Introduction
Cervical cancer is the third most common type of
gynaecological cancer and the fourth leading cause of
cancer-related mortality worldwide (1). A total of 530,000 new cases and
275,000 mortalities were estimated to be caused by cervical cancer
annually worldwide (2). Currently,
the mainstay therapy for patients with cervical cancer includes
surgical resection, radiotherapy, chemotherapy and other
comprehensive treatments (3).
Despite substantial advances in diagnostic technologies and
therapeutic methods, the therapeutic outcomes of patients with
cervical cancer remain unsatisfactory, particularly for those
diagnosed with an advanced stage of disease (4). Distant metastasis, lymph node
recurrence and cancer recurrence are primarily responsible for the
unfavourable prognosis of patients with cervical cancer (5). Persistent human papillomavirus (HPV)
infection has been identified as a primary cause of cervical cancer
(6). However, the detailed
mechanisms of the pathogenesis of cervical cancer remain largely
elusive. Therefore, further investigation into the mechanisms
underlying the tumour onset and aggressiveness of cervical cancer
is required in order to identify novel therapeutic targets that may
improve the prognosis of patients with this disease.
MicroRNAs (miRNAs) are a large group of endogenous,
single-stranded, non-coding RNA molecules comprising 18–25
nucleotides (7). miRNAs regulate
genes expression by directly interacting with the complementary
sites from the 3′-untranslated regions (3′-UTRs) of their target
genes, causing mRNA degradation and/or translation inhibition
(8). One miRNA can modulate
numerous target genes; therefore, miRNAs naturally serve as key
regulators of various physiological and pathological behaviours,
including cell proliferation, cycle, apoptosis, differentiation,
metabolism and metastasis (9).
Recent studies have revealed that miRNAs are deregulated in the
majority of human malignancies and that their deregulation is
involved in tumourigenesis and tumour development (10,11).
In cervical cancer, various miRNAs are aberrantly expressed,
including miR-379 (12), miR-383
(13), miR-466 (14) and miR-1297 (15). Furthermore, miRNAs may serve
tumour-suppressing or oncogenic roles in cervical cancer, depending
on the detailed roles of their target genes (16,17).
Therefore, miRNAs may be identified as promising therapeutic
targets for treating patients with cervical cancer.
Recent studies have reported that miR-874 is
frequently dysregulated in multiple types of human cancer, and that
it acts as a key player in tumourigenesis and tumour development
(18–21). However, the expression pattern,
impacts and underlying mechanisms of miR-874 in cervical cancer
have not been elucidated. Therefore, the present study measured
miR-874 expression, analysed the biological roles of miR-874 and
investigated its molecular mechanisms in cervical cancer cells.
Materials and methods
Tissue samples
In total, 49 pairs of cervical cancer tissues and
adjacent non-cancerous tissues were collected from patients (age
range, 46–72 years; mean age, 63 years) who underwent surgical
resection at Huzhou Central Hospital (Zhejiang, China) between May
2014 and March 2016. None of the patients had been treated with
chemotherapy or radiotherapy prior to surgery. All fresh tissues
were quickly frozen in liquid nitrogen and stored at −80°C. The
present study was approved by the Ethics Committee of Huzhou
Central Hospital, and written informed consent was provided by all
participants.
Cell culture
A total of four cervical cancer cell lines (SiHa,
HeLa, C-33A and CaSki) and a normal human cervix epithelial cell
line (Ect1/E6E7) were purchased from American Type Culture
Collection (Manassas, VA, USA). All cell lines were cultured in
Dulbecco's modified Eagle's medium (DMEM) containing 10% v/v fetal
bovine serum (FBS), 1% v/v penicillin/streptomycin mixture (all
from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), and
maintained at 37°C in an incubator with 5% CO2 and 95%
air.
Transfection assay
miR-874 mimics, negative control miRNA mimics
(miR-NC), small interfering RNA (siRNA) targeting the expression of
ETS1 (ETS1 siRNA) and negative control siRNA (NC siRNA) were
obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China). The
sequences used were as follows: miR-874 mimic,
5′-CUGCCCUGGCCCGAGGGACCGA-3′; miR-NC, 5′-UUCUCCGAACGUGUCACGUTT-3′;
ETS1 siRNA, 5′-ACUUGCUACCAUCCCGUACTT-3′; and NC siRNA,
5′-UUCUCCGAACGUGUCACGUTT-3′. In order to restore ETS1 expression,
the ETS1 overexpression plasmid pCMV-ETS1 and empty plasmid pCMV,
which were chemically synthesised by the Chinese Academy of
Sciences (Shanghai, China), were applied. For cell transfection,
cells were inoculated into 6-well plates one day prior to
transfection. miRNA mimics (100 pmol), siRNA (100 pmol) or plasmids
(4 µg) were transfected into cells using Lipofectamine 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
according to the manufacturer's protocol. At 8 h post-transfection,
the cell culture medium was discarded, and fresh DMEM containing
10% v/v FBS was added into each well.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from tissue specimens and culture cells
was isolated using the TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), according to the
manufacturer's protocol. To quantify miR-874 expression, reverse
transcription was performed using a TaqMan miRNA Reverse
Transcription kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocols. The temperature
protocol for reverse transcription was as follows: 16°C for 30 min,
42°C for 30 min and 85°C for 5 min. Next, a TaqMan miRNA PCR kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.) was adopted
for qPCR, with U6 small nuclear RNA as an internal reference. The
thermocycling conditions for qPCR were as follows: 50°C for 2 min,
95°C for 10 min; 40 cycles of denaturation at 95°C for 15 sec; and
annealing/extension at 60°C for 60 sec. To determine the level of
ETS1 mRNA, complementary DNA (cDNA) was produced from total RNA
using a PrimeScript RT reagent kit and this DNA was then subjected
to qPCR using a SYBR Premix Ex Taq™ (both Takara Biotechnology Co.,
Ltd., Dalian, China). The temperature protocol for reverse
transcription was as follows: 37°C for 15 min and 85°C for 5 sec.
qPCR was performed using the following thermocycling conditions: 5
min at 95°C, followed by 40 cycles of 95°C for 30 sec and 65°C for
45 sec. The primers were designed as follows: miR-874 forward,
5′-TGCGGCTGCCCTGGCCCGAGGGAC-3′ and reverse,
5′-CCAGTGCAGGGTCCGAGGT-3′; U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; ETS1 forward,
5′-CCACCACTACTACCGAAA-3′ and reverse, 5′-AACACTTCTGCTTGATGGC-3′;
and GAPDH forward, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse,
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′. GAPDH served as an internal control
for ETS1 mRNA expression. The relative expression of miR-874 and
ETS1 was analysed using the 2−ΔΔCq method (22).
Cell Counting kit-8 (CCK-8) assay
CCK-8 assay was performed to determine cell
proliferative ability. In brief, transfected cells were seeded into
96-well plates at a density of 3×103 cells per well and
incubated at 37°C under 5% CO2 for 0, 1, 2 or 3 days. At
indicated time points, CCK-8 assay was initiated by adding 10 µl
CCK-8 solution (Dojindo Molecular Technologies, Inc., Kumamoto,
Japan) into each well. Following incubation at 37°C for 2 h, the
absorbance value of each well was detected at a wavelength of 450
nm using a microplate reader (Bio-Rad Laboratories, Inc., Hercules,
CA, USA).
Colony formation assay
At 24 h after transfection, cells were harvested and
prepared into a single-cell suspension. Transfected cells were
plated into 6-well plates at a density of 1,000 cells/well and were
then incubated at 37°C in an incubator under 5% CO2 and
95% air for 14 days. At day 15, the cells were fixed with 95%
methanol for 20 min at room temperature and stained with methyl
violet (Beyotime Institute of Biotechnology, Shanghai, China) at
room temperature for 20 min. The number of cell colonies (>50
cells/colony) was counted under an inverted microscope (×200
magnification; IX83; Olympus Corporation, Tokyo, Japan).
Flow cytometric assay
Transfected cells were incubated at 37°C in an
incubator under 5% CO2 and 95% air for 48 h.
Subsequently, the transfected cells were collected, washed with PBS
and then subjected to cell apoptosis detection using an Annexin V
fluorescein isothiocyanate (FITC) apoptosis detection kit
(BioLegend, Inc., San Diego, CA, USA). In brief, transfected cells
were resuspended with 100 µl binding buffer. Next, the transfected
cells were stained with 5 µl Annexin V-FITC and 5 µl propidium
iodide (PI) at room temperature. Following incubation for 20 min in
the dark, the cell apoptosis rate was detected using a flow
cytometer (FACScan; BD Biosciences, Franklin Lakes, NJ, USA), and
analyzed using CellQuest software version 3.3 (BD Biosciences,
Franklin Lakes, NJ, USA).
Transwell migration and invasion
assays
Cell migration and invasion abilities were
determined using Transwell chambers (Corning Incorporated, Corning,
NY, USA) coated without or with Matrigel (BD Biosciences),
respectively. Transfected cells were collected at 48 h
post-transfection and were resuspended with FBS-free DMEM. The
upper Transwell chambers were filled with 1×105 cells in
FBS-free DMEM, and the lower chambers were filled with 500 µl DMEM
containing 10% FBS. The chambers were incubated at 37°C in an
atmosphere of 5% CO2 and 95% air for 24 h. The cells
were fixed with 95% methanol at room temperature for 15 min,
stained with 0.5% crystal violet at room temperature for 15 min and
washed with PBS. The number of migrated and invaded cells in five
randomly selected visual fields/chambers was counted under an
inverted microscope at ×200 magnification.
Bioinformatics analysis and luciferase
reporter assay
TargetScan7.1 (http://www.targetscan.org/) and microRNA.org (http://www.microrna.org/microrna/home.do) were applied
to predict the putative targets of miR-874. ETS1 was predicted as a
major potential target of miR-874. The wild-type (Wt) and mutant
(Mut) 3′-UTR of ETS1 was produced by Shanghai GenePharma Co., Ltd.
and inserted into the pGL3 luciferase vector (Promega Corporation,
Madison, WI, USA) to generate pGL3-ETS1-3′-UTR Wt and
pGL3-ETS1-3′-UTR Mut, respectively. Cells were inoculated into
24-well plates at a density of 1.0×105 cells each well.
Following overnight incubation, miR-874 mimics or miR-NC, in
combination with pGL3-ETS1-3′-UTR Wt or pGL3-ETS1-3′-UTR Mut, were
transfected into cells using Lipofectamine 2000 reagent. Luciferase
activities were detected at 48 h post-transfection using a Dual
Luciferase Reporter assay kit (Promega Corporation), according to
the manufacturer's protocol. Firefly luciferase activity was
normalized to Renilla luciferase activity.
Western blot analysis
Total protein was extracted from tissue specimens or
culture cells using radioimmunoprecipitation assay buffer, and the
concentration of total protein was quantified using a bicinchoninic
acid assay kit (both Beyotime Institute of Biotechnology, Inc.,
Shanghai, China). Subsequently, 10% SDS-PAGE was utilised to
separate equal amounts of proteins (20 µg); the separated proteins
were transferred onto polyvinylidene difluoride membranes (EMD
Millipore, Billerica, MA, USA). Subsequently, membranes were
blocked with 5% skimmed milk that was dissolved in Tris-buffered
saline containing 0.1% Tween-20 (TBST) for 1 h at room temperature.
The membranes were incubated with monoclonal anti-mouse ETS1
antibody (1:1,000 dilution; cat. no. ab96478; Abcam, Cambridge, UK)
and monoclonal anti-rabbit GAPDH (1:1,000 dilution; cat. no.
ab9484; Abcam) primary antibodies overnight at 4°C. Following
washing three times with TBST, the membranes were incubated with
corresponding horseradish peroxidase-conjugated secondary
antibodies (1:5,000 dilution; cat. no. ab6789; Abcam) at room
temperature for 2 h. The protein signals were visualised using an
enhanced chemiluminescence plus reagent (GE Healthcare, Chicago,
IL, USA). Protein expression was quantified using Quantity One
software version 4.62 (Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
Statistical analysis
Statistical analysis was performed using SPSS
software (version 18.0; SPSS, Inc., Chicago, IL, USA). Data are
expressed as the mean ± standard deviation from ≥3 independent
experiments and analysed using Student's t-test or one-way analysis
of variance (ANOVA). Student-Newman-Keuls test was applied for post
hoc analysis following ANOVA. The association between miR-874 and
the clinicopathological features of patients with cervical cancer
was analysed using χ2 test. Spearman's correlation
analysis was performed to evaluate the correlation between miR-874
and ETS1 mRNA in cervical cancer tissues. P<0.05 were considered
to indicate a statistically significant difference.
Results
miR-874 is downregulated in cervical
cancer tissues and cell lines
To clarify the expression pattern of miR-874 in
cervical cancer, miR-874 expression was initially detected in 49
pairs of cervical cancer tissues and adjacent non-cancerous
tissues. The results of RT-qPCR analysis demonstrated that miR-874
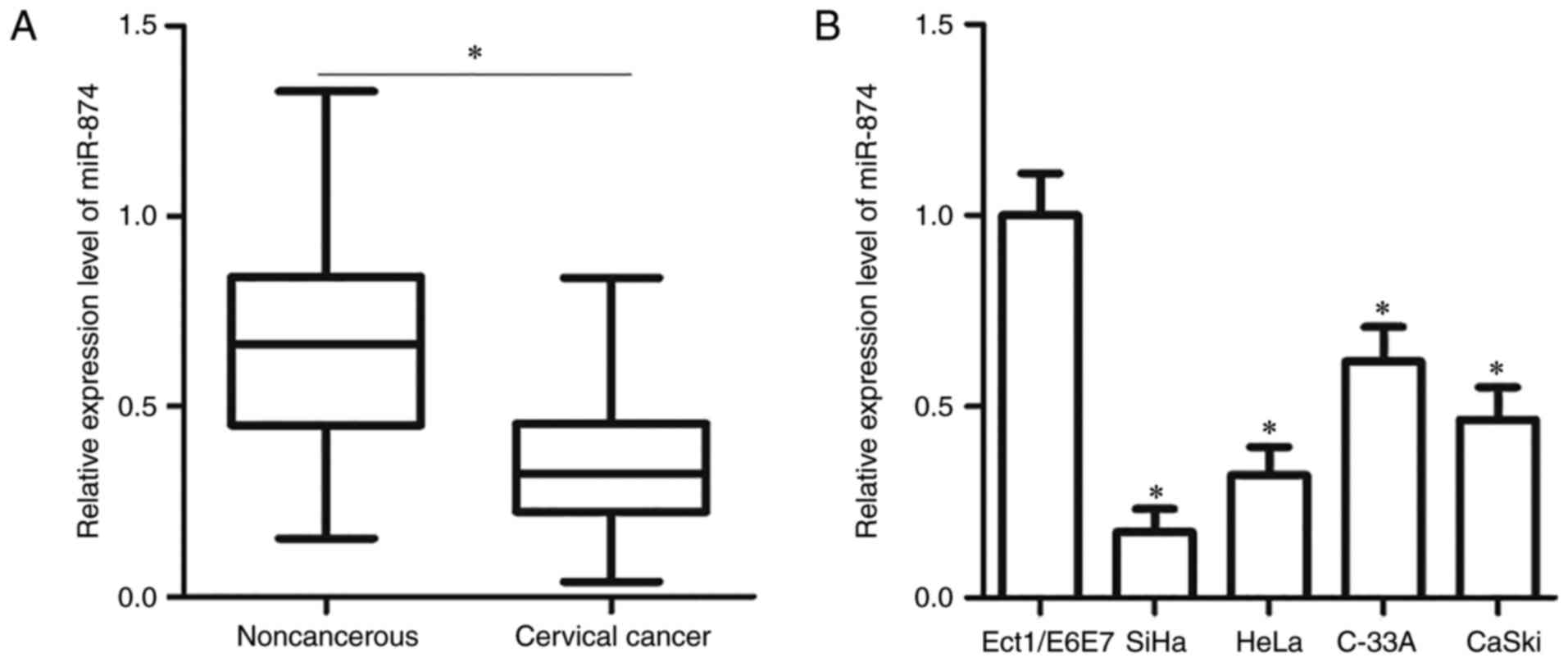
expression was downregulated in cervical cancer tissues compared
with that in adjacent non-cancerous tissues (P<0.05; Fig. 1A). To determine the clinical
significance of miR-874 in cervical cancer, the association between
miR-874 and the clinicopathological features of patients with
cervical cancer was determined. All patients were divided into
either low- or high-expression groups based on the median
expression of miR-874. The results demonstrated that reduced
miR-874 levels were correlated with the FIGO stage (P=0.007) and
lymph node metastasis (P=0.032). However, no significant
correlation was observed with other clinicopathological factors,
including age, tumour size or family history of cancer (all
P>0.05; Table I). The expression
level of miR-874 was also determined in four cervical cancer cell
lines (SiHa, HeLa, C-33A and CaSki) and a normal human cervix
epithelial cell line (Ect1/E6E7). miR-874 expression was
downregulated in all the tested cervical cancer cell lines,
compared with that in Ect1/E6E7 cells (P<0.05; Fig. 1B). These results suggested that
miR-874 is associated with cervical cancer progression.
 | Table I.Association between miR-874 and
clinicopathological features of patients with cervical cancer. |
Table I.
Association between miR-874 and
clinicopathological features of patients with cervical cancer.
|
| miR-874
expression |
|
|---|
|
|
|
|
|---|
| Clinicopathological
features | Low | High | P-value |
|---|
| Age, years |
|
| 0.478 |
|
<60 | 7 | 9 |
|
|
≥60 | 18 | 15 |
|
| Tumour size,
cm |
|
| 0.674 |
|
<4 | 11 | 12 |
|
| ≥4 | 14 | 12 |
|
| Family history of
cancer |
|
| 0.291 |
|
Yes | 5 | 8 |
|
| No | 20 | 16 |
|
| FIGO stage |
|
| 0.007 |
|
I–II | 7 | 16 |
|
|
III–IV | 18 | 8 |
|
| Lymph node
metastasis |
|
| 0.032 |
| No | 7 | 14 |
|
|
Yes | 18 | 10 |
|
miR-874 inhibits the proliferation and
promotes the apoptosis of cervical cancer cells
To illustrate the biological roles of miR-874 in
cervical cancer, the SiHa and HeLa cells were used, which exhibited
relatively lower miR-874 expression compared with the two other
cervical cancer cell lines in subsequent functional assays. SiHa
and HeLa cells were transfected with miR-874 mimics or miR-NC. The
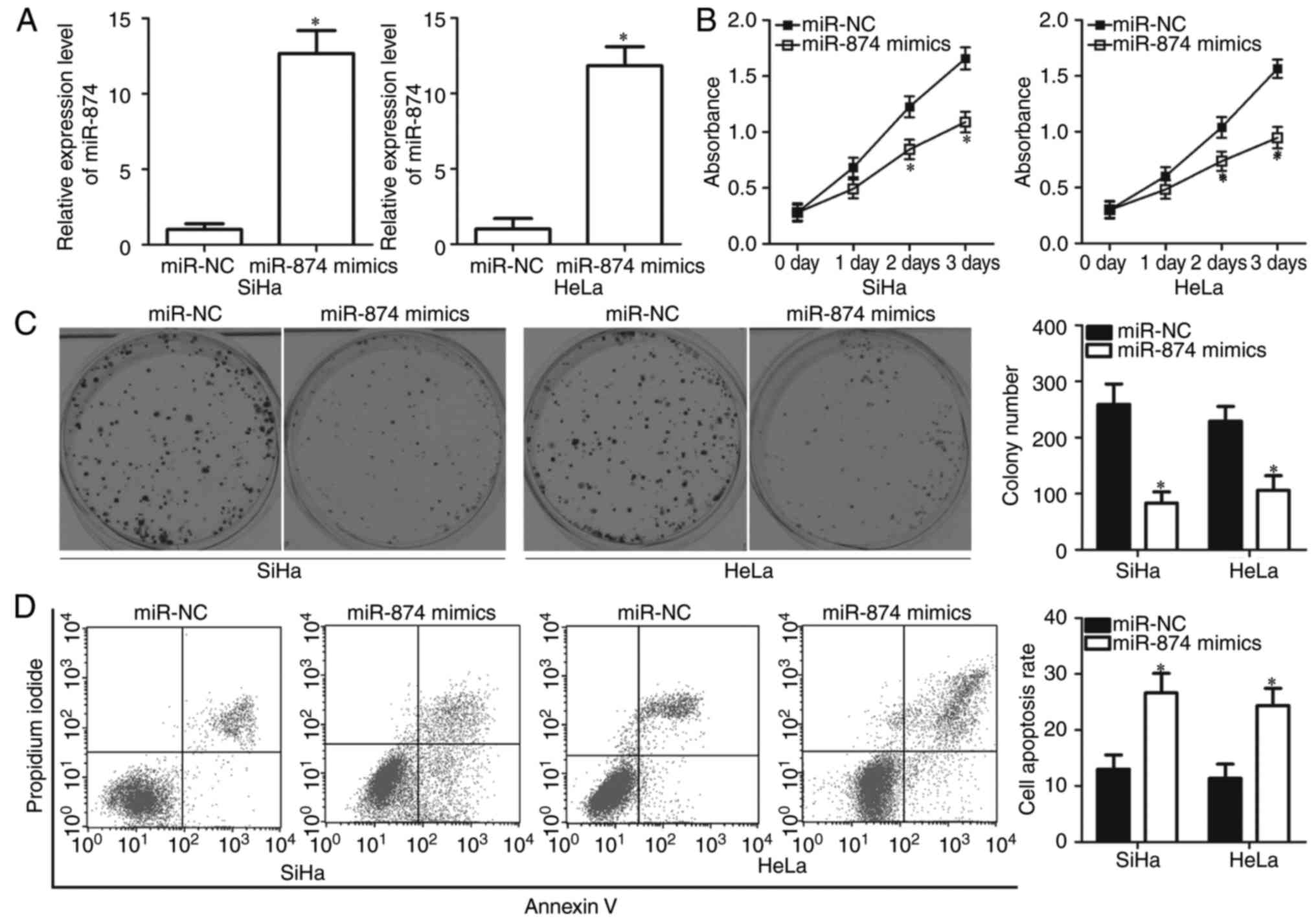
results of RT-qPCR analysis confirmed that miR-874 was markedly
overexpressed in SiHa and HeLa cells following transfection with
miR-874 mimics (P<0.05; Fig.
2A). CCK-8 assay was performed to detect the proliferation of
SiHa and HeLa cells that were transfected with miR-874 mimics or
miR-NC. Ectopic miR-874 expression significantly decreased the
proliferative abilities of SiHa and HeLa cells (P<0.05; Fig. 2B). To confirm this observation, a
colony formation assay was used to evaluate the effect of miR-874
overexpression on colony formation ability in cervical cancer.
Similarly, miR-874 upregulation significantly reduced the number
and size of surviving colonies compared with the miR-NC groups
(Fig. 2C; P<0.05). Flow
cytometric assay was employed to examine the biological functions
of miR-874 in cervical cancer cell apoptosis. The results revealed
that resumption of miR-874 expression increased the apoptotic rate
of SiHa and HeLa cells (Fig. 2D;
P<0.05). Taken together, these results suggested that miR-874
inhibits cell proliferation and promotes cell apoptosis in cervical
cancer.
miR-874 restricts cell migration and
invasion in cervical cancer
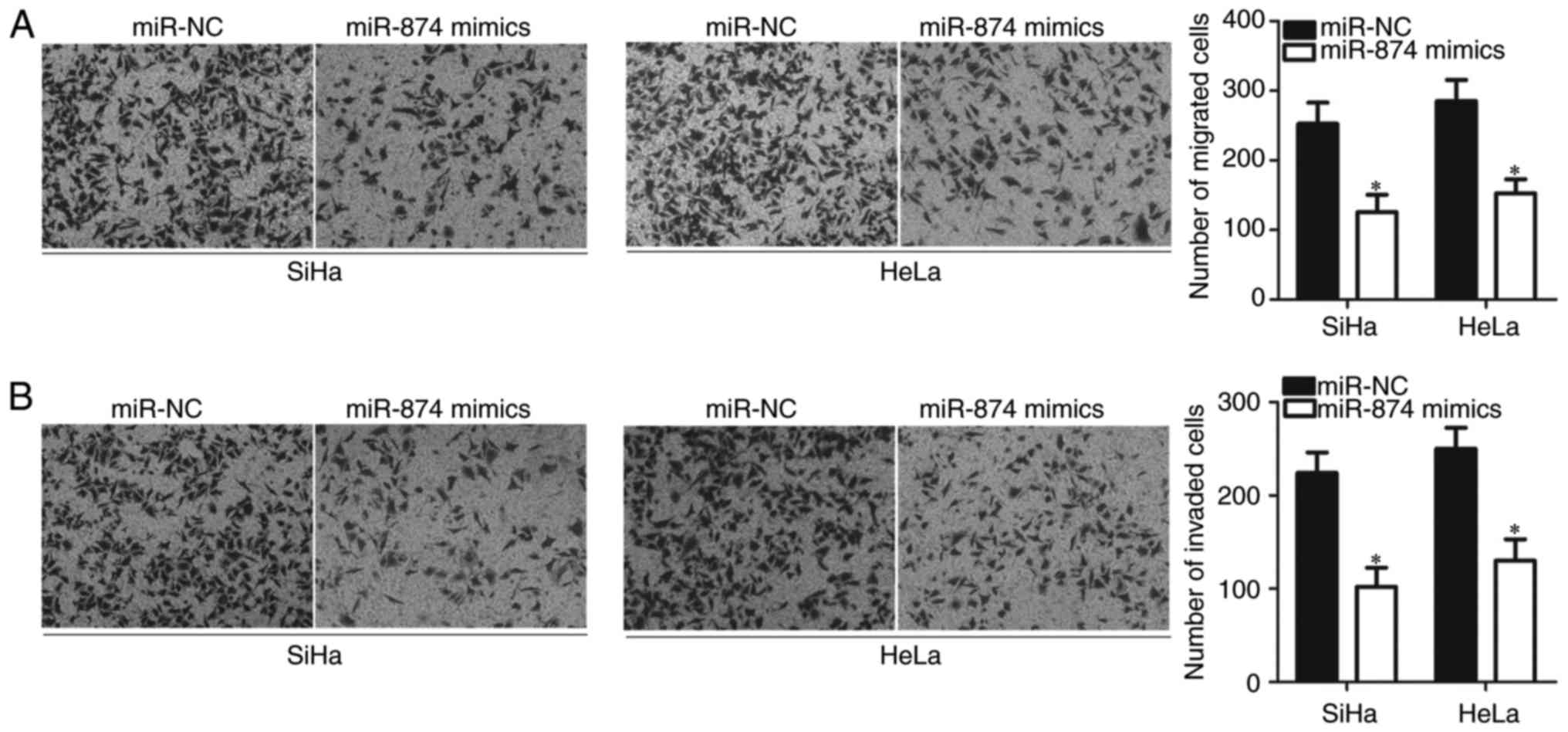
Considering the association between miR-874 and
lymph node metastasis, Transwell migration and invasion assays were
utilised to investigate the effect of miR-874 overexpression on
cervical cancer cell metastasis. SiHa and HeLa cells transfected
with miR-874 mimics exhibited significantly lower migration
(P<0.05; Fig. 3A) and invasion
(P<0.05; Fig. 3B) abilities,
compared with those transfected with miR-NC. These results
suggested that miR-874 serves a tumour-suppressive role in cervical
cancer metastasis.
miR-874 directly targets ETS1 in
cervical cancer cells
The biological roles of miRNA depend on its specific
target. Therefore, bioinformatic analysis was performed to predict
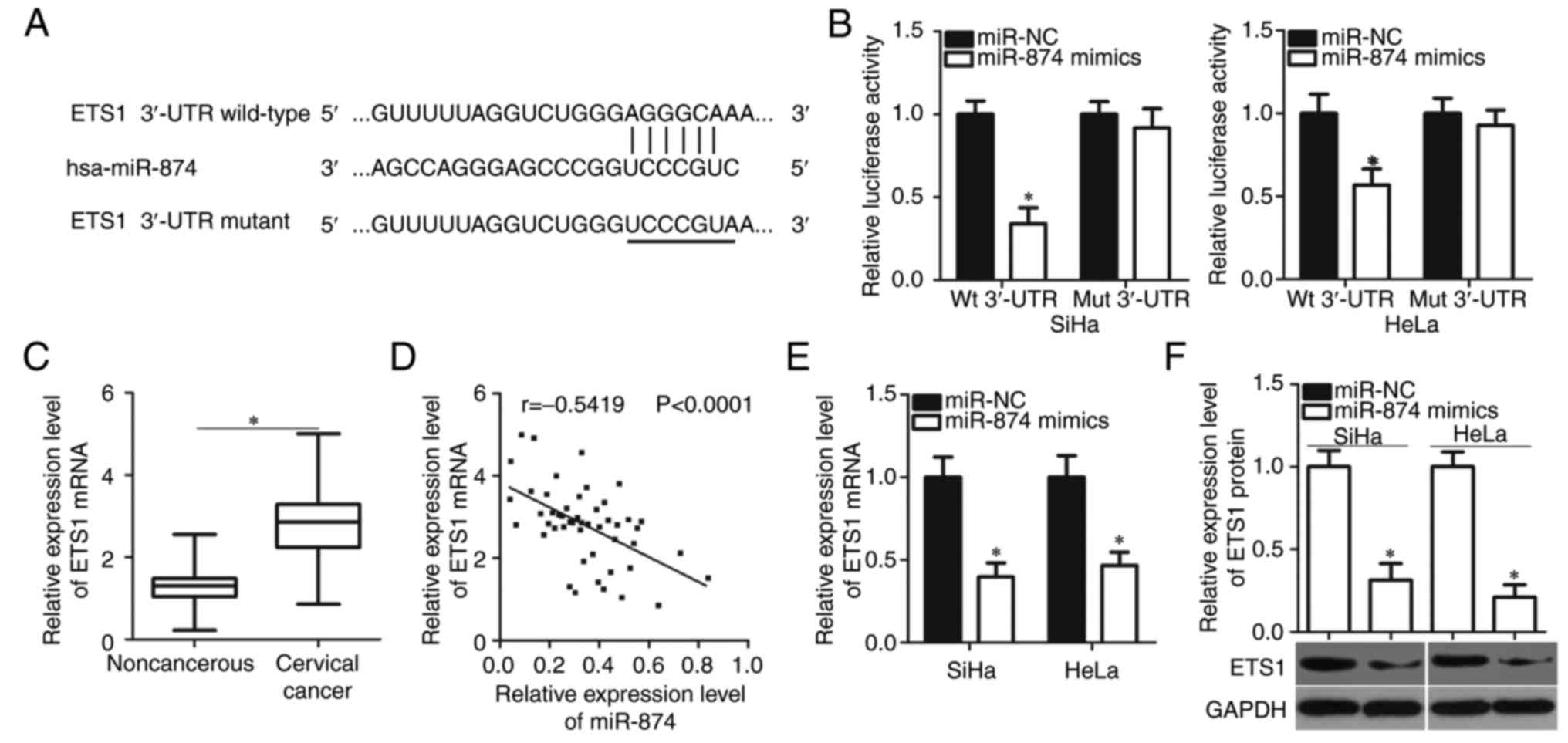
the putative target genes of miR-874. According to TargetScan7.1
and microRNA.org, ETS1 was a predicted target gene of
miR-874 (Fig. 4A). Luciferase
reporter assay was conducted to investigate whether miR-874
directly targets the 3′-UTR of ETS1 in cervical cancer cells.
miR-874 overexpression significantly suppressed the luciferase
activities of the plasmid carrying the Wt binding site in SiHa and
HeLa cells (P<0.05). However, the upregulation of miR-874 did
not serve an inhibitory role in the luciferase activities of the
plasmid harbouring the Mut binding site (Fig. 4B).
To further evaluate the association between miR-874
and ETS1 in cervical cancer, ETS1 expression was detected in 49
pairs of cervical cancer tissues and adjacent non-cancerous tissues
through RT-qPCR. Results revealed that the levels of ETS1 mRNA were
significantly higher in the cervical cancer tissues than in the
adjacent non-cancerous tissues (P<0.05; Fig. 4C). The ETS1 mRNA levels were also
identified to be inversely correlated with miR-874 expression in
cervical cancer tissues (r=−0.5419; P<0.0001; Fig. 4D). The effects of miR-874
overexpression on ETS1 mRNA and protein levels in SiHa and HeLa
cells were illustrated by RT-qPCR and western blot analysis,
respectively. Restoration of miR-874 expression decreased ETS1
expression in SiHa and HeLa cells at mRNA (P<0.05; Fig. 4E) and protein (P<0.05; Fig. 4F) levels. Taken together, these
results indicated that ETS1 is a direct target gene of miR-874 in
cervical cancer.
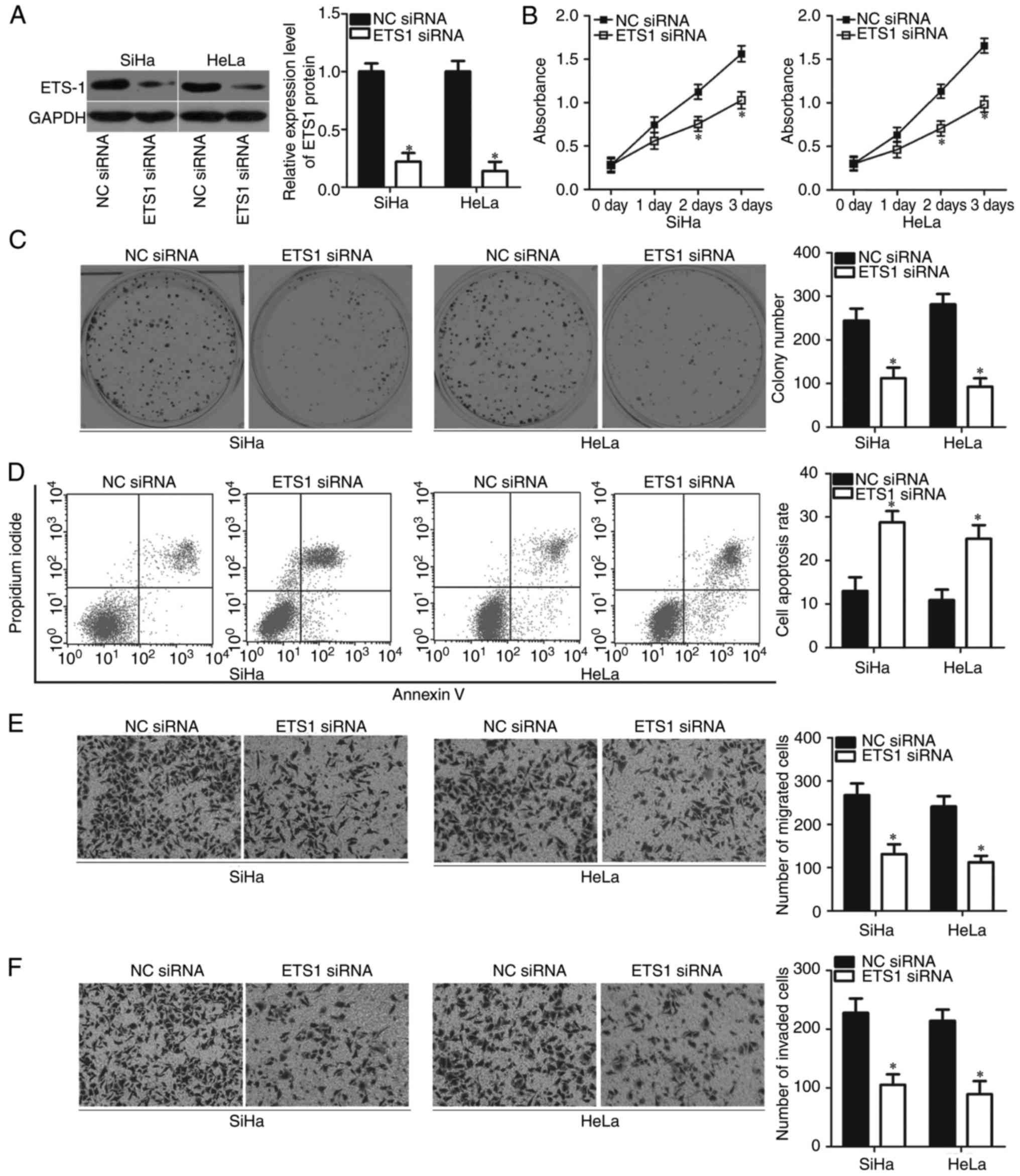
Inhibition of ETS1 simulates the
tumour-suppressive effects of miR-874 in cervical cancer cells
Considering that ETS1 is a direct target gene of
miR-874 in cervical cancer, we hypothesised that inhibiting ETS1
can mimic the biological functions of miR-874 in cervical cancer
cells. SiHa and HeLa cells were transfected with ETS1 siRNA to
knock down endogenous ETS1 expression. Following transfection,
western blot analysis revealed that ETS1 protein expression was
silenced effectively in the SiHa and HeLa cells transfected with
ETS1 siRNA (P<0.05; Fig. 5A). As
expected, inhibition of ETS1 significantly reduced the
proliferative (P<0.05; Fig. 5B)
and colony-forming abilities (P<0.05; Fig. 5C) of SiHa and HeLa cells.
ETS1-knockdown also promoted the apoptosis (P<0.05; Fig. 5D) and prohibited the metastasis
(P<0.05; Fig. 5E and F) of SiHa
and HeLa cells. These results further demonstrated that ETS1 is a
direct target gene of miR-874 in cervical cancer cells.
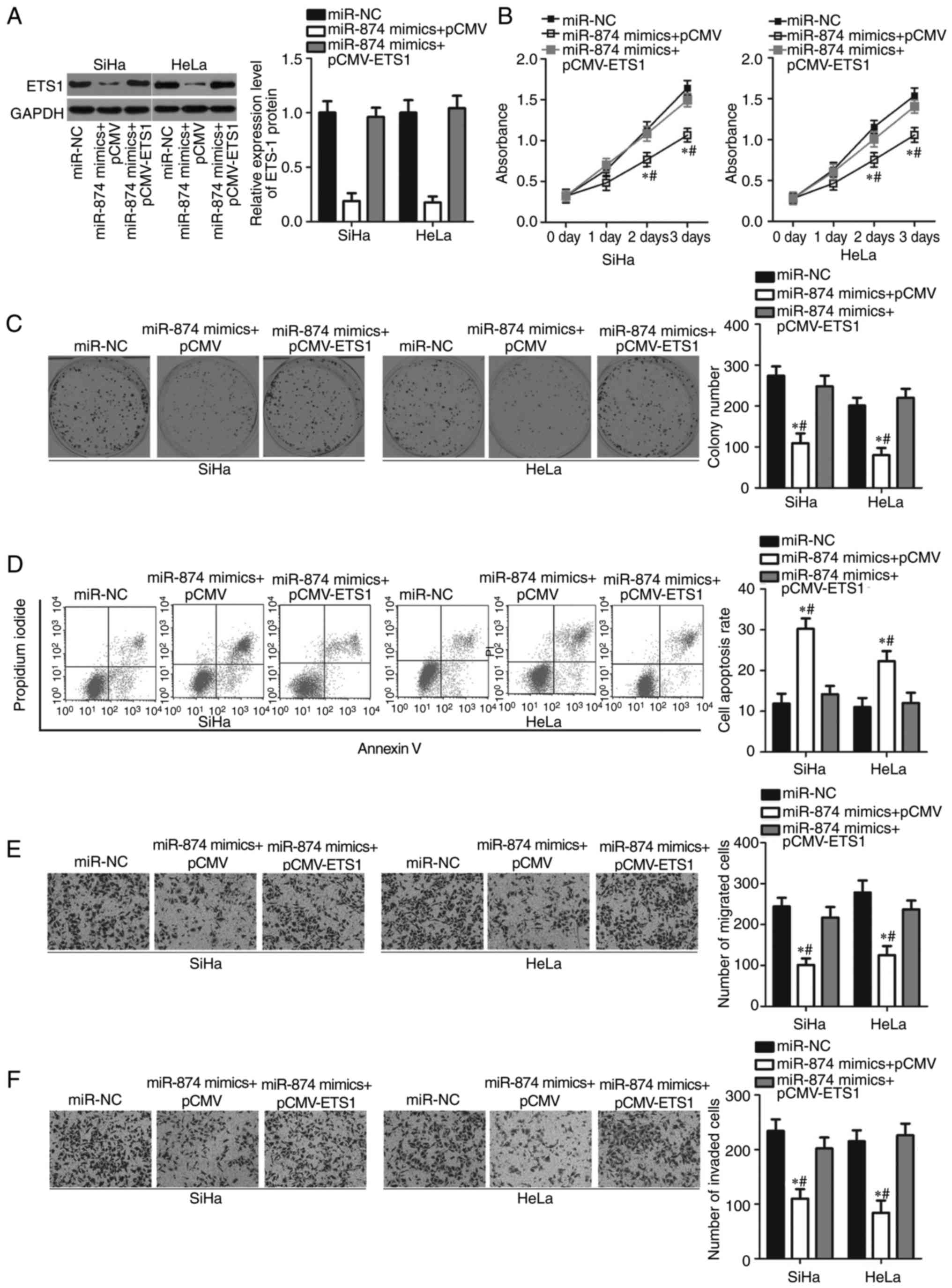
Upregulation of ETS1 reverses the
biological function of miR-874 overexpression in cervical cancer
cells
To investigate whether ETS1 is a functional mediator
of miR-874, rescue experiments were performed by transfecting
pCMV-ETS1 or pCMV into miR-874-overexpressing SiHa and HeLa cells.
Following transfection, ETS1 protein levels were detected using
western blot analysis. The decrease in ETS1 protein level induced
by miR-874 overexpression was significantly restored in the SiHa
and HeLa cells following co-transfection with pCMV-ETS1 (P<0.05;
Fig. 6A). Furthermore, recovered
ETS1 expression counteracted the effects of miR-874 overexpression
on the proliferation (P<0.05; Fig.
6B), colony formation (P<0.05; Fig. 6C), apoptosis (P<0.05; Fig. 6D), migration (P<0.05; Fig. 6E) and invasion (P<0.05; Fig. 6F) of SiHa and HeLa cells. These
findings clearly demonstrated that miR-874 may serve as a tumour
suppressor in cervical cancer progression, at least partly, by
suppressing ETS1 expression.
Discussion
An increasing number of studies have reported that
miRNAs are differentially expressed in cervical cancer and serve
critical roles in cervical oncogenesis and progression (23–25).
Therefore, identifying the aberrantly expressed miRNAs implicated
in the formation and progression of cervical cancer may provide key
clues for the development of effective therapeutic targets in
treating patients with this malignancy. In our current study,
miR-874 expression was significantly downregulated in cervical
cancer tissues and cell lines. Decreased miR-874 expression was
correlated with FIGO stage and lymph node metastasis. In addition,
resumption of miR-874 expression suppressed the proliferation,
migration and invasion, but increased the apoptosis of cervical
cancer cells. Further investigation demonstrated that ETS1 is a
novel target of miR-874 in cervical cancer. Furthermore, ETS1
expression was upregulated in cervical cancer tissues, and the
upregulation of ETS1 was inversely correlated with miR-874
expression. Furthermore, ETS1-knockdown simulated the tumour
suppressive effects of miR-874 in cervical cancer cells. Notably,
reintroduction of ETS1 expression counteracted the inhibitory
effects of miR-874 overexpression in cervical cancer cells. These
findings suggested that miR-874 may be a valuable therapeutic
target for treating patients with cervical cancer.
miR-874 is dysregulated in diverse types of human
malignancy. For example, miR-874 is downregulated in colorectal
cancer, and the downregulation of miR-874 is associated with TNM
stage and lymph node metastasis (18). In osteosarcoma, miR-874 expression
is downregulated in tumour tissues and cell lines. Low miR-874
expression is strongly correlated with TNM stage, tumour size and
lymph node metastasis in patients with osteosarcoma (19). In hepatocellular carcinoma,
decreased miR-874 expression in tumour tissues is significantly
associated with tumour size, vascular invasion, TNM stage, tumour
differentiation and inferior patient outcomes (20). In gastric cancer, miR-874 expression
is reduced in clinical samples and few cell lines. The
downregulation of miR-874 expression is correlated with certain
characteristic features of patients with gastric cancer that
indicate an unfavourable prognosis (21). These findings demonstrated that
miR-874 is frequently mildly expressed in these types of human
cancer and suggested that this miRNA has a strong diagnostic and
prognostic value.
Dysregulation of miR-874 participates in the
occurrence and development of multiple types of human cancer. For
instance, miR-874 overexpression restricts colorectal cancer cell
proliferation, inhibits colony formation, induces apoptosis and
improves the chemosensitivity to 5-FU by directly targeting XIAP
and STAT3 and inhibiting the Hippo signalling pathway (18,26,27).
Dong et al demonstrated that miR-874 targets E2F3 to impede
proliferation and motility, and to promote the apoptosis of
osteosarcoma cells (19). Multiple
studies have reported that induced expression of miR-874 suppresses
the growth, metastasis and epithelial-mesenchymal transition, but
promotes the apoptosis in vitro and reduces the
tumourigenicity in vivo of hepatocellular carcinoma cells by
inhibiting DOR, SOX12 and PIN1 expression (20,28,29).
Studies by Jiang et al (21)
and Zhang et al (30)
revealed that miR-874 re-expression represses the angiogenesis,
growth and motility in vitro, and the tumourigenicity in
vivo of gastric cancer cells via the blockade of STAT3 and
AQP3. These findings suggested that miR-874 may hold the potential
to be developed as a therapeutic target in treating patients with
these specific human malignancy types.
ETS1, a member of the ETS family of transcription
factors, is validated as a direct target gene of miR-874 in
cervical cancer. It directly binds to specific DNA sequences
containing a GGAA/T core motif and serves crucial roles in
carcinogenesis and cancer progression. ETS1 is reportedly
overexpressed in multiple types of human malignant tumour,
including ovarian cancer (31),
colorectal cancer (32), prostate
cancer (33) and gastric cancer
(34). ETS1 is also upregulated in
cervical cancer, and its upregulation is significantly correlated
with tumour differentiation and lymph node metastasis (35). Patients with cervical cancer
exhibiting high ETS1 levels exhibit a poorer prognosis than those
with low ETS1 levels (35,36). The deregulated ETS1 is implicated in
the oncogenesis and development of multiple types of human cancer
by regulating several pathological behaviours (37–39).
Taken together, these findings indicated that targeting ETS1
through miRNA-based targeted therapy has promising therapeutic
applications in treating patients with cervical cancer.
In summary, to the best of our knowledge, the
present study was the first to reveal that miR-874 inhibits the
progression of cervical cancer by directly targeting ETS1. These
findings highlighted the importance of the miR-874/ETS1 pathway in
the treatment of cervical cancer in the future.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YJ and HL designed this research. HL, YunfeiP,
YuefenP, JS, QQ and LZ performed functional experiments. WH and QW
analyzed the data of this study. All authors have read and approved
the final draft.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Huzhou Central Hospital (Huzhou, China), and was
performed in accordance with the Declaration of Helsinki and the
guidelines of the Ethics Committee of Huzhou Central Hospital.
Written informed consent was obtained from all patients for the use
of their clinical tissues.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arbyn M, Castellsagué X, de Sanjosé S,
Bruni L, Saraiya M, Bray F and Ferlay J: Worldwide burden of
cervical cancer in 2008. Ann Oncol. 22:2675–2686. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marth C, Landoni F, Mahner S, McCormack M,
Gonzalez-Martin A and Colombo N: Cervical cancer: ESMO clinical
practice guidelines for diagnosis, treatment and follow-up. Ann
Oncol. 28:iv72–iv83. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Seoud M, Tjalma WA and Ronsse V: Cervical
adenocarcinoma: Moving towards better prevention. Vaccine.
29:9148–9158. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kogo R, How C, Chaudary N, Bruce J, Shi W,
Hill RP, Zahedi P, Yip KW and Liu FF: The microRNA-218~Survivin
axis regulates migration, invasion, and lymph node metastasis in
cervical cancer. Oncotarget. 6:1090–1100. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Matsukura T and Sugase M: Pitfalls in the
epidemiologic classification of human papillomavirus types
associated with cervical cancer using polymerase chain reaction:
Driver and passenger. Int J Gynecol Cancer. 18:1042–1050. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Macfarlane LA and Murphy PR: MicroRNA:
Biogenesis, function and role in cancer. Curr Genomics. 11:537–561.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kent OA and Mendell JT: A small piece in
the cancer puzzle: MicroRNAs as tumor suppressors and oncogenes.
Oncogene. 25:6188–6196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shi X, Yuan N, Zhang S, Yuan F and Wang X:
MicroRNA-379 suppresses cervical cancer cell proliferation and
invasion by directly targeting V-crk avian sarcoma virus CT10
oncogene homolog-like. Oncol Res. Jan 2–2018.(Epub ahead of print).
doi: 10.3727/096504017X15140534417184. View Article : Google Scholar
|
|
13
|
Teng P, Jiao Y, Hao M and Tang X:
microRNA-383 suppresses the PI3K-AKT-MTOR signaling pathway to
inhibit development of cervical cancer via down-regulating PARP2. J
Cell Biochem. 119:5243–5252. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou LL, Shen Y, Gong JM, Sun P and Sheng
JH: MicroRNA-466 with tumor markers for cervical cancer screening.
Oncotarget. 8:70821–70827. 2017.PubMed/NCBI
|
|
15
|
Wang Z, He S, Guo P, Guo X and Zheng J:
MicroRNA-1297 inhibits metastasis and epithelial-mesenchymal
transition by targeting AEG-1 in cervical cancer. Oncol Rep.
38:3121–3129. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mei J, Wang DH, Wang LL, Chen Q, Pan LL
and Xia L: MicroRNA-200c suppressed cervical cancer cell metastasis
and growth via targeting MAP4K4. Eur Rev Med Pharmacol Sci.
22:623–631. 2018.PubMed/NCBI
|
|
17
|
Zhang Z, Wang J, Wang X, Song W, Shi Y and
Zhang L: MicroRNA-21 promotes proliferation, migration, and
invasion of cervical cancer through targeting TIMP3. Arch Gynecol
Obstet. 297:433–442. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Han J, Liu Z, Wang N and Pan W:
MicroRNA-874 inhibits growth, induces apoptosis and reverses
chemoresistance in colorectal cancer by targeting X-linked
inhibitor of apoptosis protein. Oncol Rep. 36:542–550. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dong D, Gong Y, Zhang D, Bao H and Gu G:
miR-874 suppresses the proliferation and metastasis of osteosarcoma
by targeting E2F3. Tumour Biol. 37:6447–6455. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Y, Wei Y, Li X, Liang X, Wang L,
Song J, Zhang X, Zhang C, Niu J, Zhang P, et al: microRNA-874
suppresses tumor proliferation and metastasis in hepatocellular
carcinoma by targeting the DOR/EGFR/ERK pathway. Cell Death Dis.
9:1302018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang B, Li Z, Zhang W, Wang H, Zhi X,
Feng J, Chen Z, Zhu Y, Yang L, Xu H and Xu Z: miR-874 inhibits cell
proliferation, migration and invasion through targeting aquaporin-3
in gastric cancer. J Gastroenterol. 49:1011–1025. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Granados-López AJ, Ruiz-Carrillo JL,
Servín-González LS, Martínez-Rodríguez JL, Reyes-Estrada CA,
Gutiérrez-Hernández R and López JA: Use of mature miRNA strand
selection in miRNAs families in cervical cancer development. Int J
Mol Sci. 18:E4072017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li J, Liu Q, Clark LH, Qiu H, Bae-Jump VL
and Zhou C: Deregulated miRNAs in human cervical cancer: Functional
importance and potential clinical use. Future Oncol. 13:743–753.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dai S, Lu Y, Long Y, Lai Y, Du P, Ding N
and Yao D: Prognostic value of microRNAs in cervical carcinoma: A
systematic review and meta-analysis. Oncotarget. 7:35369–35378.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao B and Dong AS: MiR-874 inhibits cell
growth and induces apoptosis by targeting STAT3 in human colorectal
cancer cells. Eur Rev Med Pharmacol Sci. 20:269–277.
2016.PubMed/NCBI
|
|
27
|
Que K, Tong Y, Que G, Li L, Lin H, Huang
S, Wang R and Tang L: Downregulation of miR-874-3p promotes
chemotherapeutic resistance in colorectal cancer via inactivation
of the Hippo signaling pathway. Oncol Rep. 38:3376–3386.
2017.PubMed/NCBI
|
|
28
|
Jiang T, Guan LY, Ye YS, Liu HY and Li R:
MiR-874 inhibits metastasis and epithelial-mesenchymal transition
in hepatocellular carcinoma by targeting SOX12. Am J Cancer Res.
7:1310–1321. 2017.PubMed/NCBI
|
|
29
|
Leong KW, Cheng CW, Wong CM, Ng IO, Kwong
YL and Tse E: miR-874-3p is down-regulated in hepatocellular
carcinoma and negatively regulates PIN1 expression. Oncotarget.
8:11343–11355. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang X, Tang J, Zhi X, Xie K, Wang W, Li
Z, Zhu Y, Yang L, Xu H and Xu Z: miR-874 functions as a tumor
suppressor by inhibiting angiogenesis through STAT3/VEGF-A pathway
in gastric cancer. Oncotarget. 6:1605–1617. 2015.PubMed/NCBI
|
|
31
|
Lin Z, Liu Y, Sun Y and He X: Expression
of Ets-1, Ang-2 and maspin in ovarian cancer and their role in
tumor angiogenesis. J Exp Clin Cancer Res. 30:312011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tokuhara K, Ogata Y, Nakagawa M and
Shirouzu K: Ets-1 expression in vascular endothelial cells as an
angiogenic and prognostic factor in colorectal carcinoma. Int Surg.
88:25–33. 2003.PubMed/NCBI
|
|
33
|
Xu S, Ge J, Zhang Z and Zhou W: MiR-129
inhibits cell proliferation and metastasis by targeting ETS1 via
PI3K/AKT/mTOR pathway in prostate cancer. Biomed Pharmacother.
96:634–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zheng L, Qi T, Yang D, Qi M, Li D, Xiang
X, Huang K and Tong Q: microRNA-9 suppresses the proliferation,
invasion and metastasis of gastric cancer cells through targeting
cyclin D1 and Ets1. PLoS One. 8:e557192013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu FL, Li YL, Wang ZD and Feng YJ:
Expression and significance about VEGF, KDR, MMP-1, and
transcription factor Ets-1 in human cervical carcinoma. Zhongguo Yi
Xue Ke Xue Yuan Xue Bao. 25:396–400. 2003.(In Chinese). PubMed/NCBI
|
|
36
|
Fujimoto J, Aoki I, Toyoki H, Khatun S and
Tamaya T: Clinical implications of expression of ETS-1 related to
angiogenesis in uterine cervical cancers. Ann Oncol. 13:1598–1604.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tomar S, Plotnik JP, Haley J, Scantland J,
Dasari S, Sheikh Z, Emerson R, Lenz D, Hollenhorst PC and Mitra AK:
ETS1 induction by the microenvironment promotes ovarian cancer
metastasis through focal adhesion kinase. Cancer Lett. 414:190–204.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu M, Liu X, Jin W, Li Y, Li Y, Hu Q, Chu
PK, Tang G and Ping Y: Targeting ETS1 with RNAi-based
supramolecular nanoassemblies for multidrug-resistant breast cancer
therapy. J Control Release. 253:110–121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang SY, Lu ZM, Lin YF, Chen LS, Luo XN,
Song XH, Chen SH and Wu YL: miR-144-3p, a tumor suppressive
microRNA targeting ETS-1 in laryngeal squamous cell carcinoma.
Oncotarget. 7:11637–11650. 2016.PubMed/NCBI
|