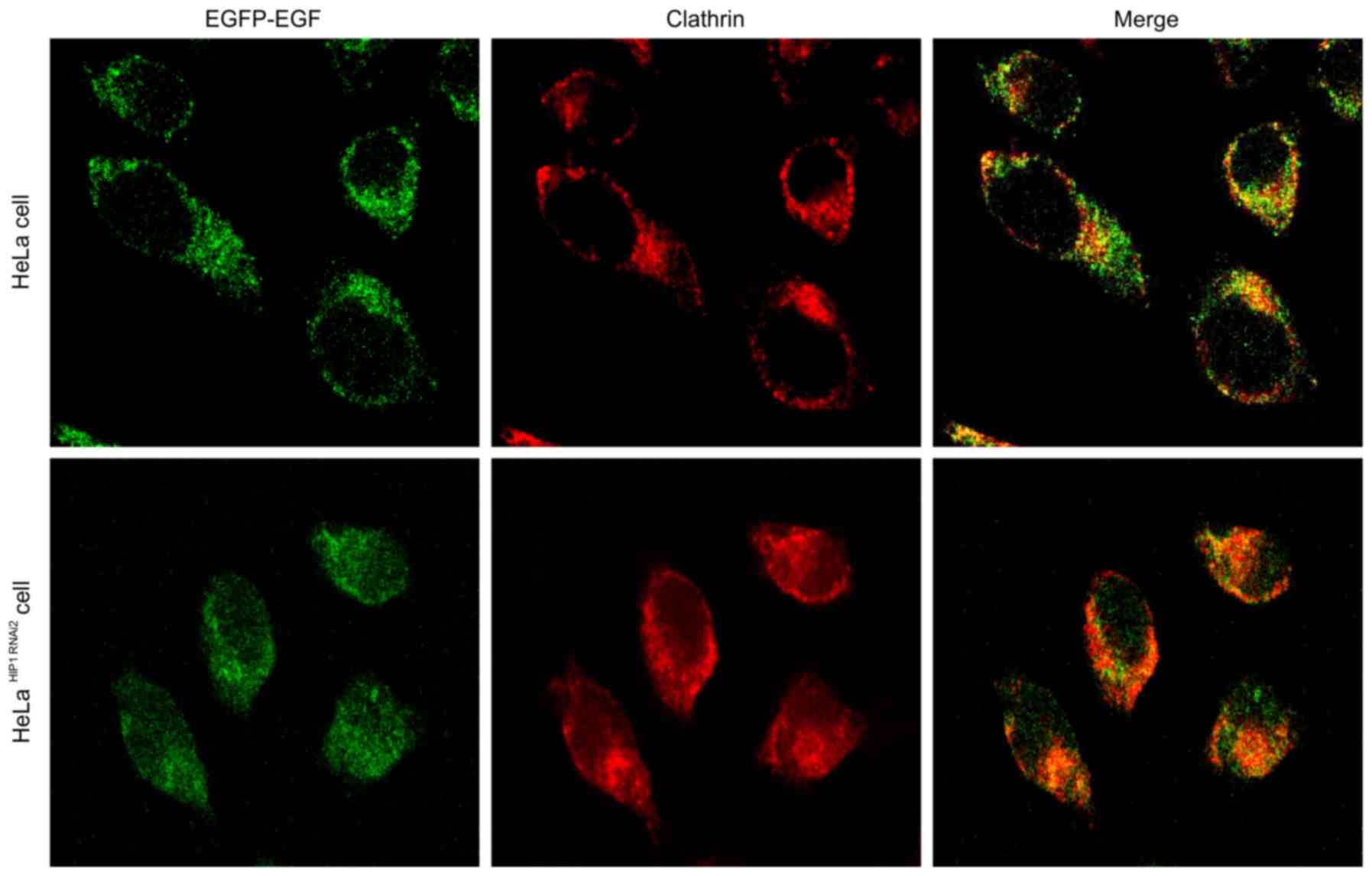
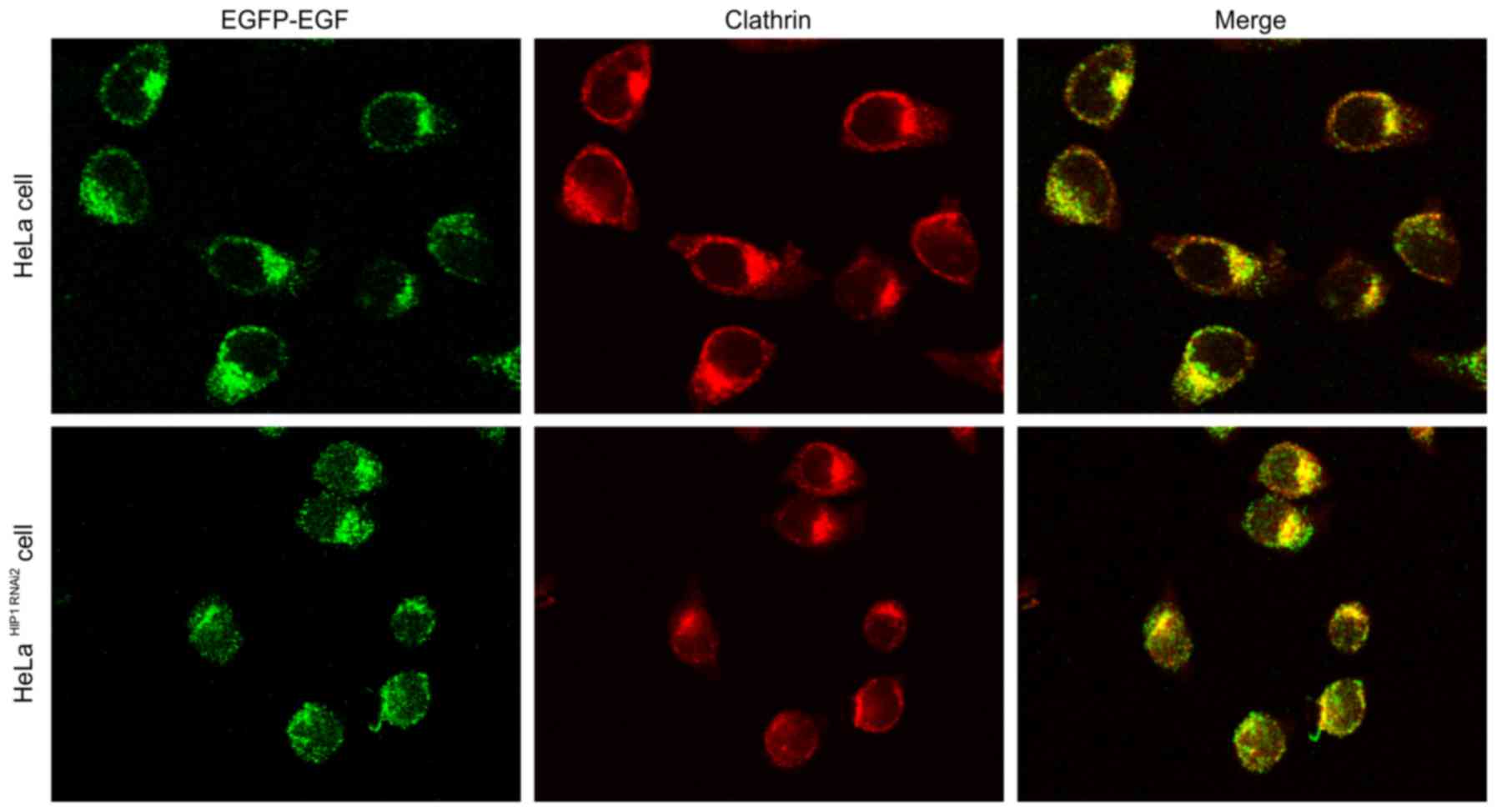
Internalization of EGF-EGFR
After stimulation with 1.5 ng/ml EGF, endocytosis of
EGF-bound EGFR was significantly accelerated after the expression
of HIP1 was blocked (Fig. 7). After
simulation with 100 ng/ml EGF, endocytosis of EGF-bound EGFR was
also significantly accelerated after the expression of HIP1 was
blocked (Fig. 8). There was no
obvious difference between 1.5 and 100 ng/ml EGF with respect to
EGFR endocytosis (Figs. 7 and
8).
Colocalization of EGF-EGFR and
clathrin
Clathrin-mediated endocytosis was also accelerated
after the expression of HIP1 was blocked, and exhibited a positive
correlation with the internalization of EGF-EGFR for both 1.5 ng/ml
(Fig. 7) and 100 ng/ml EGF
(Fig. 8). There was no obvious
difference between 1.5 and 100 ng/ml EGF with respect to clathrin
endocytosis (Figs. 7 and 8). EGFR and clathrin were colocalized in
the cytoplasm.
Discussion
Overactivation of EGFR signaling pathway is strongly
associated with carcinogenesis, and it is becoming increasingly
clear that impaired deactivation of EGFR may also be a mechanism in
cancer. A major deactivation pathway for EGFR downregulation
involves ligand-induced endocytosis of EGFR and subsequent
degradation in lysosomes; this is important in carcinogenesis,
e.g., in breast cancer (8).
N-acetylglucosaminyltransferase Va (GnT-Va) is involved in
the EGF-induced downregulation of EGFR and intracellular signaling
by inhibiting receptor endocytosis. When GnT-Va expression is
knocked down in highly invasive human breast cancer cells,
ligand-induced downregulation of EGFR expression is inhibited via
decreased EGFR endocytosis, resulting in delayed downstream signal
transduction and inhibition of EGF-induced invasiveness phenotypes
(10,11).
HIP1 may be involved in the endocytosis of EGFR.
Previous studies have demonstrated the colocalization of HIP1 and
markers of clathrin-mediated endocytosis in neuronal cells and the
enrichment of HIP1 on clathrin-coated vesicles purified from brain
homogenates (5). HIP1 binds to
clathrin adaptor protein 2 (AP2) and the terminal domain of the
clathrin heavy chain, predominantly via a small fragment at amino
acids 276–335. This region, a clathrin-box, contains consensus
clathrin- and AP2-binding sites with high binding affinity to the
terminal domain of the clathrin heavy chain and the ear domain of
the AP2 subunit, respectively, leading to efficient stimulation of
the clathrin assembly via its central helical domain by binding
directly to the clathrin light chain (3–5). These
results suggest that HIP1 has functional roles in clathrin-mediated
endocytosis.
In this experiment, we screened 17 tumor cell lines
to identify cells with high expression of both HIP1 and EGFR. HeLa
cells had obviously high expression of both HIP1 and EGFR. Various
siRNAs were designed to block the expression of HIP1 in HeLa cells
to evaluate its inhibitory effects on EGFR endocytosis. Two siRNA
sequences (taattgagcgactata cagag and acagcgatatagcaagctaaa,
efficiently blocked HIP1 expression. We confirmed these results
using NCI-H1299 cells as controls. After the blockage of HIP1
expression and stimulation with 1.5 ng/ml EGF, EGFR endocytosis was
significantly accelerated. The same results were obtained after
stimulation with 100 ng/ml EGF. The acceleration of EGFR
endocytosis was only correlated with HIP1 blockage. HIP1 can
stabilize EGFR on cell surfaces by decreasing EGFR endocytosis.
This process was correlated with clathrin endocytosis.
In the present study, we explored the role of HIP1
in the degradation of EGFR in cancer cells. This is the first
analysis of EGFR and HIP1 coexpression in a large number of cell
lines, and our results clearly demonstrated the effects of HIP1
inhibition on EGFR endocytosis. These findings may explain the
proliferative and anti-apoptotic effects of HIP1 on tumor cells.
HIP1 inhibition can accelerate EGFR endocytosis and degradation.
These results also suggest a new method to treat carcinoma with
high EGFR expression by targeting HIP1, but additional studies are
needed to evaluate the clinical potential.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (81300321), the Key Discipline
Foundation of Fujian Province (2012-149), and the Young and
Middle-Aged Personnel Training Project of Fujian Province Health
Department (2014-ZQN-ZD-9).
Glossary
Abbreviations
Abbreviations:
|
HIP1
|
Huntington-interacting protein 1
|
|
AP2
|
adaptor protein 2
|
|
GnT-Va
|
N-acetylglucosaminyltransferase
Va
|
References
|
1
|
Kalchman MA, Koide HB, McCutcheon K,
Graham RK, Nichol K, Nishiyama K, Kazemi-Esfarjani P, Lynn FC,
Wellington C, Metzler M, et al: HIP1, a human homologue of S.
cerevisiae Sla2p, interacts with membrane-associated huntingtin
in the brain. Nat Genet. 16:44–53. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Legendre-Guillemin V, Metzler M,
Charbonneau M, Gan L, Chopra V, Philie J, Hayden MR and McPherson
PS: HIP1 and HIP12 display differential binding to F-actin, AP2,
and clathrin. Identification of a novel interaction with clathrin
light chain. J Biol Chem. 277:19897–19904. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Metzler M, Legendre-Guillemin V, Gan L,
Chopra V, Kwok A, McPherson PS and Hayden MR: HIP1 functions in
clathrin-mediated endocytosis through binding to clathrin and
adaptor protein 2. J Biol Chem. 276:39271–39276. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mousavi SA, Malerød L, Berg T and Kjeken
R: Clathrin-dependent endocytosis. Biochem J. 377:1–16. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bradley SV, Holland EC, Liu GY, Thomas D,
Hyun TS and Ross TS: Huntingtin interacting protein 1 is a novel
brain tumor marker that associates with epidermal growth factor
receptor. Cancer Res. 67:3609–3615. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rao DS, Hyun TS, Kumar PD, Mizukami IF,
Rubin MA, Lucas PC, Sanda MG and Ross TS: Huntingtin-interacting
protein 1 is overexpressed in prostate and colon cancer and is
critical for cellular survival. J Clin Invest. 110:351–360. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rao DS, Bradley SV, Kumar PD, Hyun TS,
Saint-Dic D, Oravecz-Wilson K, Kleer CG and Ross TS: Altered
receptor trafficking in Huntingtin Interacting Protein
1-transformed cells. Cancer Cell. 3:471–482. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bache KG, Slagsvold T and Stenmark H:
Defective downregulation of receptor tyrosine kinases in cancer.
EMBO J. 23:2707–2712. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rosell R: Cancer and alterations in the
endocytic pathway. Future Oncol. 3:487–489. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo HB, Johnson H, Randolph M, Lee I and
Pierce M: Knockdown of GnT-Va expression inhibits ligand-induced
downregulation of the epidermal growth factor receptor and
intracellular signaling by inhibiting receptor endocytosis.
Glycobiology. 19:547–559. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mutch LJ, Howden JD, Jenner EP, Poulter NS
and Rappoport JZ: Polarised clathrin-mediated endocytosis of EGFR
during chemotactic invasion. Traffic. 15:648–664. 2014. View Article : Google Scholar : PubMed/NCBI
|