Introduction
Lung cancer is one of the most common malignancies
in humans and is the main cause of cancer-related deaths (1). Non-small cell lung cancer (NSCLC),
accounts for ~80% of all lung cancer cases and can be divided into
different histological types, including adenocarcinoma, squamous
cell carcinoma and large cell carcinoma, of which lung
adenocarcinoma (LUAD) is the most common subtype (2). Although significant progress has been
made in regards to surgery, chemotherapy, radiation therapy and
molecular-targeted therapy in the past few years, the overall
5-year survival rate of lung cancer patients is still only ~15%
(3). This is due to the lack of
effective early diagnostic methods and the limited efficacy of
current therapies. Thus, the importance of discovering simple and
effective biomarkers is not only reflected in early diagnosis, but
also in improving the prognosis of lung cancer patients.
Compared with the study of biomarkers of
protein-coding genes, human studies on non-coding RNA are
relatively few. However, the human genome contains more than 98%
non-protein coding sequences, with the vast majority transcribed
into long non-coding RNAs (lncRNAs) that are >200 bases in
length. In recent years, lncRNAs have been reported to serve as
diagnostic and prognostic markers in cancer (4–8). In
lung cancer, lncRNAs, such as MALAT-1 (9) and HOTAIR (10), have been associated with cancer
development. We also previously identified a series of
differentially expressed lncRNAs in 12 pairs of NSCLC tumors and
adjacent tumor tissues (8), and
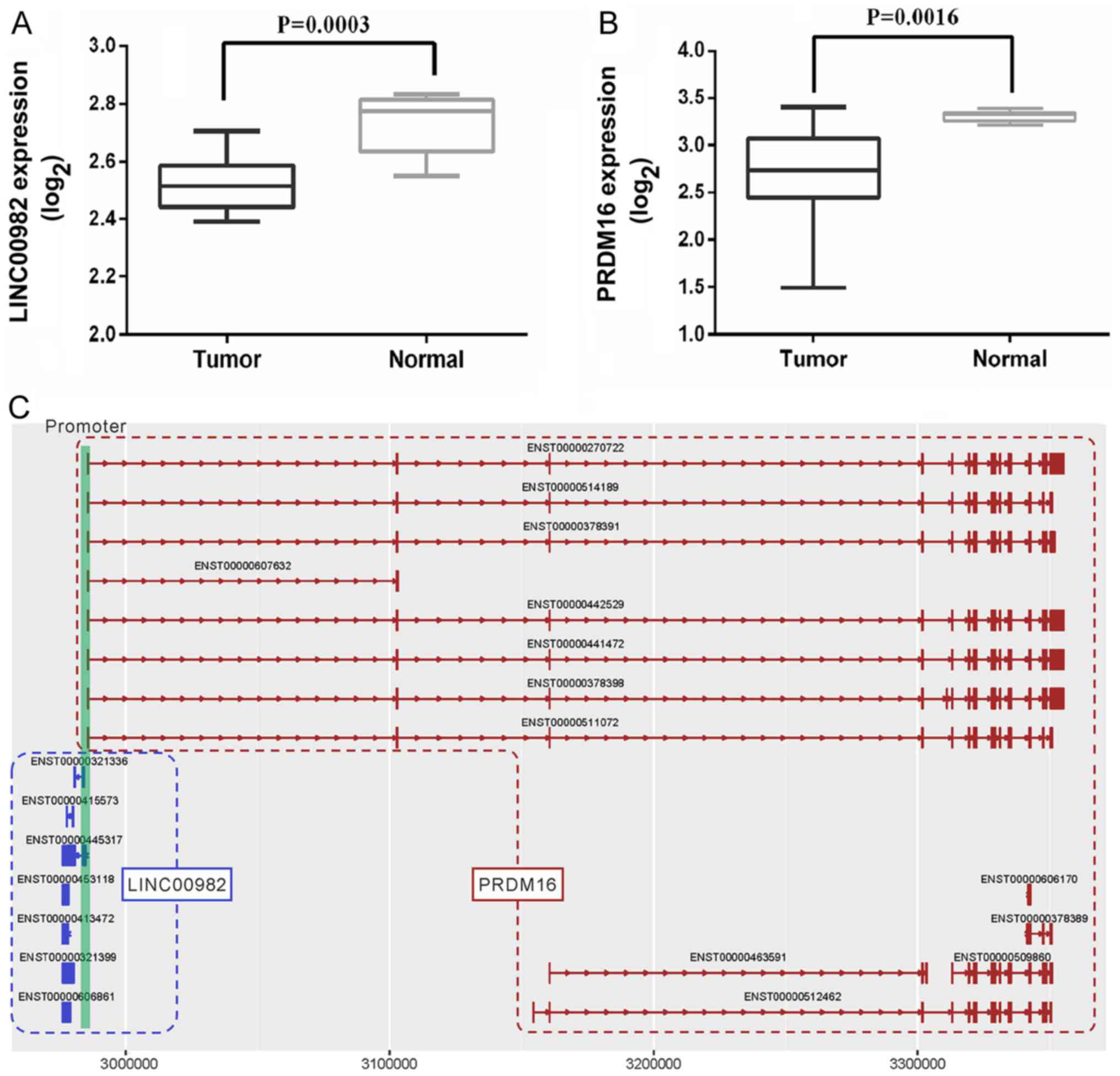
levels of long intergenic non-protein coding RNA 982
(LINC00982) and PR domain containing 16 (PRDM16) were
lower in NSCLC tumors than levels in adjacent non-tumor tissues
(Fig. 1A and B). In the present
study, we explored the expression and prognostic value of
LINC00982 in lung cancer.
LINC00982, located on chromosome 1p36.32, has
2 transcripts and has been reported to be a tumor suppressor in
gastric cancer (11,12). However, no studies are available
regarding the biological function of LINC00982 in lung
cancer. In the present study, we observed that LINC00982 and
PRDM16 share the same enhancer, ACTRT2 (enhancer ID:
GH01F003274). Dysfunction of PRDM16 has been found in many
diseases. In astrocytoma patients, poor prognosis can be predicted
by the hypomethylation status of the PRDM16 promoter
(13). Some recent studies have
reported that PRDM16 plays a significant role in the development of
cancer such as prostate (14),
colorectal (15,16) and myeloid cancers (17,18).
As in lung cancer, the PRDM16 promoter has been reported to
be methylated and upregulated PRDM16 suppressed lung cancer
cell growth (19), but the value of
this gene for diagnosis and prognosis has not been fully
explored.
Herein, we first analyzed the independent effect of
LINC00982 and PRDM16 expression on the clinical
features and survival status of LUAD patients and further explored
the combined effect of LINC00982 and PRDM16
expression on global gene expression, potentially affected pathways
and biological functions and the prognosis of LUAD patients.
Materials and methods
Data sources
LUAD transcriptome and clinical data were downloaded
from The Cancer Genome Atlas (TCGA, http://tcga-data.nci.nih.gov/) and the cBioPortal
(http://www.cbioportal.org/) database in
May 2016 (20,21). LUAD DNA promoter methylation data
were collected from MethHC (http://methhc.mbc.nctu.edu.tw/php/search.php?opt=gene).
In total, we downloaded TCGA level 3 data from 515 LUAD patients
and 59 controls. All samples had RNA sequencing on the Illumina
HiSeq 2000 version 2 platform and were normalized by the ‘RNA-Seq
by Expectation-Maximization’ (RSEM) method. Copy number data on
LINC00982 (also called FLJ42875), PRDM16 and
epidermal growth factor receptor (EGFR) were also downloaded
from cBioPortal database. We divided the LUAD patients into groups
according to the median expression of LINC00982 (3.89) and
PRDM16 (7.42). Patients in the high-PRDM16 group (≥7.42) and
high-LINC00982 group (≥3.89) were designated as the ‘both-high’
group, and those with a low expression of PRDM16 (<7.42)
and LINC00982 (<3.89) were considered as the ‘both-low’
group. In addition, we analyzed the expression profiles of
PRDM16 and LINC00982 in lung squamous cell carcinoma
(LUSC). Gene expression and clinical data on LUSC (including 501
patients and 51 controls) were also downloaded from TCGA and
analyzed in the same way as the LUAD data.
Differential expression analysis
A paired sample t-test was used to analyze the
differential gene expression and DNA methylation of
LINC00982 and PRDM16 between the 59 paired tumor
tissue and adjacent normal tissues. In order to illustrate the
association between copy number variation and gene expression of
LINC00982 and PRDM16, we matched LUAD patient IDs and
then divided these patients into high- and low-expression groups
according to the median expression of LINC00982 and
PRDM16. The Mann-Whitney U statistic was used to calculate
the differences between the two groups. We extracted expression
information on EGFR and used it as a reference. Spearman's
correlation analysis was used to explore the association of
LINC00982 and PRDM16 expression. A P-value <0.05
was considered to indicate a statistically significant
difference.
Differentially expressed genes in the ‘both-high’
and ‘both-low’ groups were analyzed using R v 3.3.3 (https://www.r-project.org/) and the bioconductor
ibrary (https://bioconductor.org/packages). The empirical
Bayes algorithm (function ‘eBayes’) in the limma package (22) was used to detect differentially
expressed genes between the ‘both-high’ and ‘both-low’ groups and
controls. We converted all gene expression values to z-scores and
used heatmaps in the ‘pheatmap’ package(https://CRAN.R-project.org/package=pheatmap) to
show the results. Significantly differentially expressed genes
(upregulated or downregulated) were considered as an absolute value
of the logarithmic transformed fold-change (log2 (FC)) ≥1 and a
false discovery rate (FDR)-adjusted P-value ≤0.05. A Venn diagram
was used to compare the upregulated and downregulated genes and
affected pathways between the ‘both-high’ and ‘both-low’ groups,
respectively.
Pathway enrichment analysis
We performed KEGG pathway enrichment analysis using
differentially expressed genes in the ‘both-high’ and ‘both-low’
groups. The following formula was used to conduct the enrichment
analysis:
P(X=k)=1-Cmk·CN-mn-kCNn
Where N is the number of all genes in the
dataset, m represents the number of differentially expressed
genes in the dataset, n is the number of all genes in the
enriched KEGG pathway and k is the number of differentially
expressed genes in the KEGG pathway. An FDR P-value ≤0.05 was
considered significantly enriched. The enrichment percentage in
each subsystem was calculated as the number of differentially
expressed genes divided by the number of all genes.
Gene co-expression with PRDM16 and
LINC00982 was defined by the Spearman's correlation
coefficient between each gene and PRDM16 and
LINC00982 expression. Genes with an absolute Spearman's
correlation coefficient >0.3 were considered to be co-expressed
with PRDM16 and LINC00982. In LUAD, the Spearman's
correlation coefficient information was downloaded from cBioPortal
(http://www.cbioportal.org/index.do).
Co-expressed genes were uploaded into the Ingenuity Pathway
Analysis software (Qiagen Redwood City Inc., Redwood City, CA, USA)
to compare enriched pathways.
Clinicopathological and survival
analysis
For clinical data analysis, categorical variables
(i.e., sex, race, residual tumors, primary site, stage and smoking
history) were given as numbers and percentages. Continuous
variables (e.g., age) are presented as the mean ± standard
deviation (SD). Student's t-tests were used to compare the means
for continuous variables in two groups, and χ2 tests
were used to compare the prevalence of categorical variables.
Kaplan-Meier survival curves were constructed to compare
differences in overall survival and disease-free survival between
the high-LINC00982 and low-LINC00982 groups and the high-PRDM16 and
low-PRDM16 groups, as well as the ‘both-low’ and ‘both-high’
groups. The log-rank test was used to assess differences in
survival between groups using the ‘survival’ package in R.
Furthermore, we analyzed the association of LINC00982 and
PRDM16 expression on overall survival and disease-free
survival stratified by tumor stage. The effect of LINC00982
and PRDM16 expression and other clinicopathological factors
(sex, age, residual tumors, primary site, stage and smoking status)
on overall survival and disease-free survival was analyzed by using
univariate Cox regression models. A multivariate Cox regression
model was used to compare the independent effect of
LINC00982 and PRDM16 expression on overall survival
and disease-free survival and adjusted for corresponding covariates
(smoking history, primary site, residual tumors and stage).
Cell lines
Human LUAD cell lines A549, H1299 and H1975 and a
normal lung epithelium cell line (BEAS-2B) were obtained from the
Chinese Academy of Sciences Committee on Type Culture Collection
Cell Bank (Shanghai, China). All cell lines were cultured in
Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(FBS; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), 25 U/ml
penicillin and 25 µg/ml streptomycin at 37°C in 5%
CO2.
Quantitative real-time PCR (RT-qPCR)
analysis
Total RNA was extracted from cell lines samples with
Invitrogen™ TRIzol reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Reverse-transcription
PCR was performed with the Prime-Script RT Reagent kit (Tiangen
Biotech, Beijing, China). Gene expression levels were determined by
RT-qPCR and normalized against an endogenous control
(β-actin) using SYBR Premix Ex Taq (ABI; Thermo Fisher
Scientific, Inc.). Data were analyzed using the ΔΔCt approach and
expressed as the target gene/β-actin ratio [2−ΔCt (target gene
- β−actin)]. The primers of the longer transcription of
LINC00982 (NR_015440.1, termed LINC00982-1) were as follows:
Forward: 5′-CCGGCCCTCTTAGCTTCAAA-3′ and reverse,
5′-GTGGAAAAGAAACCCACCGC-3′. The primers of the shorter
transcription of LINC00982 (NR_024371.1, termed LINC00982-2) were
as follows: Forward: 5′-GCTTCCCTTCCGTTCACTCA-3′ and reverse:
5′-GGCTGAGTCTTTCTGGACCC-3′. Primers for PRDM16 were as
follows: forward: 5′-GTTCTGCGTGGATGCAAATCA-3′ and reverse:
5′-GGTGAGGTTCTGGTCATCGC-3′. Data analysis was conducted using
GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA, USA). Data
were analyzed using one-way analysis of variance (ANOVA) followed
by the Least Significant Difference (LSD) method.
Results
LINC00982 and PRDM16 expression
profiles in NSCLC
The expression of LINC00982 and PRDM16
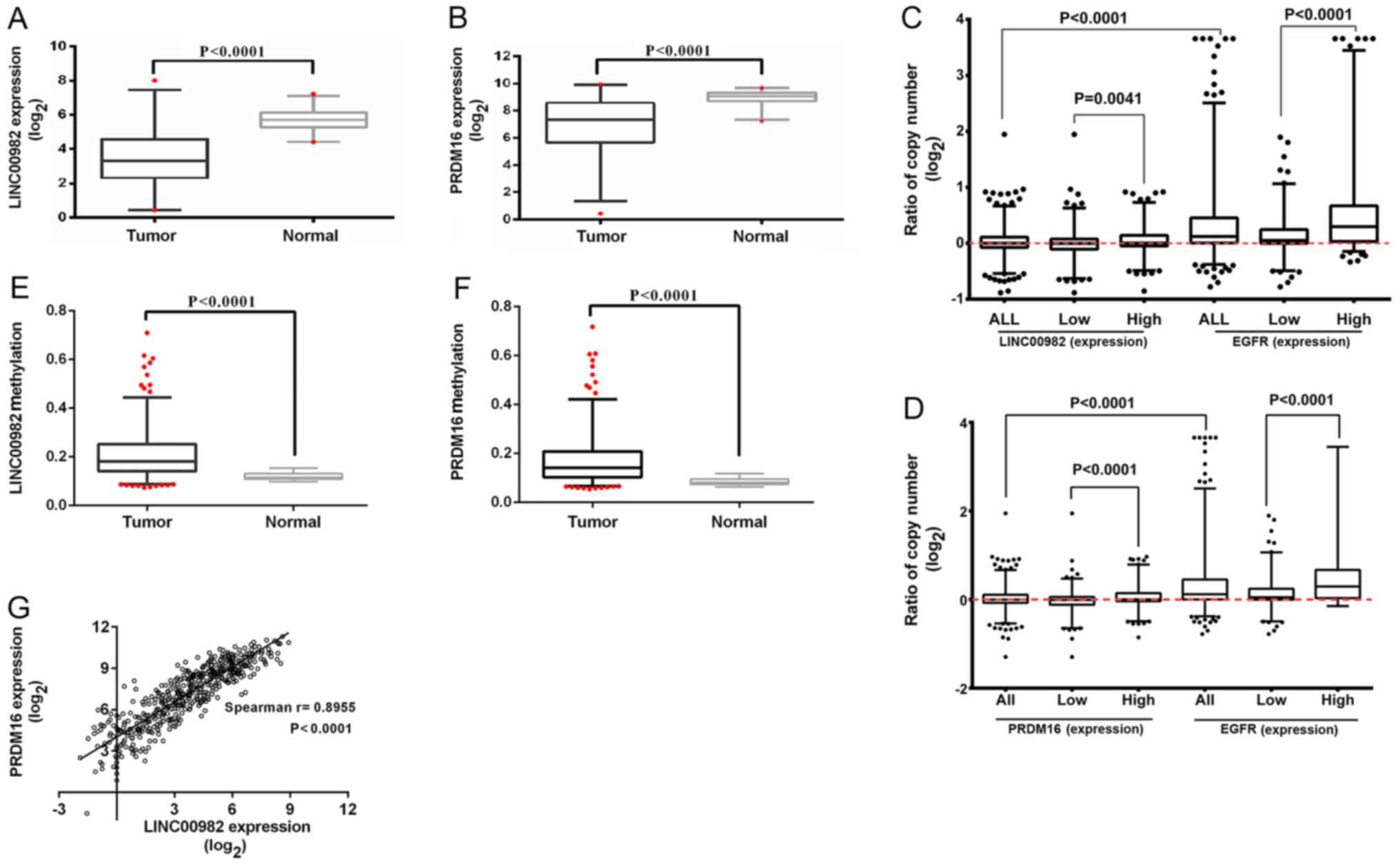
in 59 human LUAD tissues was significantly decreased compared to
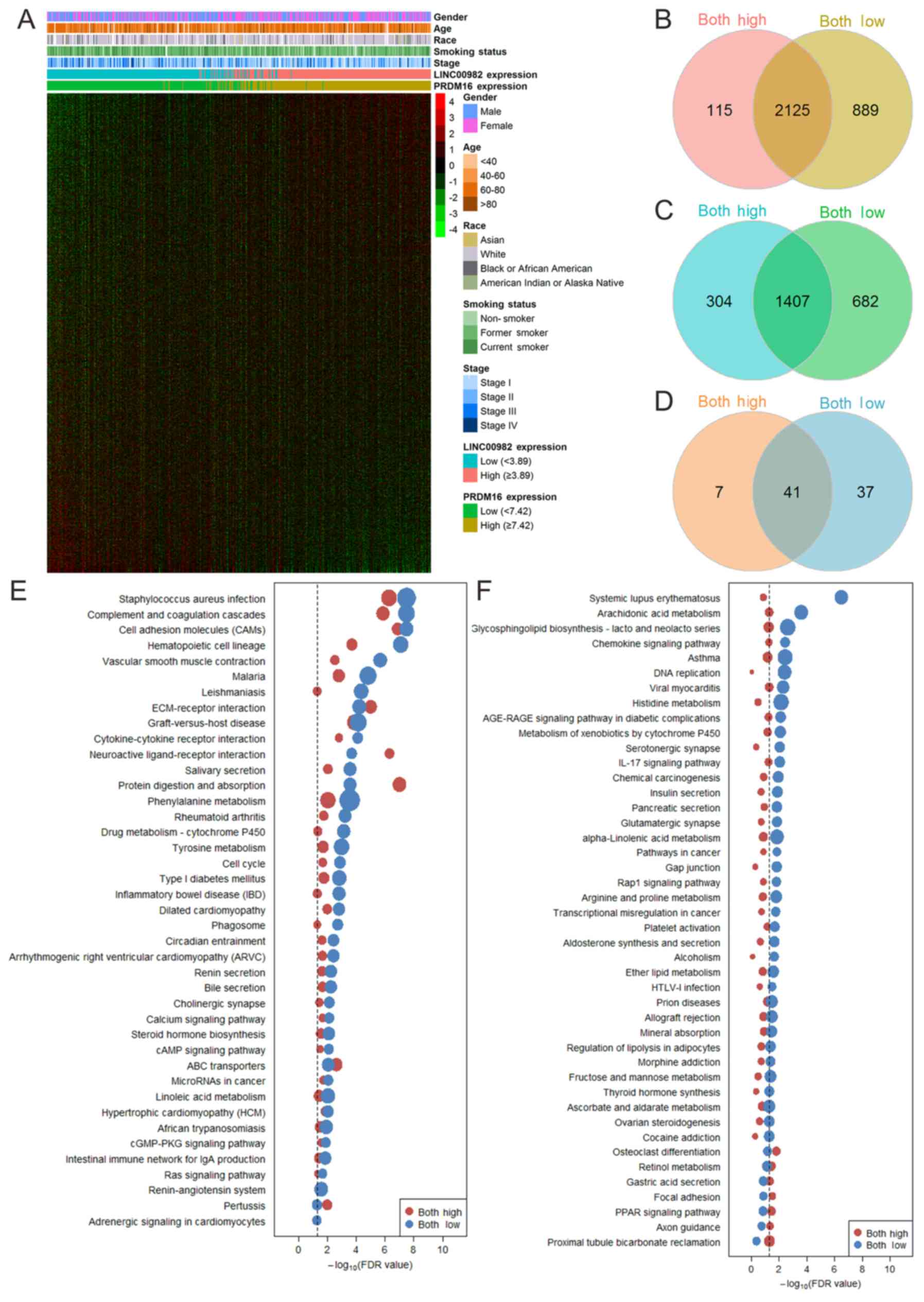
the paired adjacent normal lung tissues (Fig. 2A and B). In addition, the same trend
was observed in the LUSC tissues (data not shown). Furthermore, we
stratified LUAD patients by the median expression of
LINC00982 and PRDM16 and found that the
low-LINC00982 (<3.89) and low-PRDM16 (<7.42)
groups were decreased in tumors compared to adjacent normal lung
tissues, whereas the high-LINC00982 (≥3.89) and
high-PRDM16 (≥7.42) groups had no significant changes (data
not shown). Analysis of copy number variations revealed that the
copy number of the two genes was also lower in patients with low
gene expression (Fig. 2C and D).
EGFR expression was positively associated with gene copy
number (Fig. 2C and D) as had been
previously observed (23). In
addition, the low level of expression was consistent with the
hypermethylation of the promoter region of these two genes in tumor
samples compared with adjacent tissues (Fig. 2E and F). We analyzed the Spearman's
correlation between LINC00982 and PRDM16 expression
and found that they were positively correlated (Fig. 2G).
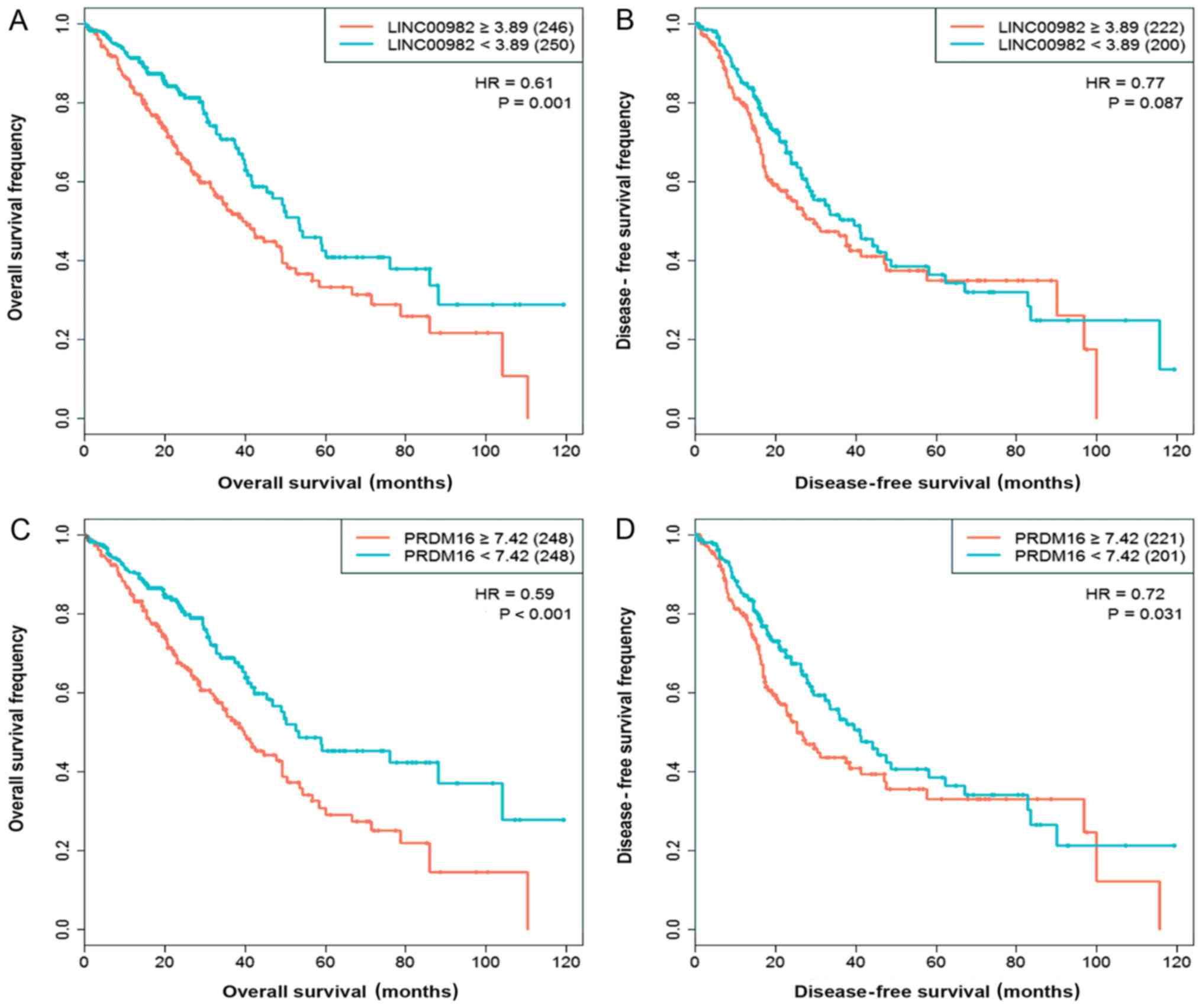
We used Kaplan-Meier curves to explore the effect of
LINC00982 and PRDM16 expression on LUAD patient
survival status. The results indicated that patients with low
expression of LINC00982 or PRDM16 showed poor overall
survival and disease-free survival than the high-expression
corresponding groups, although the effect of LINC00982 on
disease-free survival did not reach the significance threshold
(Fig. 3). We also analyzed the
effect of these two genes on survival in LUSC patients; however,
neither affected survival status (data not shown). Therefore, we
focused on the influence of LINC00982 and PRDM16 on
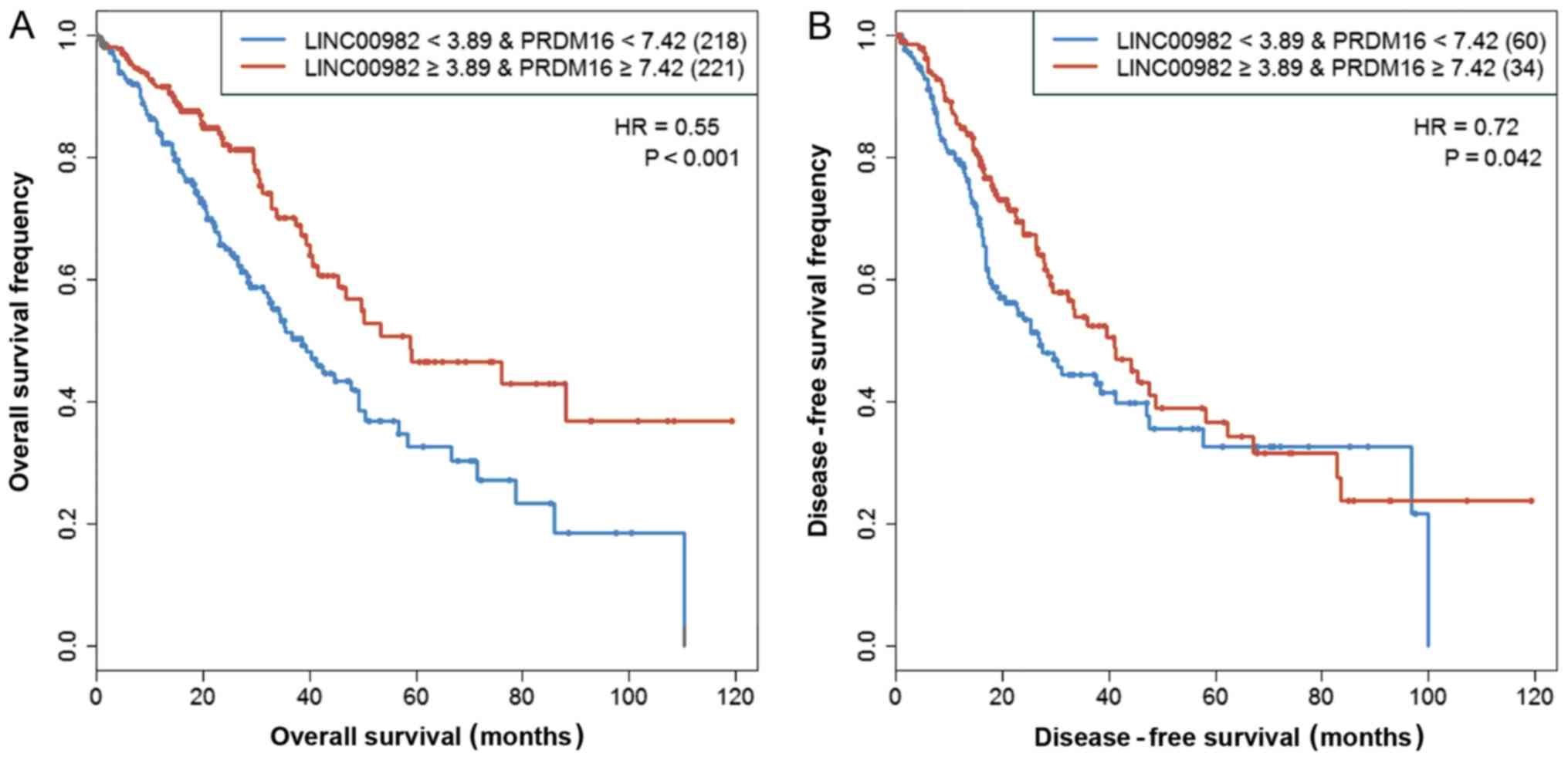
LUAD in the subsequent analyses. In order to study the combined
effect of LINC00982 and PRDM16 on patient survival,
we combined the low-LINC00982 group and low-PRDM16 group into the
‘both-low’ group, as well as combining the high-LINC00982 group and
high-PRDM16 group into the ‘both-high’ group. The Kaplan-Meier
curves revealed that compared with the ‘both-low’ group, patients
with high expression of these two genes presented with
significantly prolonged overall survival (HR=0.55, P<0.001) and
disease-free survival (HR=0.72, P=0.042) (Fig. 4). We also observed a consistent
trend in patients with early-stage disease (I/II) (data not
shown).
Gene expression and pathway analysis
of LINC00982 and PRDM16 in LUAD
In Fig. 5 the gene
expression profiles and KEGG pathway enrichment results in LUAD
patients are displayed. Genes with an expression value of zero were
removed. In total, we assessed the gene expression of 19,606 genes
in 515 LUAD patients and 59 controls (data not shown). The global
gene expression in the high-LINC00982 group and low-LINC00982 group
revealed a relatively large difference, as well as the high-PRDM16
and low-PRDM16 patients. Other characteristics (sex, age, race,
smoking status and stage) were approximately randomly distributed,
indicating that these variables contributed less to gene expression
changes. A Venn diagram of differentially expressed genes in the
‘both-high’ and ‘both-low’ groups compared with adjacent normal
tissues is shown in Fig. 5B and C.
In total, 3,951 and 5,103 differentially expressed genes were
observed in the ‘both-high’ group (data not shown) and the
‘both-low’ group (data not shown), respectively. Furthermore, there
were 2,125 common downregulated and 1,407 common upregulated genes
between the ‘both-high’ and ‘both-low’ groups (Fig. 5B and C). From the KEGG enrichment
results, there were 48 significantly enriched pathways in the
‘both-high’ group (data not shown) and 78 significantly enriched
pathways in the ‘both-low’ group (data not shown). There were 41
commonly and 44 differentially enriched KEGG pathways between the
two groups (Fig. 5D). The
enrichment profiles indicated that most of the pathways resulted in
serious damage in the ‘both-low’ group as compared to the ‘both
high’-group (Fig. 5E and F). We
also analyzed the biological pathway enrichment of the genes
co-expressed with LINC00982 and PRDM16 using
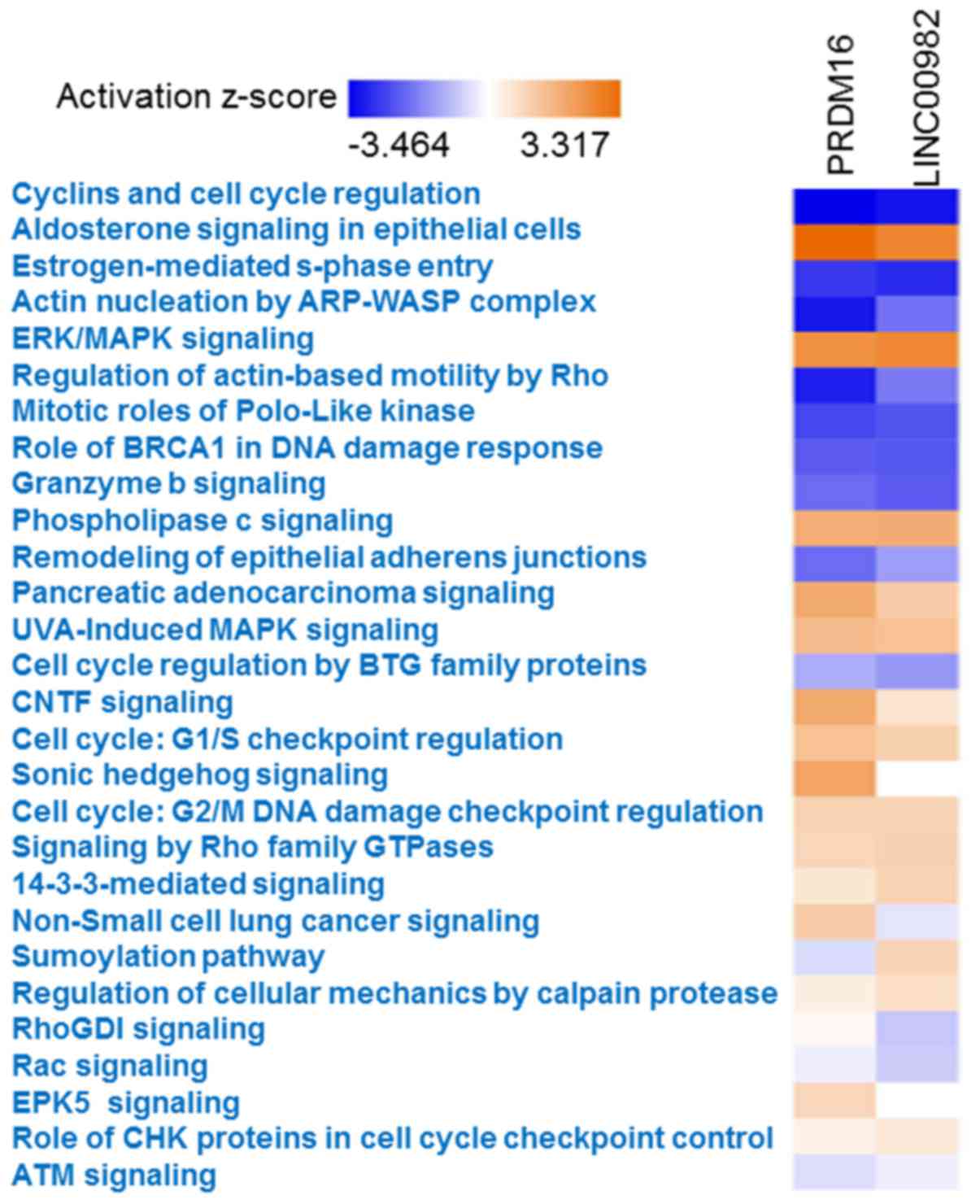
Ingenuity Pathway Analysis (Fig.
6). There were several common pathways associated with
co-expression of LINC00982 and PRDM16, such as
cyclins and cell cycle regulation, NSCLC signaling and ERK/MAPK
signaling.
LINC00982 and PRDM16 expression,
clinicopathological variables and patient survival
Table I depicts the
515 LUAD patient characteristics in the high- and low-expressed
LINC00982 and PRDM16 groups. We observed that females
exhibited higher expression of LINC00982 (P=0.043) and
PRDM16 (P=0.002) compared with males. There were no
differences in age, race, residual tumor or primary site between
the high-LINC00982 and low-LINC00982 groups, or between the
high-PRDM16 and low-PRDM16 groups. Furthermore, patients in the
low-LINC00982 group showed higher smoking frequency (P=0.002) and
more serious disease stage (P=0.009) compared with the
high-LINC00982 group. We also observed more patients who currently
smoke in the low-PRDM16 group compared with the high-PRDM16 group
(P<0.001). The results for smoking status, stage and gene
expression of LINC00982 and PRDM16 are not shown.
 | Table I.Lung adenocarcinoma patient
characteristics stratified by LINC00982 and PRDM16
expression. |
Table I.
Lung adenocarcinoma patient
characteristics stratified by LINC00982 and PRDM16
expression.
|
| LINC00982
expression |
| PRDM16
expression |
|
|---|
|
|
|
|
|
|
|---|
| Patient
characteristics | High (≥3.89) | Low (<3.89) | P-value | High (≥7.42) | Low (<7.42) | P-value |
|---|
| Sex, n (%) |
|
Male | 107 (41.5) | 130 (50.8) | 0.043 | 101 (39.1) | 136 (53.1) | 0.002 |
|
Female | 151 (58.5) | 126 (49.2) |
| 157 (60.9) | 120 (46.9) |
|
| Mean age,
years | 65.8±9.4 | 64.9±10.1 | 0.297 | 65.9±9.1 | 64.8±10.3 | 0.221 |
| Race, n (%) |
|
Asian | 5 (2.2) | 3 (1.4) | 0.768 | 4 (1.7) | 4 (1.8) | 0.541 |
|
White | 199 (87.3) | 189 (85.5) |
| 204 (87.9) | 184 (84.8) |
|
|
Black/African American | 23 (10.1) | 28 (12.7) |
| 22 (9.5) | 29 (13.4) |
|
|
American Indian/Alaska
Native | 1 (0.4) | 1 (0.5) |
| 2 (0.9) | 0 |
|
| Residual tumor, n
(%) |
| R0 | 163 (94.7) | 181 (95.8) | 0.896 | 159 (94.1) | 185 (96.4) | 0.549 |
| R1 | 7 (4.1) | 6 (3.2) |
| 8 (4.7) | 5 (2.5) |
|
| R2 | 2 (1.2) | 2 (1.0) |
| 2 (1.2) | 2 (1.0) |
|
| Primary site, n
(%) |
|
L-lower | 46 (18.3) | 32 (12.9) | 0.245 | 40 (16) | 38 (15.3) | 0.626 |
|
L-upper | 53 (21.1) | 70 (28.2) |
| 56 (22.4) | 67 (26.9) |
|
|
R-lower | 50 (19.9) | 46 (18.5) |
| 51 (20.4) | 45 (18.1) |
|
|
R-middle | 12 (4.8) | 9 (3.6) |
| 13 (5.2) | 8 (3.2) |
|
|
R-upper | 90 (35.9) | 91 (36.7) |
| 90 (36) | 91 (36.5) |
|
| Smoking, n (%) |
| Never
smoked | 49 (19.7) | 26 (10.5) | 0.002 | 49 (19.6) | 26 (10.6) | <0.001 |
| Current
smoker | 47 (19) | 72 (29) |
| 46 (18.4) | 73 (29.7) |
|
| Former
smoker | 152 (61.3) | 150 (60.5) |
| 155 (62) | 147 (59.8) |
|
| Stage, n (%) |
| I | 152 (60.6) | 123 (48.2) | 0.009 | 149 (59.4) | 126 (49.4) | 0.076 |
| II | 59 (23.5) | 63 (24.7) |
| 59 (23.5) | 63 (24.7) |
|
|
III | 29 (11.6) | 55 (21.6) |
| 33 (13.1) | 51 (20.0) |
|
| IV | 11 (4.4) | 14 (5.5) |
| 10 (4.0) | 15 (5.9) |
|
We used univariate Cox proportional hazards models
to analyze the effect of LINC00982 and PRDM16
expression, as well as other clinicopathological variables, on
patient survival status (Table
II). We found that high expression of LINC00982 in the
continuous and categorical models all showed prolonged overall
survival and disease-free survival (all P<0.05). Furthermore,
the expression of PRDM16 in the continuous and categorical
models also associated with survival (all P<0.05). Other
clinicopathological variables such as residual tumors and stage
also showed significant association with survival status.
Therefore, we used multivariate Cox proportional hazards models
adjusting for covariates including residual tumor and stage to
verify the effect of LINC00982 and PRDM16 expression
on patient survival status (Table
III). The results indicated that the LINC00982 and
PRDM16 low-expression groups were both associated with
decreased overall survival (all P<0.05). However, the expression
of these two genes did not affect disease-free survival. The above
analysis showed that LINC00982 and PRDM16
independently affected overall survival.
 | Table II.Association of LINC00982 and
PRDM16 expression, clinicopathological characteristics and
survival status. |
Table II.
Association of LINC00982 and
PRDM16 expression, clinicopathological characteristics and
survival status.
|
|
| Overall
survival | Disease-free
survival |
|---|
|
|
|
|
|
|---|
| Variable | Total N | Hazard ratio (95%
CI) | P-value | Hazard ratio (95%
CI) | P-value |
|---|
| LINC00982
expression (continuous) | 515 | 0.87
(0.81–0.94) | <0.001 | 0.93
(0.86–1.00) | 0.038 |
| PRDM16
expression (continuous) | 515 | 0.89
(0.83–0.95) | <0.001 | 0.92
(0.86–0.98) | 0.011 |
| LINC00982
expression (categorical, above or below 3.88) | 515 | 0.57
(0.42–0.76) | <0.001 | 0.74
(0.55–0.99) | 0.040 |
| PRDM16
expression (categorical, above or below 7.41) | 515 | 0.58
(0.43–0.79) | <0.001 | 0.71
(0.53–0.95) | 0.022 |
| Sex (male vs.
female) | 514 | 0.94
(0.70–1.26) | 0.672 | 0.97
(0.73–1.30) | 0.846 |
| Age
(continuous) | 495 | 1.01
(0.99–1.02) | 0.333 | 1.00
(0.99–1.02) | 0.354 |
| Residual tumor (R0
vs. R1 or R2) | 361 | 2.22
(1.37–3.60) | 0.001 | 3.64
(1.83–7.23) | <0.001 |
| Primary site
(L-site vs R-site) | 499 | 1.04
(0.77–1.40) | 0.814 | 1.11
(0.82–1.51) | 0.493 |
| Smoking (never
smoker vs. current smoker or former smoker) | 496 | 0.92
(0.61–1.38) | 0.672 | 1.04
(0.68–1.58) | 0.873 |
| Stage (stage I or
II vs. stage III or IV) | 506 | 2.65
(1.95–3.62) | <0.001 | 1.73
(1.21–2.47) | 0.003 |
 | Table III.Multivariate survival model of
LINC00982 and PRDM16 expression on survival
status. |
Table III.
Multivariate survival model of
LINC00982 and PRDM16 expression on survival
status.
| Variable | Hazard ratio (95%
CI) |
P-valuea |
|---|
| Overall
survival |
|
LINC00982 expression
(continuous) | 0.89
(0.82–0.98) | 0.015 |
|
LINC00982 expression
(categorical, above or below 3.88) | 0.66
(0.46–0.95) | 0.023 |
|
PRDM16 expression
(continuous) | 0.91
(0.84–0.98) | 0.019 |
|
PRDM16 expression
(categorical, above or below 7.41) | 0.62
(0.44–0.89) | 0.008 |
| Disease-free
survival |
|
LINC00982 expression
(continuous) | 0.95
(0.87–1.04) | 0.288 |
|
LINC00982 expression
(categorical, above or below 3.88) | 0.75
(0.53–1.08) | 0.123 |
|
PRDM16 expression
(continuous) | 0.94
(0.87–1.03) | 0.177 |
|
PRDM16 expression
(categorical, above or below 7.41) | 0.76
(0.53–1.08) | 0.124 |
RT-qPCR validation
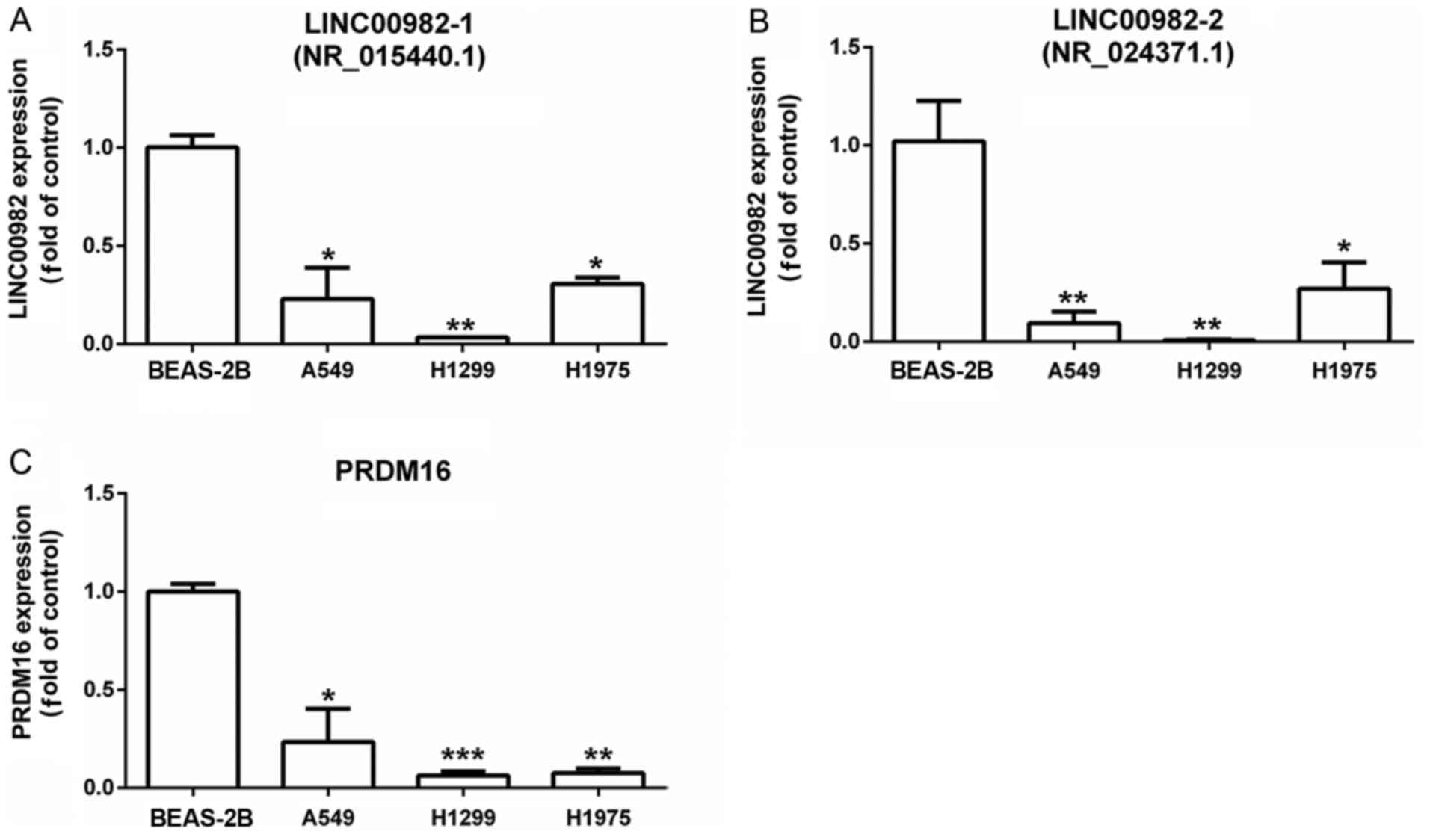
We explored LINC00982-1, LINC00982-2 and
PRDM16 expression in LUAD cell lines (A549, H1299 and H1975)
and a normal lung epithelium cell line (BEAS-2B) by RT-qPCR
(Fig. 7). We found that
LINC00982-1, LINC00982-2, the two transcripts of
LINC00982 and PRDM16 expression were significantly
decreased in LUAD cell lines (A549, H1299 and H1975) compared to
normal cell lines (BEAS-2B).
Discussion
Our results revealed that the combined effect of
LINC00982 and PRDM16 expression was a risk factor
that affected global gene expression, altered cancer-related
pathways and biological functions, and decreased patient survival
in LUAD. Additionally, our experimental results revealed that
LINC00982 and PRDM16 transcripts were downregulated
in LUAD cell lines compared with the normal BEAS-2B cell line.
LUAD is a complex disease that is associated with
altered gene expression, DNA methylation, protein modification and
non-coding RNA dysfunction (24).
In particular, lncRNAs play a significant role in cellular
homeostasis and tumorigenesis and often serve as markers of
prognosis and diagnostic targets for therapy (25). A recent study suggested that
CCAT2, a LUAD-specific lncRNA, promoted invasion and
metastasis of LUAD (26). In a
previous study, Fei et al reported that LINC00982 is
dysregulated in gastric cancer patients, and the high expression of
LINC00982 was related to better overall survival (11). LINC00982 is located about 0.5
kb telomeric at the 5′ untranslated region of PRDM16, which
suggests that these transcripts have the same enhancer, ACTRT2
(27). The PRDM16 gene is
not only associated with myelodysplastic syndrome (MDS) and acute
myeloid leukemia (AML) (28), but
also solid tumors such as lung cancer. Some studies have indicated
that PRDM16 expression is downregulated in lung cancer cells
due to the methylation of its promoter (19), which was consistent with our
results.
In the present study, compared with adjacent normal
tissues, LINC00982 and PRDM16 expression were
significantly decreased in tumor samples. Considering the increased
methylation level of the promoter region and the decreased copy
number in LUAD patients, we speculated that the dysregulation of
LINC00982 and PRDM16 may be caused by DNA methylation
and gene copy number disorders. Furthermore, we used three survival
analysis models to show that high expression of LINC00982
and PRDM16 was associated with higher patient survival,
especially overall survival. In addition, patients with high
expression of LINC00982 and PRDM16 demonstrated
better overall survival and disease-free survival than patients
with low expression of these two genes. Notably, these associations
were consistent in patients with early tumor stages (stage I and
II), combined with the evidence that high expression of
LINC00982 and PRDM16 were related to low TNM stage,
which may aid the early diagnosis of LUAD and improve the prognosis
of affected patients, especially with a combination of changes in
their expression.
Through pathway enrichment analysis, we found that
PRDM16-associated genes were enriched in many canonical pathways,
which are consistent with LINC00982-associated gene enriched
pathways. However, the extents of the impact are differential, such
as cyclins and cell cycle regulation, aldosterone signaling in
epithelial cells, and estrogen-mediated S-phase entry. We observed
that LINC00982 and PRDM16 were negatively associated
with cyclins and cell cycle regulation in LUAD. Tumors are
characterized by malignant cell growth and proliferation.
Abnormalities in cell proliferation, differentiation and apoptosis
are involved in the development and progression of tumors, and cell
cycle disorder is the most important mechanism of tumor growth
(29). The cell cycle is a highly
orderly process. As a regulatory factor, cyclin overexpression is
associated with carcinogenesis (30–32).
In many tumor cells and proliferating cells, cyclin is
overexpressed, and many tumor-suppressor genes such as p53
(29), BRCA1 (33) and Rb (34) play crucial roles in blocking the
cell cycle. Consistent with our findings, a recent study reported
that LINC00982 inhibited cell proliferation and rendered
cell cycle arrest in gastric cancer cells (11) and PRDM16 was also reported to
alter cell cycle distribution in stem (35), indicating that LINC00982 and
PRDM16 may impede the occurrence and development of LUAD by
mediating cell cycle arrest.
In recent years, precision medicine has
increasingly been used in the treatment of cancer, especially in
exploring and identifying biomarkers (36). The treatment of LUAD is typically
carried out with multiple targeted therapies. Therefore, a better
understanding of both coding genes and non-coding RNAs will help to
improve the diagnosis and prognosis of human LUAD (37,38).
In the present study, we identified LINC00982 and
PRDM16 gene markers for predicting overall and disease-free
survival based on RNA-Seq data that was obtained from TCGA.
Additionally, after correcting for covariates, low expression of
both LINC00982 and PRDM16 remained associated with
reduced overall survival by Cox analysis models. Furthermore, by
stratified analysis, low expression of both LINC00982 and
PRDM16 was associated with poor overall survival and
disease-free survival in stage I and II patients. In addition, we
found that the risk ratio of LINC00982 or PRDM16
expression was lower than both LINC00982 and PRDM16
expression based on survival analysis. We therefore concluded that
the interaction of LINC00982 and PRDM16 may play a
significant role in the prognosis of LUAD patients than single
LINC00982 or PRDM16 expression, and it was better to
use these two genes as prognostic markers than using only one gene.
However, we observed no association between the expression of
LINC00982 and PRDM16 with patient survival in LUSC.
This difference may be due to tumor heterogeneity if the genes that
drive LUAD and LUSC are different (39). Finally, we observed that
LINC00982 and PRDM16 were substantially decreased in
human LUAD cell lines compared with a normal cell line. Therefore,
we hypothesized that a combination of LINC00982 and
PRDM16 expression may help to facilitate the prognosis of
LUAD.
In conclusion, in the present study we found that
LINC00982 and PRDM16 had low expression in tumor
samples compared with adjacent normal tissues, and their expression
levels were associated with their methylation status and copy
number variations. Furthermore, patients with low expression of
LINC00982 and PRDM16 were associated with more
altered gene expression and influenced pathways compared with
high-expression groups. In addition, independently and jointly, low
expression of LINC00982 and PRDM16 was associated
with poor patient survival, revealing that this combination had
prognostic and diagnostic value. Our findings may also provide
useful information to obtain a better understanding of LUAD.
However, there are also several limitations in this study. Firstly,
the biological functions of LINC00982 and PRDM16 need
to be validated in cell and animal experiments. Secondly, it was a
retrospective study, and as these findings are based on the
reanalysis of TCGA data, prospective random population studies are
needed to confirm these promising results. Lastly, data concerning
drug therapy and prognosis of LUAD patients are not available and
limit the analysis of outcomes in our study. Given the limitations
of this study, further large-sample and in-depth studies are
required to confirm these results.
Acknowledgements
Not applicable.
Funding
The present study was financially supported by the
National Natural Science Foundation of China (grant no. 81602929),
the Scientific Project of Shanghai Health and Planning Commission
(grant no. 20164Y0198) and the Shanghai Municipal Planning
Commission of Science and Research Fund (grant no. 20144Y0249).
Availability of data and materials
LUSC and LUAD transcriptome and clinical data were
downloaded from The Cancer Genome Atlas (TCGA, http://tcga-data.nci.nih.gov/) and the
cBioPortal (http://www.cbioportal.org/) database. LUAD DNA
promoter methylation data were collected from MethHC (http://methhc.mbc.nctu.edu.tw/php/search.php?opt=gene).
The other datasets used during the present study are available from
the corresponding author upon reasonable request.
Authors' contributions
BQ, XY and WLv designed this study. XY and WLv
performed data collection. WLv, WLi, TF and XY conducted data
analysis. XY, NF, YW and HL performed the experiment. All authors
wrote the manuscript. WLv, WLi, XY and BQ revised the manuscript.
The final version of the manuscript has been read and approved by
all authors, and each author believes that the manuscript
represents honest work.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gazdar AF: Should we continue to use the
term non-small-cell lung cancer? Ann Oncol. 21 Suppl
7:vii225–vii229. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zonderman AB, Ejiogu N, Norbeck J and
Evans MK: The influence of health disparities on targeting cancer
prevention efforts. Am J Prev Med. 46 3 Suppl 1:S87–S97. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kang YH, Kim D and Jin EJ: Down-regulation
of phospholipase D stimulates death of lung cancer cells involving
up-regulation of the long ncRNA ANRIL. Anticancer Res.
35:2795–2803. 2015.PubMed/NCBI
|
|
5
|
Liu X, Xiao ZD, Han L, Zhang J, Lee SW,
Wang W, Lee H, Zhuang L, Chen J, Lin HK, et al: LncRNA NBR2
engages a metabolic checkpoint by regulating AMPK under energy
stress. Nat Cell Biol. 18:431–442. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Y, Guo Q, Zhao Y, Chen J, Wang S, Hu
J and Sun Y: BRAF-activated long non-coding RNA contributes to cell
proliferation and activates autophagy in papillary thyroid
carcinoma. Oncol Lett. 8:1947–1952. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhao Y, Guo Q, Chen J, Hu J, Wang S and
Sun Y: Role of long non-coding RNA HULC in cell proliferation,
apoptosis and tumor metastasis of gastric cancer: A clinical and in
vitro investigation. Oncol Rep. 31:358–364. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Feng N, Ching T, Wang Y, Liu B, Lin H, Shi
O, Zhang X, Zheng M, Zheng X, Gao M, et al: Analysis of microarray
data on gene expression and methylation to identify long non-coding
RNAs in non-small cell lung cancer. Sci Rep. 6:372332016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schmidt LH, Spieker T, Koschmieder S,
Schäffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D,
Marra A, et al: The long noncoding MALAT-1 RNA indicates a poor
prognosis in non-small cell lung cancer and induces migration and
tumor growth. J Thorac Oncol. 6:1984–1992. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Loewen G, Jayawickramarajah J, Zhuo Y and
Shan B: Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol.
7:902014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fei ZH, Yu XJ, Zhou M, Su HF, Zheng Z and
Xie CY: Upregulated expression of long non-coding RNA LINC00982
regulates cell proliferation and its clinical relevance in patients
with gastric cancer. Tumour Biol. 37:1983–1993. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cao WJ, Wu HL, He BS, Zhang YS and Zhang
ZY: Analysis of long non-coding RNA expression profiles in gastric
cancer. World J Gastroenterol. 19:3658–3664. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lei Q, Liu X, Fu H, Sun Y, Wang L, Xu G,
Wang W, Yu Z, Liu C, Li P, et al: miR-101 reverses hypomethylation
of the PRDM16 promoter to disrupt mitochondrial function in
astrocytoma cells. Oncotarget. 7:5007–5022. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu S, Xu Y, Song M, Chen G, Wang H, Zhao
Y, Wang Z and Li F: PRDM16 is associated with evasion of apoptosis
by prostatic cancer cells according to RNA interference screening.
Mol Med Rep. 14:3357–3361. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Burghel GJ, Lin WY, Whitehouse H, Brock I,
Hammond D, Bury J, Stephenson Y, George R and Cox A: Identification
of candidate driver genes in common focal chromosomal aberrations
of microsatellite stable colorectal cancer. PLoS One. 8:e838592013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Momi N, Wali RK, Chhaparia A, Calderwood
AH, Tiwari AK, Ledbetter SE, DeLaCruz M and Roy HK: Su2000 PRDM16
is a novel proto-oncogene in colorectal cancer (CRC): Modulation of
the early warburg effect in field carcinogenesis. Gastroenterology.
150:S6062016. View Article : Google Scholar
|
|
17
|
Matsuo H, Goyama S, Kamikubo Y and Adachi
S: The subtype-specific features of EVI1 and PRDM16 in acute
myeloid leukemia. Haematologica. 100:e116–e117. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li L, Zhao CT, Cui BL, Wu SL, Liu XD, Su
Z, Yang J, Wang W, Cui ZG and Zhao HG: Expression of HOXB4, PRDM16
and HOXA9 in patients with acute myeloid leukemia and its clinical
significance. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 24:326–331.
2016.(In Chinese). PubMed/NCBI
|
|
19
|
Tan SX, Hu RC, Liu JJ, Tan YL and Liu WE:
Methylation of PRDM2, PRDM5 and PRDM16 genes in lung cancer cells.
Int J Clin Exp Pathol. 7:2305–2311. 2014.PubMed/NCBI
|
|
20
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hirsch FR, Varella-Garcia M, Bunn PA Jr,
Di Maria MV, Veve R, Bremmes RM, Barón AE, Zeng C and Franklin WA:
Epidermal growth factor receptor in non-small-cell lung carcinomas:
Correlation between gene copy number and protein expression and
impact on prognosis. J Clin Oncol. 21:3798–3807. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cancer Genome Atlas Research Network:
Comprehensive molecular profiling of lung adenocarcinoma. Nature.
511:543–550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Castillo J, Stueve TR and Marconett CN:
Intersecting transcriptomic profiling technologies and long
non-coding RNA function in lung adenocarcinoma: Discovery,
mechanisms, and therapeutic applications. Oncotarget.
8:81538–81557. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L
and Yin R: CCAT2 is a lung adenocarcinoma-specific long non-coding
RNA and promotes invasion of non-small cell lung cancer. Tumour
Biol. 35:5375–5380. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lavallée VP, Lemieux S, Boucher G, Gendron
P, Boivin I, Girard S, Hébert J and Sauvageau G: Identification of
MYC mutations in acute myeloid leukemias with NUP98-NSD1
translocations. Leukemia. 30:1621–1624. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Duhoux FP, Ameye G, Montano-Almendras CP,
Bahloula K, Mozziconacci MJ, Laibe S, Wlodarska I, Michaux L,
Talmant P, Richebourg S, et al: PRDM16 (1p36) translocations define
a distinct entity of myeloid malignancies with poor prognosis but
may also occur in lymphoid malignancies. Br J Haematol. 156:76–88.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vermeulen K, Van Bockstaele DR and
Berneman ZN: The cell cycle: A review of regulation, deregulation
and therapeutic targets in cancer. Cell Prolif. 36:131–149. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pagano M and Draetta G: Cyclin A, cell
cycle control and oncogenesis. Prog Growth Factor Res. 3:267–277.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yue W, Zhao X, Zhang L, Xu S, Liu Z, Ma L,
Jia W, Qian Z, Zhang C, Wang Y and Yang X: Cell cycle protein
cyclin Y is associated with human non-small-cell lung cancer
proliferation and tumorigenesis. Clin Lung Cancer. 12:43–50. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lamb R, Lehn S, Rogerson L, Clarke RB and
Landberg G: Cell cycle regulators cyclin D1 and CDK4/6 have
estrogen receptor-dependent divergent functions in breast cancer
migration and stem cell-like activity. Cell Cycle. 12:2384–2394.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jhanwar-Uniyal M: BRCA1 in cancer, cell
cycle and genomic stability. Front Biosci. 8:s1107–s1117. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Paternot S, Bockstaele L, Bisteau X,
Kooken H, Coulonval K and Roger PP: Rb inactivation in cell cycle
and cancer: The puzzle of highly regulated activating
phosphorylation of CDK4 versus constitutively active CDK-activating
kinase. Cell Cycle. 9:689–699. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chuikov S, Levi BP, Smith ML and Morrison
SJ: Prdm16 promotes stem cell maintenance in multiple tissues,
partly by regulating oxidative stress. Nat Cell Biol. 12:999–1006.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wishart DS: Emerging applications of
metabolomics in drug discovery and precision medicine. Nat Rev Drug
Discov. 15:473–484. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang EB, Yin DD, Sun M, Kong R, Liu XH,
You LH, Han L, Xia R, Wang KM, Yang JS, et al: P53-regulated long
non-coding RNA TUG1 affects cell proliferation in human non-small
cell lung cancer, partly through epigenetically regulating HOXB7
expression. Cell Death Dis. 5:e12432014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang X, Song JH, Cheng Y, Wu W, Bhagat T,
Yu Y, Abraham JM, Ibrahim S, Ravich W, Roland BC, et al: Long
non-coding RNA HNF1A-AS1 regulates proliferation and migration in
oesophageal adenocarcinoma cells. Gut. 63:881–890. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jamal-Hanjani M, Wilson GA, McGranahan N,
Birkbak NJ, Watkins TB, Veeriah S, Shafi S, Johnson DH, Mitter R,
Rosenthal R, et al: Tracking the evolution of non-small-cell lung
cancer. N Engl J Med. 376:2109–2121. 2017. View Article : Google Scholar : PubMed/NCBI
|