Introduction
Cervical cancer (CC) remains one of the most common
cancers diagnosed worldwide, and is the third leading cause of
cancer-related deaths in women. Approximately 527,600 new patients
were diagnosed with CC and ~265,700 deaths occurred in 2012
(1). Standard treatment depends on
the clinical stage. However, radiotherapy either alone or with
adjuvant chemotherapy remains the standard treatment. Indeed,
radiation therapy is utilized for over 60% of patients with CC and
is the first treatment choice in ~52% of all patients with cancer
(2).
Unfortunately, nearly 50% of all patients with CC do
not respond to standard treatment due to acquired radioresistance,
which is considered the main cause of related deaths associated
with treatment failure in patients with CC (3). Thus, radioresistance could be defined
as the ability of tumor cells to survive and repair the molecular
damage caused by ionizing radiation and its effectors such as free
radicals.
MicroRNAs or miRNAs are a group of endogenous, small
non-coding RNAs, which are ~21–25 nucleotides in length. miRNAs
play a critical role in post-transcriptional gene regulation by
degrading or preventing the translation of their target messenger
RNA (mRNA). Recently, miRNAs have been called ‘the master
regulators’ of gene expression, due to the fact that they have been
implicated in a wide variety of cellular processes. To date, few
miRNA expression profiles have been related to the radioresistance
of patients with CC and CC-derived cell lines; for instance,
miR-630, miR-1246, miR-1290, miR-3138, miR-31-3p and miR-3676 were
found upregulated, whereas miR-1271, miR-15b*, miR-19b-1*,
miR-378*, miR-95, miR-100-5p, miR-200a-5p, miR-320, and miR-342
were found downregulated (4,5).
Recently, we showed that patients with locally advanced CC who do
not respond to conventional treatment have a specific miRNA
signature, with miR-125 highlighted in that latter signature
(6). In the present study, we
hypothesized the importance of miR-125a as a potential key
regulator for the treatment response of CC patients.
The miR-125 family consists of 3 homologous members:
miR-125a; miR-125b-1 and miR-125b-2. These miRNA family members
have been linked to several tumors and other chronic degenerative
diseases, playing a role as tumor suppressors or oncogenes
(7–9).
In the present study, we showed that miR-125a is
downregulated in patients with CC who do not respond to standard
chemotherapy and radiotherapy treatment. Then, we employed a
radioresistance in vitro model employing SiHa, CaSki and
HeLa cell lines established by fractionated radiation in order to
elucidate the role of miR-125 in the induction of radioresistance.
Finally, we demonstrated that overexpression of miR-125a
significantly decreased radioresistance through the negative
regulation of CDKN1A in CC cells.
Materials and methods
Patient selection and CC samples
We selected 62 tumor samples from the National
Cancer Institute of Mexico (INCan) Tumor Bank; 30 fresh-frozen
cancer samples were analyzed for miR-125a expression by means of
qRT-PCR (15 patients were non-responders and 15, complete
responders). Additionally, 32-paraffin-embedded tissues were
employed to assess p21 expression by immunohistochemistry (IHC)
analysis. All patients included accepted to participate in the
study and signed informed consent; the Institutional Ethics and
Scientific Board Committees approved the protocol in accordance
with The Code of Ethics of the World Medical Association (WMA)
(Declaration of Helsinki). All patients were histologically and
clinically diagnosed with locally advanced CC [stages IB2-IVA
according to the International Federation of Gynecology and
Obstetrics (FIGO) classification]. Samples were obtained from
patients diagnosed between 2011 and 2014 at the Department of
Obstetrics and Gynecology, INCan. All patients had a median of 55
months of clinical follow-up. After sample-taking, the patient
samples were categorized into 2 groups depending on their clinical
response to standard treatment. Complete response (CR) was defined
as the disappearance of all signs of cancer in response to
treatment, and no response (NR) was defined as patients with
partial, progressive or stable disease. The tumor samples employed
in the present study contained at least 80% of tumor cells on
pathological examination. Standard treatment for patients with
locally advanced CC consists of 5 cycles of 40 mg/m2 of
CDDP (cis-diamminedichloroplatinum II) and a total of 55 Gy
of radiotherapy and 30 Gy of internal brachytherapy.
Cell lines
Human CC cell lines SiHa (HTB-35), CaSki (CRL-1550)
and HeLa (CCL-2) were purchased from the American Type Culture
Collection (ATCC; Rockville, MD, USA) and cultured according to
cell-line specifications. CaSki and HeLa cell lines were cultured
in RPMI-1640 medium, although the SiHa cell line was cultured in
EMEM medium. All cell lines were maintained with 100 U/ml of
penicillin and 100 mg/ml of streptomycin, 10% fetal bovine serum,
and incubated at 37°C in a 5% CO2 atmosphere. All cancer
cell lines employed in the present study were authenticated by
means of the Authentifiler PCR Amplification kit (cat. no. 4479566;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) on a 3500 Genetic
Analyzer (cat. no. 4440462; Applied Biosystems, Foster City, CA,
USA) following the International Cell Line Authentication Committee
(ICLAC) guidelines.
Establishment of a radioresistant in
vitro model
The human CC cell lines SiHa, CaSki and HeLa were
employed to establish a radioresistant cell line model by
fractionated radiation. The parental cell lines were grown to 80%
confluence, and then the cell lines were trypsinized and divided
into 2 subcultures: one for irradiation (RR, radioresistant cells),
and the remaining subculture for the non-irradiated condition (RS,
radiosensitive cells). When RR culture cells reached 60%
confluence, they were irradiated with 2 Gy of X-ray irradiation
using an X-ray Linear Accelerator (CL2100C/D; Varian Medical
Systems, Palo Alto, CA, USA); immediately after irradiation, the
cells were returned to the incubator. Then, after 24 h, the
irradiated cell lines were trypsinized and subcultured into new
flasks. When they again reached 60% confluence, the irradiation
protocol was repeated until the cells reached a total dose of 56
Gy. RS cells were treated under the same conditions as the RR
culture, but without irradiation.
Determination of lethal dose 50 of
radiation
Lethal dose 50 (LD50) was determined by
colony formation assay (CFA). The parental cell lines were
incubated at 37°C for 24 h and subsequently were irradiated at
different doses ranging from 0–10 Gy. The cells were harvested and
counted 24 h after irradiation. Subsequently, 3×103
cells were plated in 6-well culture plates and incubated under
standard conditions for 2 weeks. The colonies formed were fixed and
stained with glutaraldehyde 6.0% (vol/vol) and crystal violet 0.5%
(wt/vol) in water. Finally, colonies consisting of 50 cells or more
were counted using an optical microscope and the surviving fraction
was determined.
RNA isolation from tumor samples and
cell lines
Total RNA from CC tissues and cell lines were
extracted and purified with the miRNeasy Mini kit (cat. no. 217004;
Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's
instructions. RNA quantification was performed using an Epoch
spectrophotometer (BioTek Instruments, Inc., Winooski, VT,
USA).
Relative quantification of miR-125 by
qRT-PCR
The expression of miR-125 was assessed using the
TaqMan MicroRNA assay (Applied Biosystems, Carlsbad, CA, USA)
according to the manufacturer's protocol. Briefly, 100 ng of total
RNA was subjected to reverse transcription reaction using
miRNA-specific RT primers and the TaqMan miRNA reverse
transcription kit. The 15-µl reactions were incubated according to
the manufacturer's protocol. Real-time qPCR was performed using
TaqMan Universal Master Mix II no UNG in a StepOne qPCR instrument
(Applied Biosystems, Carlsbad, CA, USA). Relative expression of
miR-125a was calculated utilizing the comparative 2−∆∆Ct
method. RNU-44 and RNU-6b expression were employed as endogenous
control. All of the qPCR reactions were assessed in 3 independent
experiments and each reaction was performed in triplicate.
Western blot analysis
Total protein from cell lysates was extracted using
RIPA buffer (sc-24948; Santa Cruz Biotechnology, Inc., Santa Cruz,
TX, USA). Then, 50 µg of protein was separated by
SDS-polyacrylamide gel electrophoresis (PAGE) and transferred onto
a polyvinylidene difluoride (PVDF) membrane (GE Healthcare,
Milwaukee, WI, USA) in a Trans-Blot Turbo (Bio-Rad) semi-dry
chamber at 25 V and 1 mA for 30 min. After blocking with 5% non-fat
milk for 2 h, the membrane was incubated with the specific antibody
overnight at 4°C on a rocking platform, washed, and then incubated
with the corresponding secondary antibody for 2 h at room
temperature. The blot was visualized using the Super Signal West
Femto Chemiluminescent substrate (Pierce, Rockford, IL, USA) in a
C-Digit scanner (LI-COR)™ employing Image Studio (LI-COR
Biosciences, Lincoln, NE, USA) software. The primary antibodies
were purchased from Santa Cruz Biotechnology, Inc.: anti-p21 (187)
(1:2,000; sc-817). All secondary antibodies were obtained from the
Cell Signaling Technology, Inc. (Beverly, MA, USA); anti-mouse
(1:5,000; #7076S). β-actin (C4) (1:5,000; sc-47778; Santa Cruz
Biotechnology, Inc.) was utilized as an internal control.
Luciferase reporter assays
Reporter plasmids were constructed by ligation of
synthetic oligonucleotide duplexes (IDT) containing one of the 3
putative miR-125a target regions in the CKN1A mRNA 3′UTR, including
region 1, 5′-CTAGTGCACTGGGGAGCCCGTCTCAGTGTA-3′ and
AGCTTACACTGAGACGGGCTCCCCAGTGCA; region 2,
5′-CTAGTACACAAGGGCACCCTAGTTCTACCTCAGGCAA-3′; and region
5′-AGCTTTGCCTGAGGTAGAACTAGG'-GTGCCCTTCTTGTGTA-3′; and finally,
region 3, 5′-CTAGTAGACTGTAAACCTCTCGAGGGCA-3′; and region,
5′-AGCTTGCCCTCGAGAGGTTTACAGTCTA-3′. All putative regions were
obtained from microRNA.org and cloned into the
pMIR-REPORT plasmid (Ambion Inc., Austin, TX, USA) (10). Each construction was co-transfected
with miR-125a mirVana miRNA mimic (Applied Biosystems) and the
pMIR-REPORT β-gal control plasmid (Ambion) into HeLa cells.
Luciferase activity was analyzed 48 h after transfection utilizing
the Dual-Luciferase Reporter Assay System (Applied Biosystems) in a
GloMax 96 Microplate Luminometer (Promega, Madison, WI, USA).
Luciferase activity was normalized to β-gal activity for each
transfected well; each experiment was performed in triplicate.
Transfection of miRNA mimics and
inhibitors
miR-125a mimics and inhibitors were purchased from
Ambion and assessed according to the manufacturer's instructions.
The pre-miR negative control and scramble oligonucleotide for the
miRNA transfection experiments were not homologous to any human
miRNA sequences and can be obtained from the Pre-miR miRNA Starter
kit (cat. no. Am1540; Thermo Fisher Scientific, Inc.).
Oligonucleotide transfection was performed using the Lipofectamine
RNAiMAX transfection reagent (cat. no. 13778150; Thermo Fisher
Scientific, Inc.). All experiments were replicated in 6-well plates
with a final concentration of 25 pmol of each oligonucleotide, and
7.5 µl of Lipofectamine RNAi/MAX was used for each
transfection.
P21 expression in CC tissues
IHC was performed on serial sections using p21 (187)
(sc-817; Santa Cruz Biotechnology, Inc.) antibody in 32
paraffin-embedded blocks from patients with CC (non-responders, 15;
and complete responders, 17). Serial sections, 5-µm thick, were
immunostained using the biotin-streptavidin-peroxidase method.
Briefly, the sections were deparaffinized in xylene and rehydrated
with ethanol. Then, the samples were hydrated by autoclave
pretreatment in 10 mM citrate buffer (pH 6.0) for 5 min. Endogen
peroxidase was quenched with 3% hydrogen peroxidase for 10 min at
room temperature. Next, the slides were incubated with the primary
antibody at a 1:200 dilution in Tris-buffered solution (50 mM
Tris-HCl, 150 mM NaCl, pH 7.4) for 2 h. Finally, the antibody was
visualized with 3,3-diaminobenzidine tetrahydrochloride. The
fraction of positive cells was estimated using a four-tiered scale
(≤5%, 1; 6–25%, 2; 51–75%, 3 and ≥75%, 4). Staining intensity was
also scored on a 4-tiered scale (negative, 0; low-intensity
positive staining, 1; moderate-intensity positive staining, 2; and
strong-intensity positive staining, 3. Two independent pathologists
evaluated the stained sections and the average score for each slide
was used for statistical analysis.
Statistical analysis
Data are expressed as the mean ± standard error of
the mean (SEM) of at least 3 separate experiments performed in
triplicate. The differences between groups were analyzed using a
double-sided Student's t-test when only 2 groups were present, and
the null hypothesis was rejected at 0.01 levels unless otherwise
specified.
Results
Radioresistant cell model
We established a cancer cell line radioresistant
model by fractionated irradiation. First, we determined the
LD50 of radiation for HeLa, CaSki and SiHa cell lines by
CFA. The previously mentioned cell lines were irradiated at
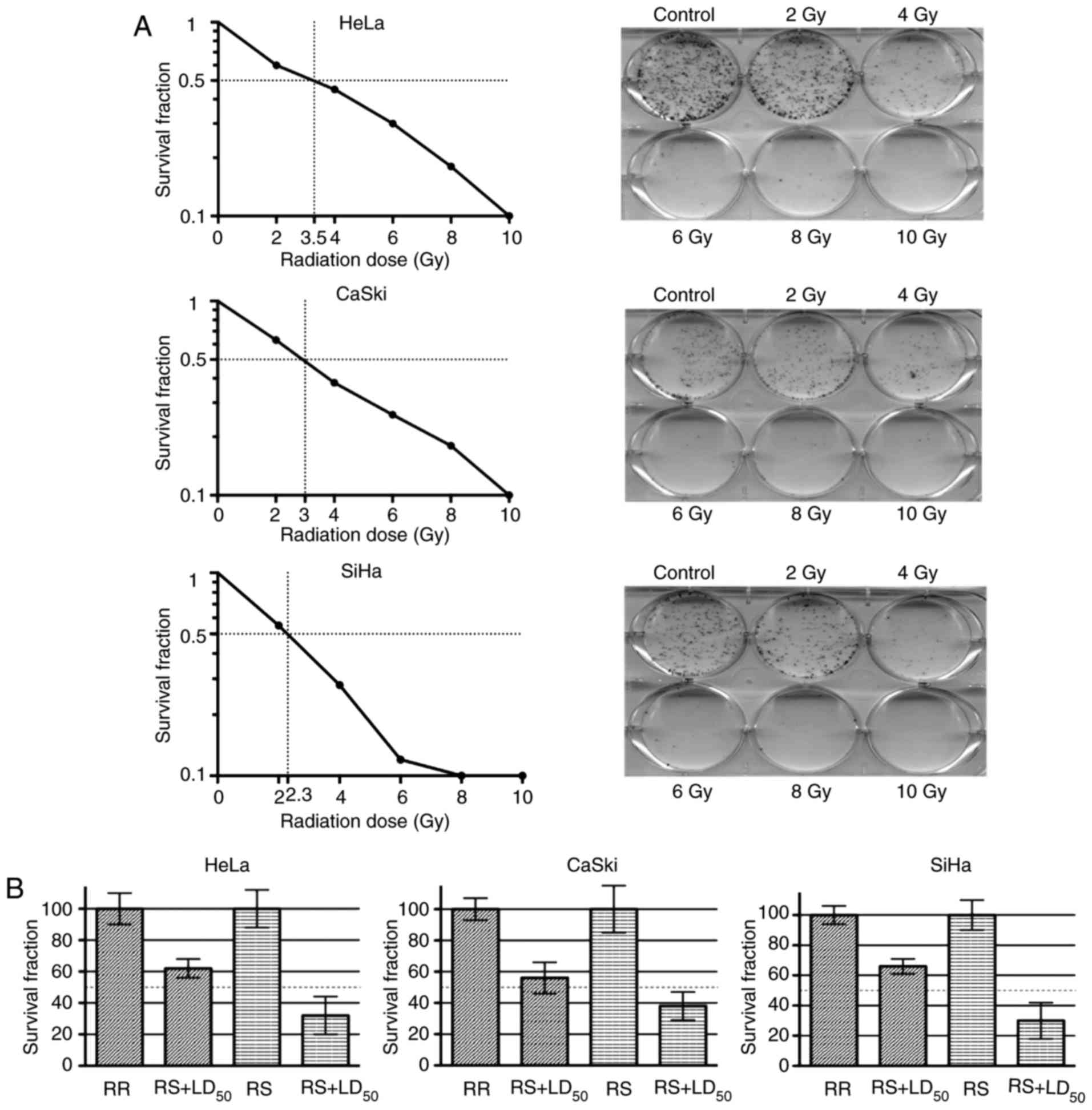
different doses (0–10 Gy) and survival curves were determined.
Therefore, we calculated a ‘parental’ LD50 for each cell
line. Hence, HeLa cells exhibited a LD50 higher than
that of the CaSki and SiHa cells (CaSki, 3.0 Gy; HeLa, 3.5 Gy; and
SiHa, 2.3 Gy) (Fig. 1A).
After 28 episodes of irradiation, cell lines
HeLa-RR, SiHa-RR and CaSki-RR reached a total of 56 Gy, and stable
radioresistant cells were obtained from the surviving fraction of
parental irradiated cells. Next, LD50 for parental cell
lines were employed to confirm the radioresistant phenotype in the
irradiated cell lines. After parental cell lines were irradiated at
LD50-calculated doses, as expected, all of the RR
subcultures had a higher survival rate than the RS cultures.
Accordingly, the survival fractions of the HeLa-RR, SiHa-RR and
CaSki-RR cells were 58, 62 and 64%, respectively (Fig. 1B). Therefore, we confirmed the
establishment of a radioresistant phenotype in the RR
subcultures.
CC samples
The patients with CC enrolled in the present study
had a clinical and pathological diagnosis of locally advanced CC
(LACC). All cervical samples were histologically analyzed to
confirm a minimum of 80% tumor cells. A summary of the clinical and
pathological characteristics of all patients is presented in
Table I. Median age of patients at
diagnosis was 52 years (range, 31–68 years). As expected, the most
prevalent HPV genotypes identified were HPV-16 (46.8%), HPV-18
(25.8%) and HPV-45 (24.2%).
 | Table I.Clinical and pathological
characteristics of the cervical cancer patients enrolled in the
present study |
Table I.
Clinical and pathological
characteristics of the cervical cancer patients enrolled in the
present study
| Characteristics | Patients, n (%) |
|---|
| Total number | 62 |
| Age (years) |
|
|
Median | 52 |
|
Range | 31–68 |
| Histological
type |
|
| Squamous
cell carcinoma | 54 (87.1) |
|
Adenocarcinoma | 8 (12.9) |
| Tumor size (cm) |
|
|
<4 | 11 (17.8) |
| ≥4 | 51 (82.2) |
| Without
data | 0 (0.0) |
| Clinical stage
(FIGO) |
|
| IIA | 0 (0.0) |
| IIB | 37 (59.9) |
| IIIA | 16 (25.6) |
|
IIIB | 9 (14.5) |
|
IVA | 0 (0.0) |
| HPV
genotypification |
|
| Type
16 | 29 (46.8) |
| Type
18 | 16 (25.8) |
| Type
45 | 15 (24.2) |
|
Others | 2 (3.2) |
| Clinical
response |
|
|
Complete response | 32 (51.6) |
| No
response | 30 (48.4) |
Relative quantification of miR-125a in
CC samples and the radioresistant cell model
We recently published an expression profile of miRNA
associated with clinical response in patients with CC under
standard treatment of chemotherapy and radiotherapy; one of these
identified miRNA was miR-125a (6).
Based on this evidence, we aimed to elucidate the functional role
of miR-125a in the radioresistance phenotype in CC. Hence, we
assessed the expression level of miR-125a in 30 dichotomized CC
samples (NR=15 and CR=15). Relative expression of miR-125a was
significantly lower in the 15 NR samples with regard to the CR
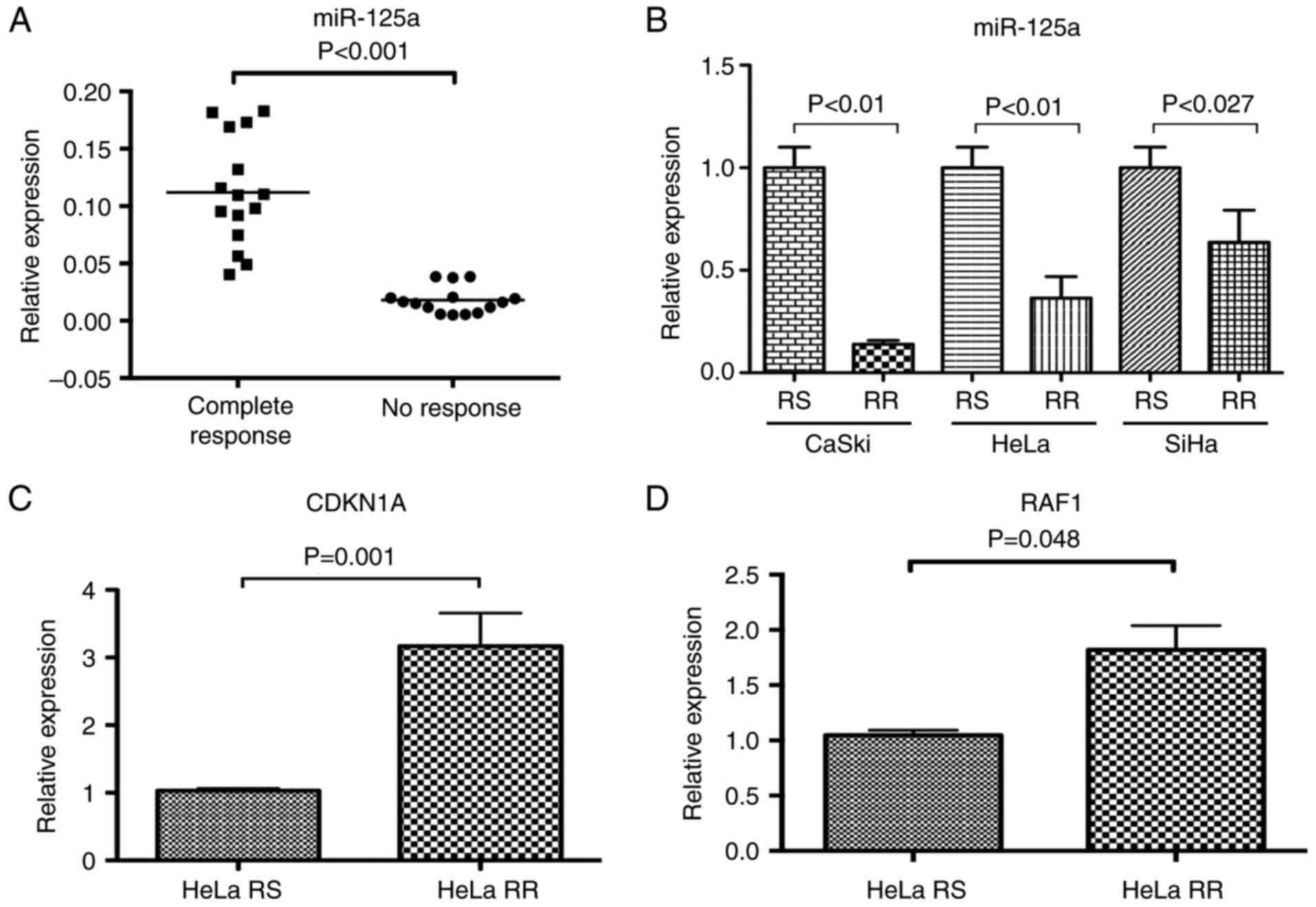
samples (P≤0.0001) (Fig. 2A).
We quantified the expression level of miR-125a in
HeLa-RR, SiHa-RR and CaSki-RR subcultures (Fig. 2B). miR-125a was underexpressed in
all RR subcultures with respect to RS cell lines.
It is noteworthy that miR-125a was found 5-fold
underexpressed in the CaSki RR cell line with respect to the CaSki
RS cell line (P=0.0001), whereas in HeLa-RR and SiHa-RR cell lines,
miR-125a was found up to 2.5-fold underexpressed (P=0.001 and
P=0.027, respectively). We conducted the following experiments on
the HeLa cell line in order to achieve a better performance of
transfection assays, such as that reported by Asgharian et
al (11).
Molecular targets of miR-125a
After confirming that miR-125a was underexpressed in
resistant CC samples and radioresistant (RR) CC cell lines, we
conducted an exhaustive search on the most citable bioinformatics
algorithms for predicting miR-125a targeted mRNA (microRNA.org, TargetScan and miRDB). Five hypothetical
target genes of miR-125 were obtained by means of this method
(CDKN1A, SP1, E2F7, AKT1 and RAF1), which were tested by qRT-PCR to
suggest a possible regulation by miR-125a. We found an
overexpression of CDKN1A and RAF1 (Fig.
2C and D) transcripts in the HeLa cell line (P=0.01 and
P=0.048), respectively, whereas, differences in SP1, E2F7 and AKT1
mRNA were not statistically significant (data not shown).
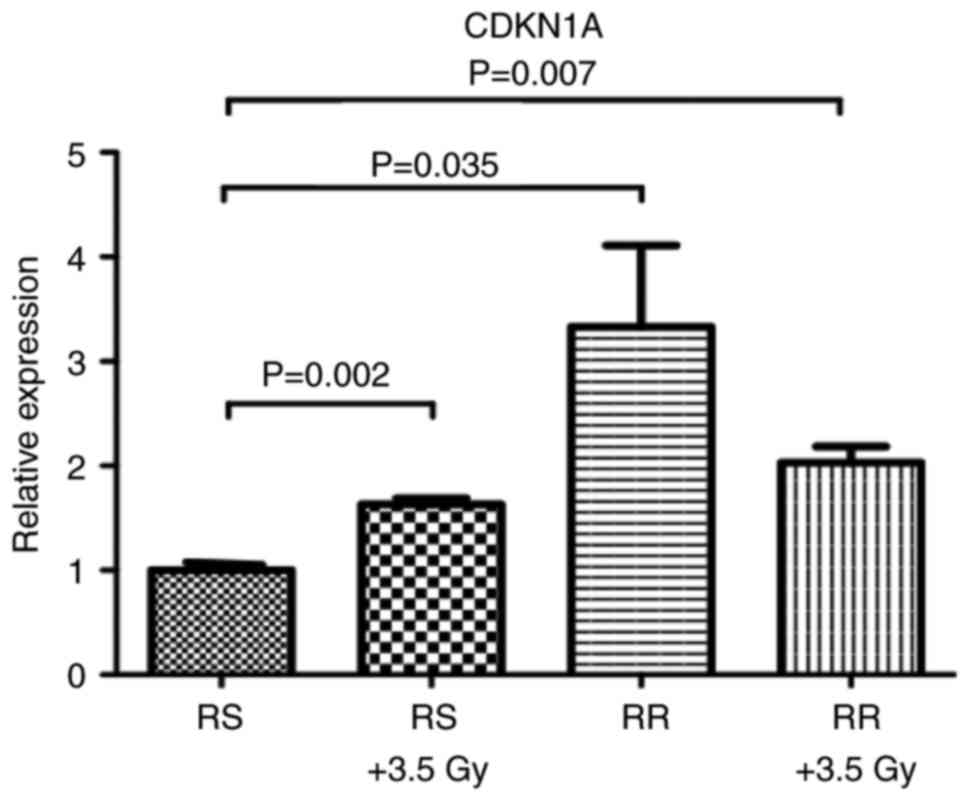
Additionally, we demonstrated that CDKN1A is overexpressed after
LD50 irradiation doses, in both RS and RR cells
(Fig. 3), suggesting a possible
role of CDKN1A in the radioresistant phenotype.
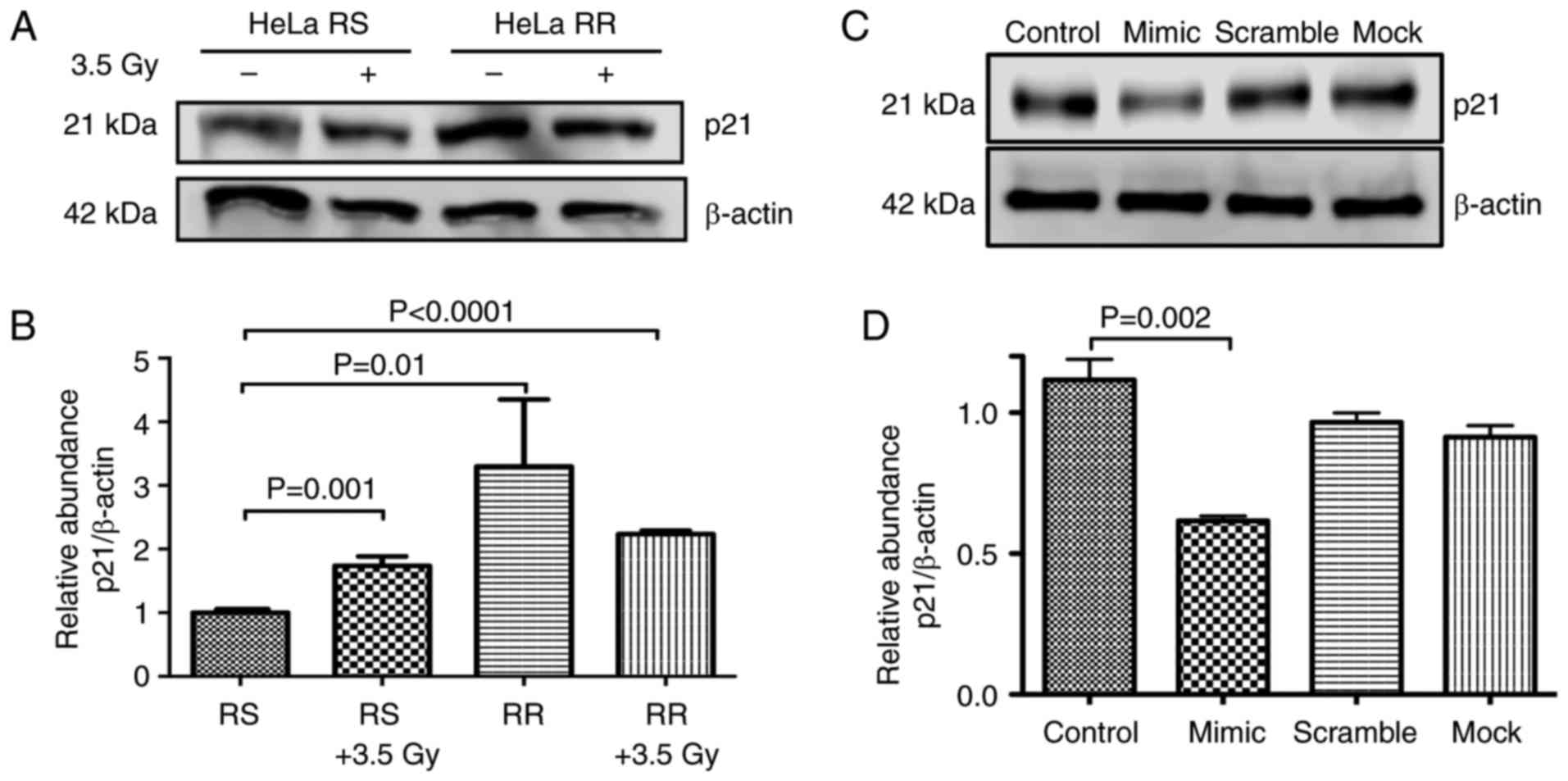
Western blot analysis of p21
The protein levels of p21 (CDKN1A) were assessed in
the HeLa-RS and HeLa-RR cells. Our results confirmed that p21 was
overexpressed by up to 3-fold in the HeLa-RR cells with regard to
the HeLa RS cells (P=0.019). In addition, the radiation increased
p21 protein levels 2-fold in the RS cells (P=0.001). It was
encouraging to note that in all cases where the cells were
irradiated, p21 was overexpressed (Fig.
4A and B). RR cells had higher levels of CDKN1A, probably due
to a downregulation in the expression of miR-125a. In order to test
this hypothesis, we restored the expression of miR-125a by means of
a mimic sequence transfected into RR cells. As observed in Fig. 4C and D, restoration of miR-125a by a
mimic sequence decreased the protein levels of p21, suggesting a
direct role for CDKN1A regulation exerted by miR-125a.
miR-125a regulates the expression of
CDKN1A
To validate whether CDKN1A mRNA is a molecular
target of miR-125a, we performed a luciferase reporter assay. Three
putative binding sites for miR-125a in the CDKN1A 3′UTR were cloned
into the pMIR-REPORT plasmid. The binding regions between miR-125a
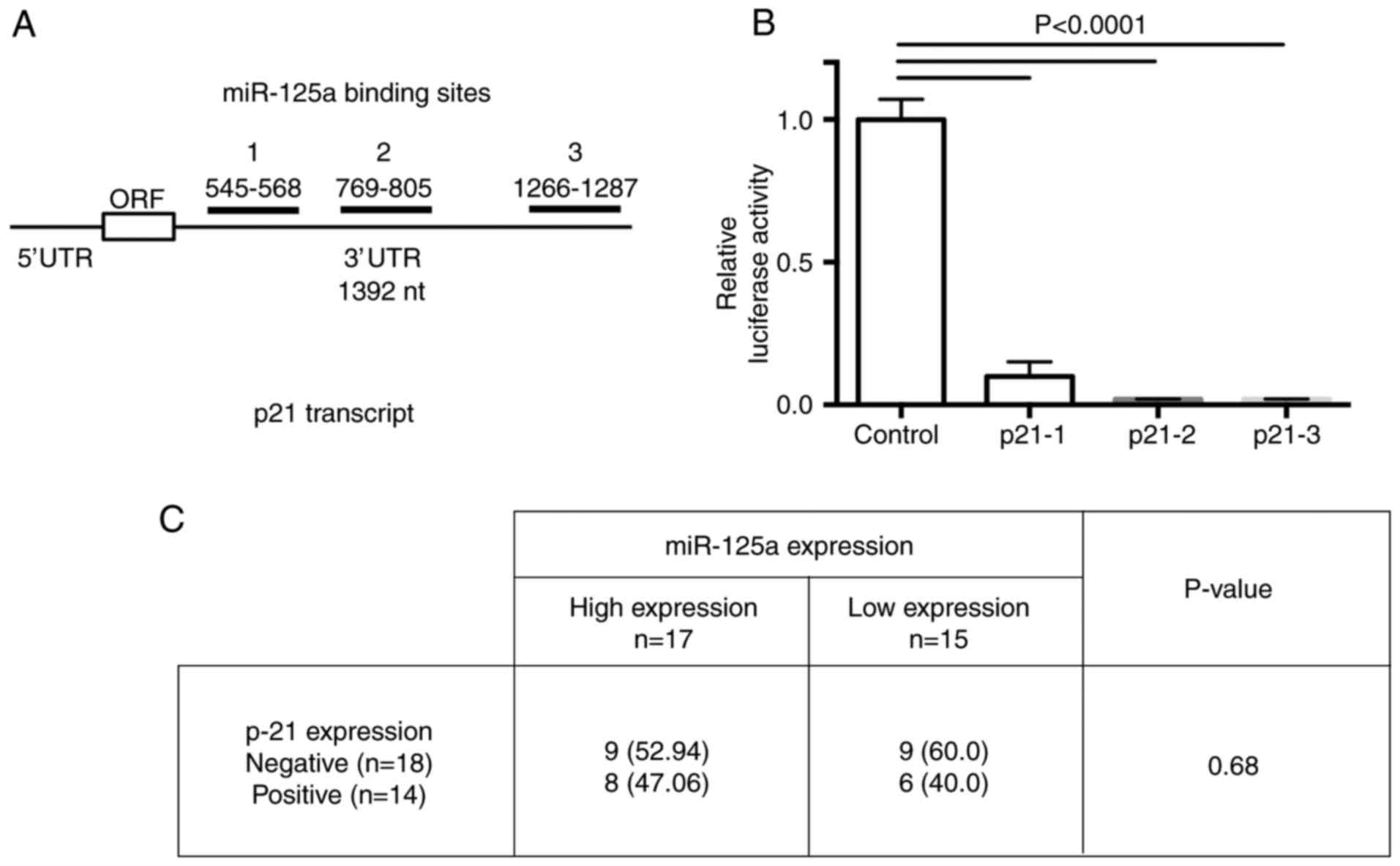
and CDKN1A mRNA comprise the following: 1, 545–568; 2, 769–805, and
3, 1266–1287 nucleotides into the 3′UTR of the p21 transcript
(Fig. 5A). After normalization with
the β-gal control, luciferase activity was suppressed in all 3
binding sites cloned. These results demonstrated that the 3 cloned
miR-125a-binding sequences are usable for inhibition of p21
transcript expression by miR-125a (Fig.
5B).
P-21 expression in CC tissues
In order to confirm that p-21 could be a RR marker
in CC tissues; we assessed protein expression by means of IHC in 32
paraffin-embedded tissues (in 15 CR and in 17 NR tissue samples).
We observed a slight difference in the p21 protein expression level
between both sample groups without a statistically significant
significance (Fig. 5C). We
hypothesized that the number of samples analyzed by IHC did not
allow corroborating a higher level of the p21 protein in patients
diagnosed as non-responders to standard chemotherapy and
radiotherapy treatment.
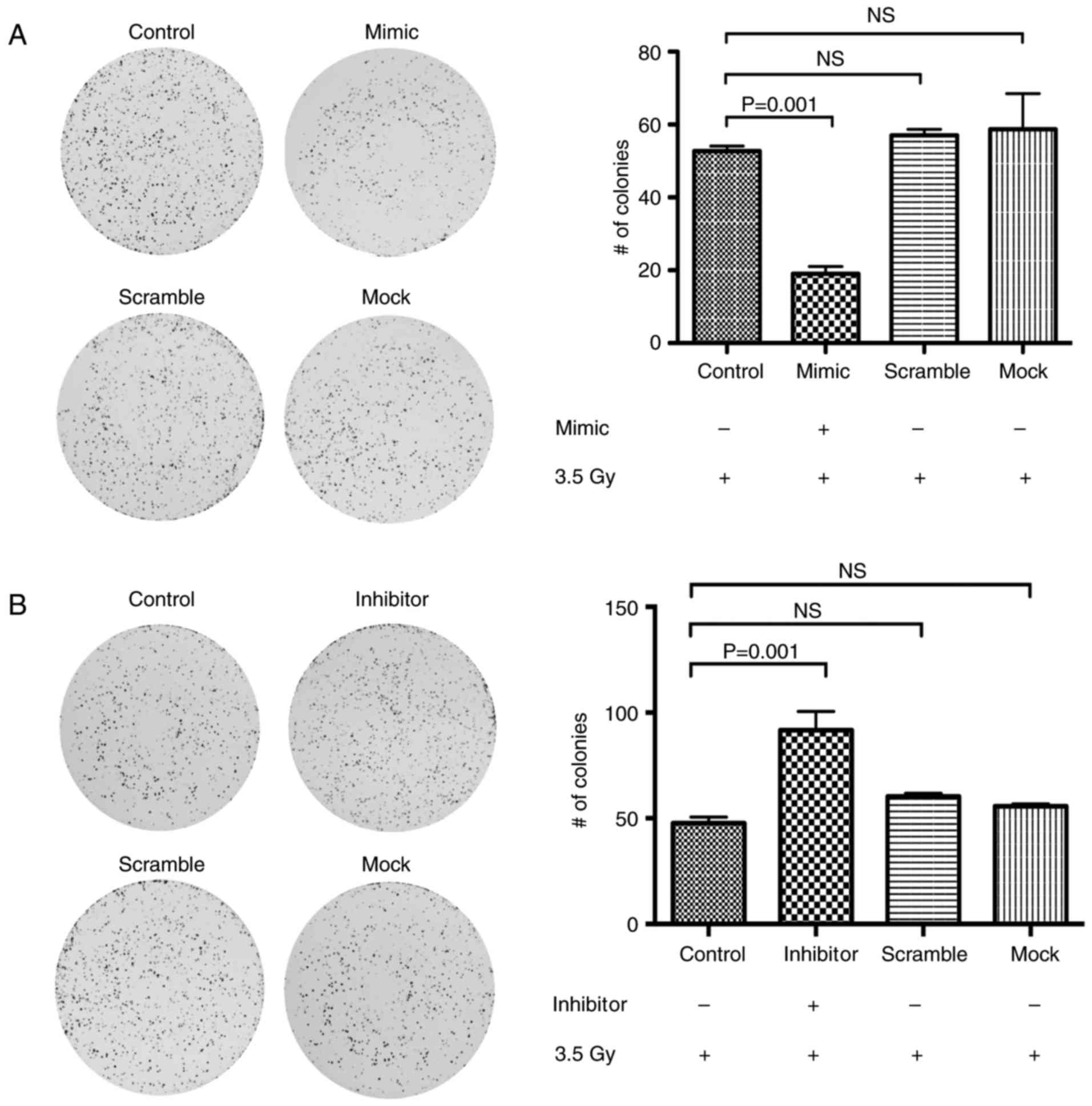
miR-125a sensitizes CC cell lines to
radiation therapy
To elucidate whether overexpression of miR-125a
sensitizes CC cells to irradiation treatment, we transfected
miR-125a mimics and inhibitors into HeLa cells and determined the
sensitivity of transfected cells after the irradiation dose. First,
we corroborated the level of expression of p21 in the transfected
cells (Fig. 4C). Then, after 24 h,
we performed a post-radiation CFA and cell survival fractions were
calculated after 2 weeks of incubation. Notably, we found a
significant decrease in survival fractions in the miR-125
mimic-transfected cells (Fig. 6A)
compared to their control cell lines (negative and scramble
oligonucleotide conditions) (P=0.005). When cells were transfected
with the miR-125 inhibitor we found a higher number of colonies
(Fig. 6B; P=0.001) in respect to
their respective negative and scramble control cells. These
findings strongly suggest that miR-125a is a regulator of acquired
radioresistance in CC.
Discussion
Recently, several research groups have reported that
the radioresistance phenotype in cell lines and tumor samples could
be explained by the expression profile of miRNAs. Notably, these
studies have listed some miRNAs as possible regulators of the
radioresistant phenotype (12–14).
However, only a few of these have been assessed by functional
assays to demonstrate a possible regulatory role. In the present
study, we demonstrated that miR-125a regulates the expression of
p21 and that this event is related to the sensitization of CC cells
to radiation therapy. The acquisition of radioresistance is a
complex process in which a wide number of genes and several
signaling pathways are involved (15–17). A
single miRNA regulates several hundreds of genes that could be
implicated in the radioresistance phenotype. Due to this, miRNAs
possess evident advantages for their consideration as
radioresistance modulators based on their ability to control
multiple targets in specific molecular pathways.
Here we showed that miR-125a is downregulated in
patients with cervical cancer diagnosed as non-responders (NRs) to
conventional therapy (Fig. 2A).
Moreover, these results were validated in vitro employing
radioresistant (RR) and radiosensitive (RS) HeLa, SiHa and CaSki
cervical cancer cell lines (Fig.
2B). This evidence indicates a correlation between the
expression level of miR-125a and the radioresistant phenotype in
cervical cancer.
Recently, Moskwa et al identified a subset of
miRNAs associated with radioresistance in glioblastoma cell lines.
Among the identified miRNAs there were the following: miR-1;
miR-150; miR-425; and notably, miR-125a (18). In addition, the authors
demonstrated, by functional assays, that miR-125a promotes
radioresistance in LN229 and U251 cell lines. In this same research
line, Shiiba et al demonstrated the participation of
miR-125b (an miR-125a homologous miRNA) in the acquisition of
radioresistance in oral squamous cell carcinoma cells to radiation
therapy (19). In CC, Fan et
al showed that downregulation of miR-125a is correlated with
preoperative metastasis and poorer progression-free survival (PFS)
and overall survival (OS) rates in patients with CC (20). In a subsequent study published by
Fan et al, the authors demonstrated that miR-125a is one of
the most significantly subexpressed miRNAs in HeLa and CaSki CC
cell lines resistant to cisplatin. Through functional assays, they
demonstrated that miR-125a depletion sensitizes chemoresistant CC
cell lines to paclitaxel in vitro and in vivo, and to
cisplatin in vivo through the regulation of STAT3 (21). These previously published data
strongly support our findings that the downregulation of miR-125a
is associated with non-response to conventional treatment in
patients with CC and could be involved in the regulation of
radioresistance and chemoresistance in CC cell lines.
CDKN1A is a validated target of miR-125a; in the
present study we showed that CDKN1A is overexpressed by
radioresistant cervical cancer cell lines. The CDKN1A gene is
located on chromosome 6p21.2 and encodes for the cyclin-dependent
kinase inhibitor 1 protein, better known as p21. Depending on its
cellular localization, p21 engages in several functions. Hence,
nuclear p21 inhibits the activity of cyclin-dependent kinases Cdk1
and Cdk2, blocking G1/S transition. Cytoplasmic p21 inhibits the
function of caspase-3, accomplishing anti-apoptotic functions.
Various studies have shown that p21 is deregulated by the effect of
ionizing radiation and is associated with the acquisition of
radioresistance (22,23). In the present study, we demonstrated
that the p21 protein is overexpressed in radioresistance cell
lines, confirming previous studies (Fig. 4A and B).
Additionally, we noted that CDKN1A mRNA and p21
protein expression is upregulated when the cells are exposed to a
sublethal radiation dose (3.5 Gy). Taken together, our results
showed that the expression of miR-125a is associated with the
response to radioresistance in patients with CC and, additionally,
they are correlated the decreased expression of p21 in
radioresistant cells. To corroborate the aforementioned findings,
we demonstrated that the ectopic expression of miR-125 decreased
>40% of the expression of the p21 protein in HeLa cells 48 h
post-transfection, and that this severely affects its cellular
ability to respond to radiation therapy (Fig. 6A and B).
The identification of specific miRNAs associated
with the regulation of radioresistance may lead to improved,
efficient treatments for patients with CC. In this respect, new
experimental methodologies should be able to corroborate the role
of miRNA candidates through functional assays. Moreover,
best-candidate miRNAs should be able to be detected in serum
fractions as useful predictor biomarkers of the radioresistance
phenotype. As demonstrated in the present study, the miR-125a mimic
sensitizes cervical cancer cells to radiation therapy, and
regulation of their target genes directly impacts the molecular
mechanisms associated with the acquisition of radioresistance.
These findings and those previously published by
other authors support the idea that miRNAs could function as
central regulators of radioresistance acquisition by cancer cells.
Our findings determined a specific role for miRNA-125a in the
radioresistance of cervical cancer, and it may be considered for
future therapeutic strategies for this neoplasia.
Acknowledgements
The present study was partially supported by the
CONACyT (SALUD-2014-1-233733 and Cátedras-2425).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancerstatistics, 2012. CA
Cancer J Clin. Mar;65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Delaney G, Jacob S, Featherstone C and
Barton M: The role of radiotherapy in cancer treatment: Estimating
optimal utilization from a review of evidence-based clinical
guidelines. Cancer. 104:1129–1137. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Benedet JL, Odicino F, Maisonneuve P,
Beller U, Creasman WT, Heintz AP, Ngan HY and Pecorelli S:
Carcinoma of the cérvix uteri. Int J Gynaecol Obstet. 83 Suppl
1:S41–S78. 2003. View Article : Google Scholar
|
|
4
|
Zhang B, Chen J, Ren Z, Chen Y, Li J, Miao
X, Song Y, Zhao T, Li Y, Shi Y, et al: A specific miRNA signature
promotes radioresistance of human cervical cancer cells. Cancer
Cell Int. 13:1182013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang CX, Zhang SM, Li J, Yang B, Ouyang W,
Mei ZJ, Chen J, Dai J, Ke S, Zhou FX, et al: MicroRNA-320 regulates
the radiosensitivity of cervical cancer cells C33AR by targeting
β-catenin. Oncol Lett. 12:4983–4990. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pedroza-Torres A, Fernández-Retana J,
Peralta-Zaragoza O, Jacobo-Herrera N, Cantú de Leon D, Cerna-Cortés
JF, Lopez-Camarillo C and Pérez-Plasencia C: A microRNA expression
signature for clinical response in locally advanced cervical
cancer. Gynecol Oncol. 142:557–565. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bousquet M, Harris MH, Zhou B and Lodish
HF: MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA.
107:pp. 21558–21563. 2010; View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hulanicka M, Garncarz M,
Parzeniecka-Jaworska M and Jank M: Plasma miRNAs as potential
biomarkers of chronic degenerative valvular disease in Dachshunds.
BMC Vet Res. 10:2052014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cowden Dahl KD, Dahl R, Kruichak JN and
Hudson LG: The epidermal growth factor receptor responsive miR-125a
represses mesenchymal morphology in ovarian cancer cells.
Neoplasia. 11:1208–1215. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and expression.
Nucleic Acids Res. 36:D149–D153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Asgharian A, Banan M and Najmabadi H:
Optimizing a lipocomplex-based gene transfer method into HeLa cell
line. Cell J. 15:372–377. 2014.PubMed/NCBI
|
|
12
|
Li G, Qiu Y, Su Z, Ren S, Liu C, Tian Y
and Liu Y: Genome-wide analyses of radioresistance-associated miRNA
expression profile in nasopharyngeal carcinoma using next
generation deep sequencing. PLoS One. 8:e844862013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo W, Xie L, Zhao L and Zhao Y: mRNA and
microRNA expression profiles of radioresistant NCI-H520 non-small
cell lung cancer cells. Mol Med Rep. 12:1857–1867. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
McDermott N, Meunier A, Wong S, Buchete V
and Marignol L: Profiling of a panel of radioresistant prostate
cancer cells identifiesde regulation of key miRNAs. Clin Transl
Radiat Oncol. 2:63–68. 2017. View Article : Google Scholar
|
|
15
|
Maier P, Hartmann L, Wenz F and Herskind
C: Cellular pathways in response to ionizin gradiation and their
target ability for tumor radio sensitization. Int J Mol Sci.
17:1022016. View Article : Google Scholar :
|
|
16
|
Ishigami T, Uzawa K, Higo M, Nomura H,
Saito K, Kato Y, Nakashima D, Shiiba M, Bukawa H, Yokoe H, et al:
Genes and molecular pathways related to radioresistance of oral
squamous cell carcinoma cells. Int J Cancer. 120:2262–2270. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Skvortsova I, Skvortsov S, Stasyk T, Raju
U, Popper BA, Schiestl B, Guggenberg Ev, Neher A, Bonn GK, Huber LA
and Lukas P: Intracellular signaling pathways regulating
radioresistance of human prostate carcinoma cells. Proteomics.
8:4521–4533. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Moskwa P, Zinn PO, Choi YE, Shukla SA,
Fendler W, Chen CC, Lu J, Golub TR, Hjelmeland A and Chowdhury D: A
functional screen identifies miRs that induce radioresistance in
glioblastomas. Mol Cancer Res. 12:1767–1778. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shiiba M, Shinozuka K, Saito K, Fushimi K,
Kasamatsu A, Ogawara K, Uzawa K, Ito H, Takiguchi Y and Tanzawa H:
MicroRNA-125b regulates proliferation and radioresistance of oral
squamous cell carcinoma. Br J Cancer. 108:1817–1821. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fan Z, Cui H, Xu X, Lin Z, Zhang X, Kang
L, Han B, Meng J, Yan Z, Yan X and Jiao S: MiR-125a suppresses
tumor growth, invasion and metastasis in cervical cancerbytargeting
STAT3. Oncotarget. 6:25266–25280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fan Z, Cui H, Yu H, Ji Q, Kang L, Han B,
Wang J, Dong Q, Li Y, Yan Z, et al: MiR-125a promotes paclitaxel
sensitivity in cervical cancer through altering STAT3 expression.
Oncogenesis. 5:e1972016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kraus A, Gross MW, Knuechel R, Münkel K,
Neff F and Schlegel J: Aberrant p21 regulation in radioresistant
primary glioblastoma multiforme cells bearing wild-type p53. J
Neurosurg. 93:863–872. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kokunai T, Urui S, Tomita H and Tamaki N:
Overcoming of radioresistance in human gliomas by p21WAF1/CIP1
antisense oligonucleotide. J Neurooncol. 51:111–119. 2001.
View Article : Google Scholar : PubMed/NCBI
|