Introduction
Gastric cancer is one of the most common types of
malignancy worldwide and the second biggest health burden in China
(1–3). Helicobacter pylori infection is
the most important cause of proximal gastric cancer (4). Altered cell apoptosis, proliferation
and certain modifications to tumor suppressor genes, which may lead
to inflammation, also contribute to gastric oncogenesis (5). Surgery remains the only curative
therapy for gastric cancer. Peri-operative and adjuvant
chemotherapy as well as chemoradiation therapy may improve the
outcome for patients with resectable gastric cancer (6). Cisplatin and fluoropyrimidine-based
chemotherapy with the addition of trastuzumab is widely used in
human epidermal growth factor receptor 2-positive patients that are
fit for chemotherapy (7).
Tumors that comprise a mass of malignant epithelial
cells are surrounded by multiple non-cancerous cell populations,
including fibroblasts, vascular endothelial cells, adipocytes and
immune regulatory cells (8–10). Fibroblasts were first described as
non-vascular, non-epithelial and non-inflammatory cells of the
connective tissue, which are embedded in the fibrillar matrix of
the connective tissue (11).
Fibroblasts are responsible for the deposition of the extracellular
matrix (ECM), regulation of epithelial differentiation and
inflammation, as well as wound healing (12). Activated fibroblasts in the tumor
microenvironment are known as tumor-associated fibroblasts (TAFs)
(13,14). TAFs have recently been identified to
have important roles in the proliferation, metastasis and
radio-/chemoresistance of cancer cells (10,15,16).
Gastric cancer-associated TAFs are major regulators of the tumor
microenvironment in this malignancy (17,18).
1-(Diaminomethylidene)-3,3-dimethylguanidine
(metformin) is an oral anti-diabetic drug, which decreases the risk
of cancer in patients with type 2 diabetes. Metformin exerts
antitumor effects on multiple cancer cell lines, including ovarian
(19), prostate (20), breast (21) and lung cancer cells (22). However, the mechanisms underlying
the antitumor effect of metformin are complex, which include
adenosine monophosphate-activated protein kinase, the
phosphatidylinositol-3-kinase/Akt pathway, mammalian target of
rapamycin, lipogenesis and apoptosis (23–25). A
previous study by our group reported that metformin suppresses the
progression of gastric cancer through the hypoxia-inducible
factor-1α/pyruvate kinase isoenzyme M2 pathway (26). However, the effect of metformin on
TAFs and the tumor microenvironment has remained elusive.
The present study focused on the antitumor effects
of metformin on TAFs regarding their subsequent interaction with
cancer cells. Using a proteomics analysis, calmodulin-like protein
3 (Calml3) was identified as one of the most upregulated factors in
the culture medium of gastric TAFs after treatment with metformin.
Although Calml3 overexpression and knockdown models have previously
demonstrated that Calml3 is able to inhibit the genesis and
initiation of metastasis in hepatocellular carcinoma (HCC)
(27), it has remained elusive
whether Calml3 secreted from TAFs exerts any antitumor effect on
gastric cancer cells. The present study was designated to reveal
the antitumor effect of metformin by regulating the tumor
microenvironment and provide a better understanding of the roles of
Calml3.
Materials and methods
Reagents
Metformin was purchased from Sangon Biotech
(Shanghai, China). Bovine serum albumin (BSA) was purchased from
Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). A Cell Counting Kit
(CCK)-8 and trypsin were purchased from Beyotime Institute of
Biotechnology (Haimen, China). Recombinant human Calml3 protein was
purchased from Sino Biological (Beijing, China).
Isolation of TAFs from gastric cancer
tissues
TAFs were isolated from surgically resected gastric
cancer specimens from six patients treated at the First People's
Hospital of Xuzhou (Suzhou, China) from January to September 2015.
The six patients were male with and age of 55–60 years (median age,
57 years). Normal gastric tissues were obtained from 6 age- and
gender-matched trauma patients. All patients provided written
informed consent for their tissues to be used for scientific
research. Ethical approval of the study was obtained from the First
People's Hospital of Xuzhou (Suzhou, China). TAFs were isolated as
reported previously (28) and were
pooled together. Gastric cancer samples were placed in RPMI-1640
medium (HyClone; GE Healthcare, Little Chalfont, UK) containing 4%
meropenem (Haibin Pharmaceutical Co., Ltd, Shenzhen, China).
Tissues were cut into pieces of ~3 mm3 in volume with a
scalpel, placed in 25-cm2 culture flasks and covered
with 1 ml fetal bovine serum (FBS; HyClone; GE Healthcare). Next,
the flasks were inverted for 48 h. After 48 h, RPMI-1640 medium
supplemented with 20% FBS (HyClone; GE Healthcare) was added to the
flasks, and the flasks were inverted again. Culture was performed
in an incubator with humidified air containing 5% CO2 at
37°C. Following tissue attachment (2–3 days), the culture medium
[Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) with 10% FBS] was changed twice
per week for the next 2–3 weeks. Under these conditions,
fibroblasts were explanted from tissue fragments, whereas other
types of cell were mostly retained within the tissue. The
fibroblasts formed multiple dense colonies that spread out on the
culture dish. After 10–14 days, the cultured cells were briefly
trypsinized in an incubator for 5 min and re-seeded into new flasks
(passage 1). After reaching confluence, which occurred every 3–4
days, the cultured cells were split at a ratio of 1:2. All
fibroblasts used in the experiments of the present study were
between passages 4 and 9.
Cell culture
The SGC-7901, BGC823, GES-1 and MGT-803 (29) human gastric cancer cell lines and
the human gastric TAFs were cultured in DMEM supplemented with 10%
FBS, 1% penicillin-streptomycin and 1 mM sodium pyruvate. These
cells were grown at 37°C in a humidified atmosphere with 5%
CO2.
Cell viability assay
After centrifugation and re-suspension, cells were
seeded in 96-well plates at 4,000 cells/well. The cells were
co-cultured in a Transwell chamber with TAFs pre-treated with or
without different concentrations of metformin. After 48 h, CCK-8
solution (Beyotime Institute of Biotechnology) was added and the
plates were cultured for an additional 2 h, according to the
manufacturer's instructions. The optical density was determined at
450 nm using a microplate reader (Synergy™ Neio; Biotek, Winooski,
VT, USA). The cell viability of each group was determined in
triplicate.
Immunofluorescence assay
Cells were washed with PBS, fixed with 4%
formaldehyde and blocked with 1% BSA in PBS for 1 h at room
temperature. Cells were then incubated overnight at 4°C with a
primary antibody against α-smooth muscle actin (1:1,000 dilution;
cat. no. ab5694; Abcam, Cambridge, UK) and subsequently with
cyanine 3-conjugated secondary antibodies (1:2,000 dilution; cat.
no. A0507; Beyotime Institute of Biotechnology, Haimen, China) for
1 h at room temperature. Nuclei were stained using DAPI and images
were captured using an FV1200 confocal microscope (Olympus, Tokyo,
Japan).
Protein extraction and tryptic
digest
Gastric TAF cells were treated with 0.2 mM metformin
or PBS as a control for 12 h. Equal quantities of cells from
triplicate wells were mixed to generate one sample for each group.
Culture medium of TAFs treated with or without metformin was
collected individually. The protein solution was reduced with 10 mM
DTT for 1 h at 37°C and alkylated with 20 mM iodoacetic acid
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 45 min at room
temperature in the dark. The protein sample was diluted with 100 mM
TEAB buffer to reach a final urea concentration of <2 M.
Finally, trypsin was added at a 1:50 trypsin-to-protein ratio for
the first digestion overnight at 37°C, and 1:100 trypsin-to-protein
ratio for a second 4-h digestion at 37°C. From each sample, ~100 µg
protein was digested with trypsin for subsequent analysis.
Tandem mass tag (TMT) labeling
After digestion, tryptic peptides were desalted
using a Strata X C18 SPE column (Phenomenex, Torrance, CA, USA) and
vacuum-dried. Peptides were reconstituted in 0.5 M TEAB and
processed with a 6-plex TMT kit according to the manufacturer's
protocol. One unit of each TMT reagent (defined as the amount of
reagent required to label 100 µg of protein) was thawed,
reconstituted in 24 µl acetonitrile (ACN), and added to each
sample. Next, the peptide mixtures were incubated for 2 h at room
temperature. The mixtures were pooled, desalted and dried by vacuum
centrifugation.
High-performance liquid chromatography
(HPLC) fractionation
The labeled peptide sample was then fractionated by
high-pH reverse-phase HPLC using an Agilent 300 Extend C18 column
(5-µm particles, 4.6 mm inner diameter and 250 mm length; Agilent
Technologies, Inc., Santa Clara, CA, USA). Peptides were separated
first, with a gradient of 2–60% ACN in 10 mM ammonium bicarbonate
in water (pH 10) over 80 min, and the eluate was collected in 80
fractions. Next, the peptides were combined into 18 fractions and
dried by vacuum centrifugation.
Quantitative proteomic analysis by
liquid chromatography tandem mass spectrometry (LC-MS/MS)
Peptides were dissolved in 0.1% formic acid (FA) and
directly loaded onto a reverse-phase pre-column (Acclaim PepMap
100; Thermo Fisher Scientific, Inc.). Next, these peptides were
separated using a reverse-phase analytical column (cat. no. 164568;
Thermo Fisher Scientific, Inc.). The gradient began with an
increase from 8 to 22% in solvent B (0.1% FA in ACN, HPLC grade)
over 26 min, followed by a further increase to 35% over 6 min,
rising up to 80% over 4 min and this concentration remaining
constant for the last 4 min, all at a steady flow rate of 300
nl/min on an EASY-nLC 1000 ultra (U)PLC system (Thermo Fisher
Scientific, Inc.). Spectra were obtained using a Q Exactive™ plus
hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher
Scientific, Inc.).
The peptides were subjected to nano spray ionization
source ionization (LTQ Oribtrap; Thermo-Fisher Scientific, Inc.)
followed by MS/MS in a Q Exactive™ plus that was coupled online to
the UPLC. Intact peptides were detected at a resolution of 70,000.
Peptides were fragmented for MS/MS using a nomalized collision
energy of 33%, and the resulting ion fragments were detected at a
resolution of 17,500. A top-20 data-dependent method was applied
for the top 20 precursor ions above a threshold ion count of
2×104 in the MS survey scan, with 30.0-sec dynamic
exclusion. The electrospray voltage applied was 2.0 kV. The
automatic gain control setting of 5×104 ions was used to
prevent overfilling of the ion trap. For MS scans, the m/z scan
range was 350–1,800. A fixed first mass was set at and m/z ratio of
100.
Database search
The resulting spectra were processed using Mascot
search engine (v.2.3.0; http://data-pc/mascot/home.html). Tandem mass spectra
were searched against the UniProt Homo sapiens database
(20,274 sequences; http://www.uniprot.org/). Trypsin/proline was
specified as a cleavage enzyme allowing up to two missing
cleavages. The mass error was set to 10 ppm for precursor ions and
0.02 Da for fragment ions. Carbamidomethyl on Cys, TMT-6plex
(N-term) and TMT-6plex (K) were specified as fixed modifications,
and oxidation on Met was specified as a variable modification. As
cut-off criteria, a false discovery rate of <1% and a peptide
ion score of >20 were used.
First, the mass error of all the identified peptides
was checked. The distribution of the mass error was near zero, and
usually <0.02 Da. Most of the identified peptides were between 8
and 16 amino acids in length, which agrees with the expected length
of tryptic peptides and indicates that tryptic digestion was
efficient.
Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) annotation
GO terms may be assigned to three domains: i)
Cellular component, ii) Molecular function and iii) Biological
process. GO annotation of the proteome was derived from the
UniProt-GOA database (www.http://www.ebi.ac.uk/GOA/). The identified protein
IDs were first converted to UniProt IDs and mapped to GO IDs in the
UniProt database. Proteins lacking a UniProt-GOA annotation were
analyzed using InterProScan (http://www.ebi.ac.uk/interpro/interproscan.html) to
annotate the protein's functional sites based on protein sequence
alignment. The proteins were then classified by GO and KEGG
annotation.
Cell clonogenic assay
To perform a standard clonogenic assay, cells were
seeded in 6-well plates at 1,000 cells/well. After treatment with
medium containing different concentrations of recombinant Calml3
for 48 h, cells were grown for 7–10 days to allow for colony
formation, subsequently fixed with pure methanol for 15 min at
37°C, and then stained with crystal violet (0.1%) for 15 min at
37°C. Clusters consisting of ≥50 cells were considered as
colonies.
Statistical analysis
Values are expressed as the mean ± standard error of
the mean of at least three independent experiments. The results
were evaluated by one-way analysis of variance followed by the
Student-Neuman-Keuls post hoc test, or Student's t-test to
determine statistical significance. The statistical analyses were
performed using GraphPad Prism 6 software (GraphPad, Inc., La
Jolla, CA, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Co-culture with gastric TAFs increases
the viability of gastric cancer cells
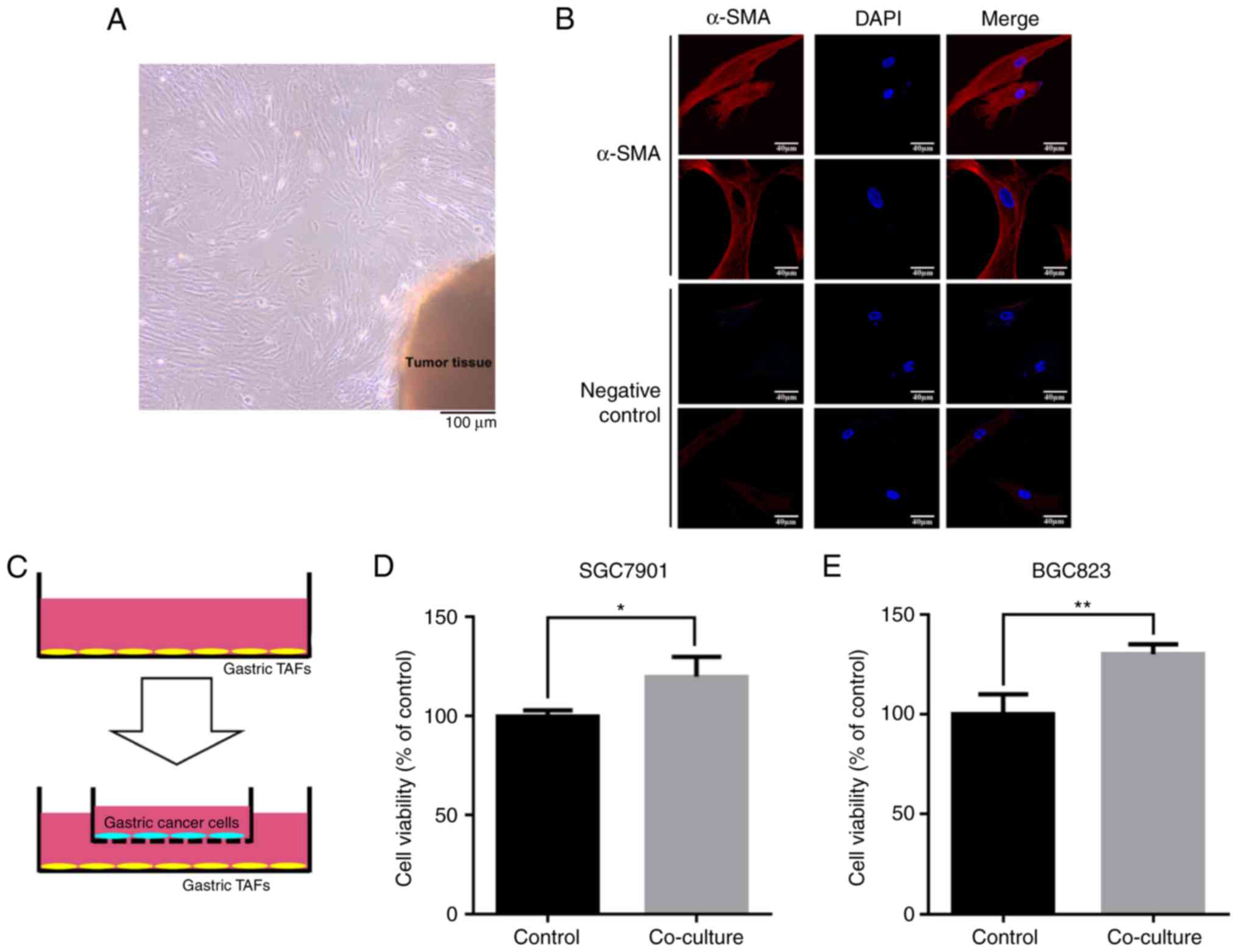
TAFs were isolated from gastric cancer tissue
(Fig. 1A). A TAF-specific marker,
α-SMA, was detected by immunocytochemistry to confirm the identity
of the TAFs (Fig. 1B). The
fibroblasts from normal gastric tissues did not express α-SMA at
detectable levels (data not shown). Gastric cancer cells were
seeded into a Transwell chamber and co-cultured with the isolated
gastric TAFs (Fig. 1C). The cell
viability assay demonstrated that gastric cancer cells grew faster
when co-cultured with gastric TAFs than normally cultured gastric
cancer cells (Fig. 1D and E). This
suggests that gastric cancer TAFs promote the proliferation of
gastric cancer cells, probably through the modulation of the cancer
microenvironment.
Metformin decreases the effect of TAFs
on the proliferation of co-cultured gastric cancer cells
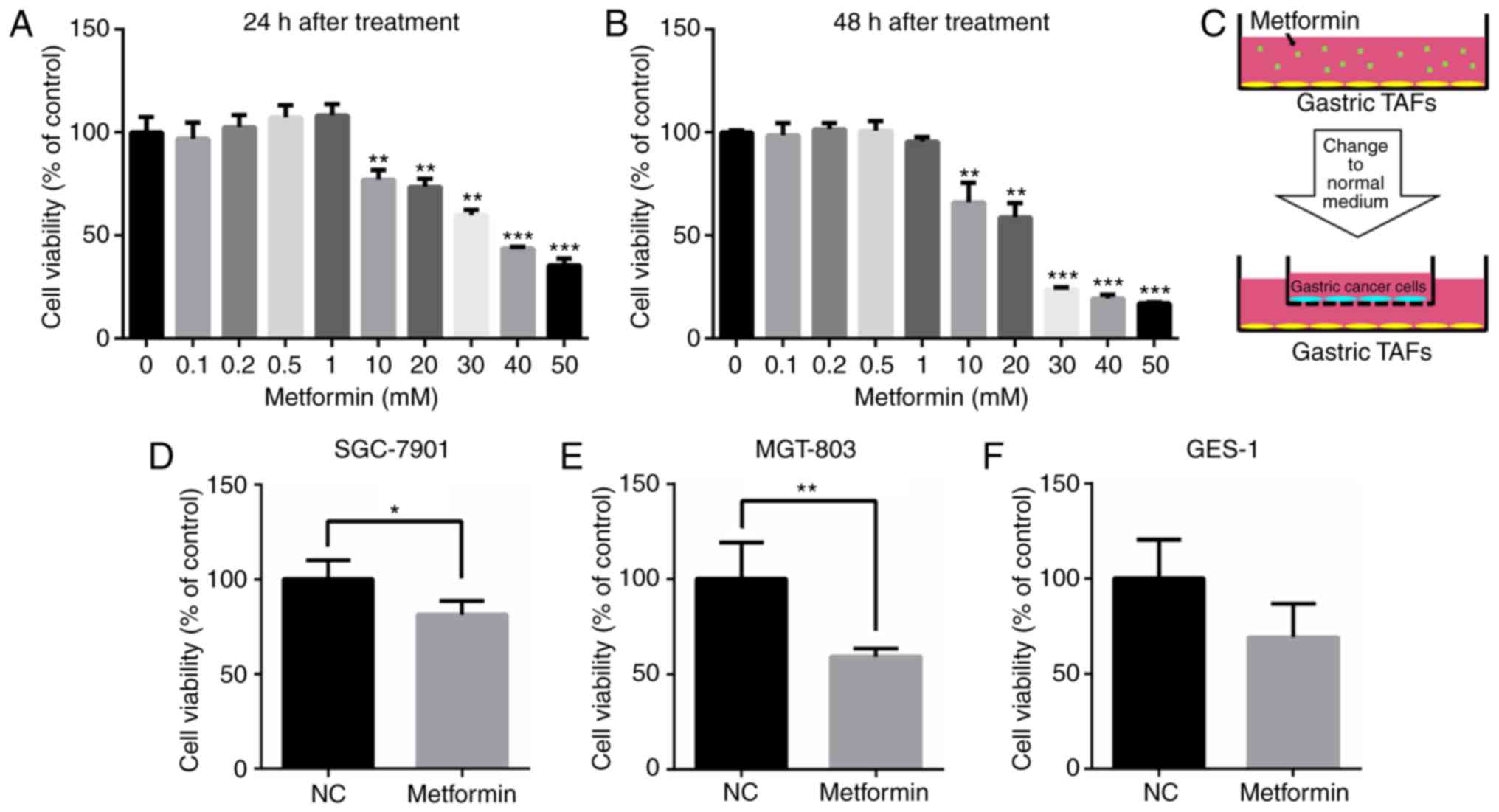
Next, it was investigated whether metformin affected
the tumor-promoting role of TAFs isolated from gastric cancer.
Gastric TAFs were treated with different concentrations of
metformin for 24 or 48 h. At concentrations of up to 1 mM,
metformin only slightly influenced the viability of the TAFs
(Fig. 2A and B). Next, gastric
cancer cells were co-cultured with gastric TAFs in a Transwell
chamber (Fig. 2C). Co-culture with
TAFs that had been pre-treated with 0.2 mM metformin for 48 h,
significantly decreased the proliferation of gastric cancer
SGC-7901 and MGT-803 cells, compared with gastric cancer cells
co-cultured with metformin-untreated TAFs (Fig. 2D and E). Although the proliferation
of GES-1 was decreased when co-cultured with TAFs pre-treated with
metformin, the difference was not significant (Fig. 2F). It was observed that metformin
reduced the stimulatory effect of TAFs on the proliferation of
gastric cancer cells, probably by altering the extracellular
proteins secreted by gastric TAFs into the culture medium.
Proteomic analysis of proteins
secreted from TAFs after metformin treatment
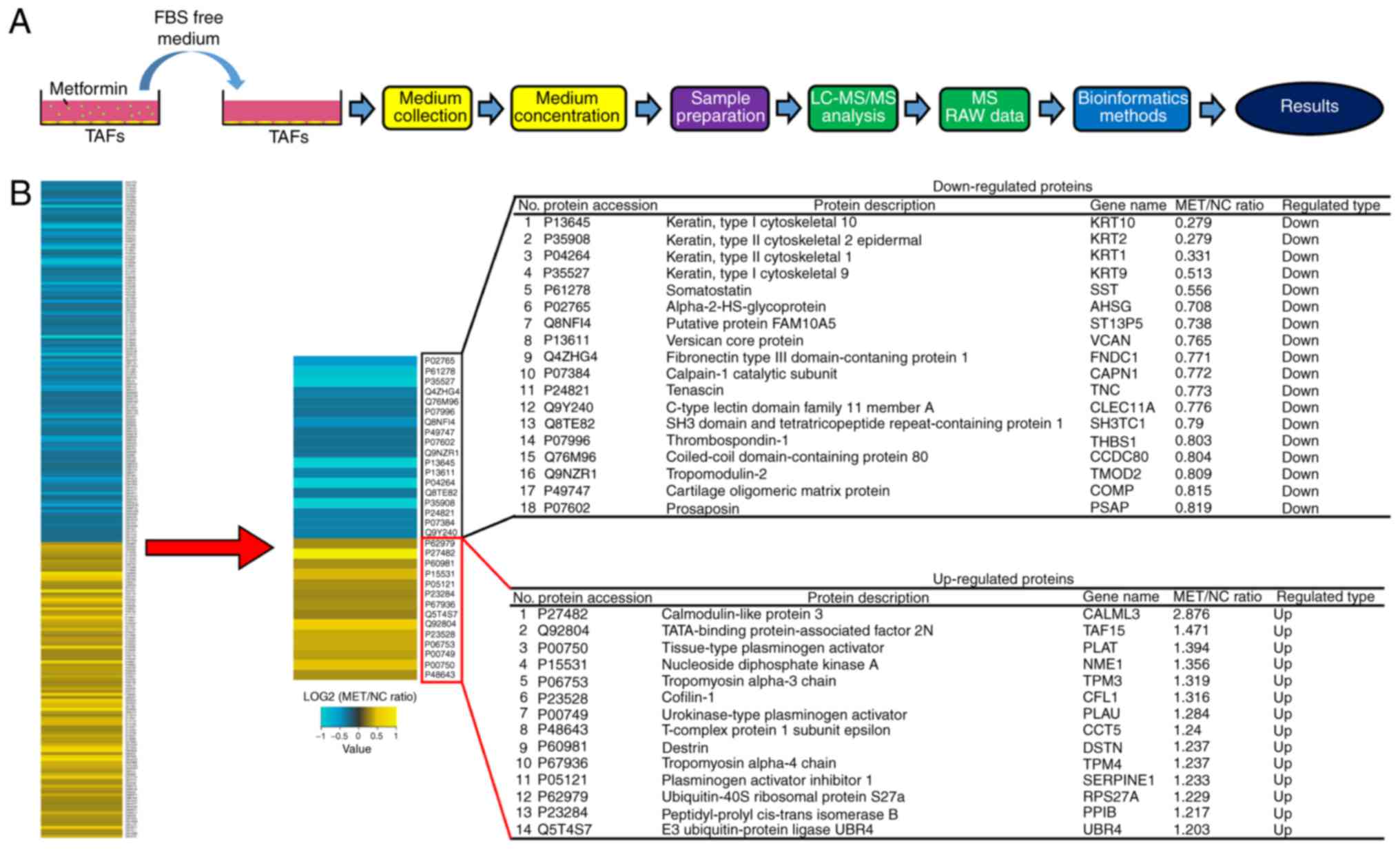
To investigate the proteomic changes in secreted
proteins from gastric TAFs following metformin treatment, the
proteomic profile of TAF-secreted proteins was investigated by
TMT-based protein quantification (Fig.
3A). In metformin-treated TAFs, >360 differentially
expressed proteins compared with those in untreated TAFs were
successfully identified, including 14 significantly upregulated and
18 significantly downregulated proteins (Fig. 3B). The upregulated proteins included
calmodulin-like protein 3 (Calml3), tropomyosin α-3 chain (TPM3),
tissue-type plasminogen activator (PLAT) and nucleoside diphosphate
kinase A (NME1).
To further determine the function and features of
the identified proteins, GO annotation was performed, including
protein domain, pathway and subcellular localization enrichment
analysis. The differentially expressed proteins were summarized for
each GO category. The proportion of differentially expressed
proteins representing each subcellular location is summarized in
Fig. 4A. Among the upregulated
proteins, 37% were extracellular proteins, 29% were cytosolic
proteins and 14% were nuclear proteins. Among the downregulated
proteins, 33% were extracellular proteins, 28% were nuclear
proteins and 17% were mitochondrial proteins. The results of the
protein domain enrichment analysis and KEGG pathway enrichment
analysis suggested that transcriptional dysregulation was
significantly enhanced in cancer, whereas the extracellular
matrix-receptor interaction pathway was the most significantly
downregulated pathway (Fig. 4B and
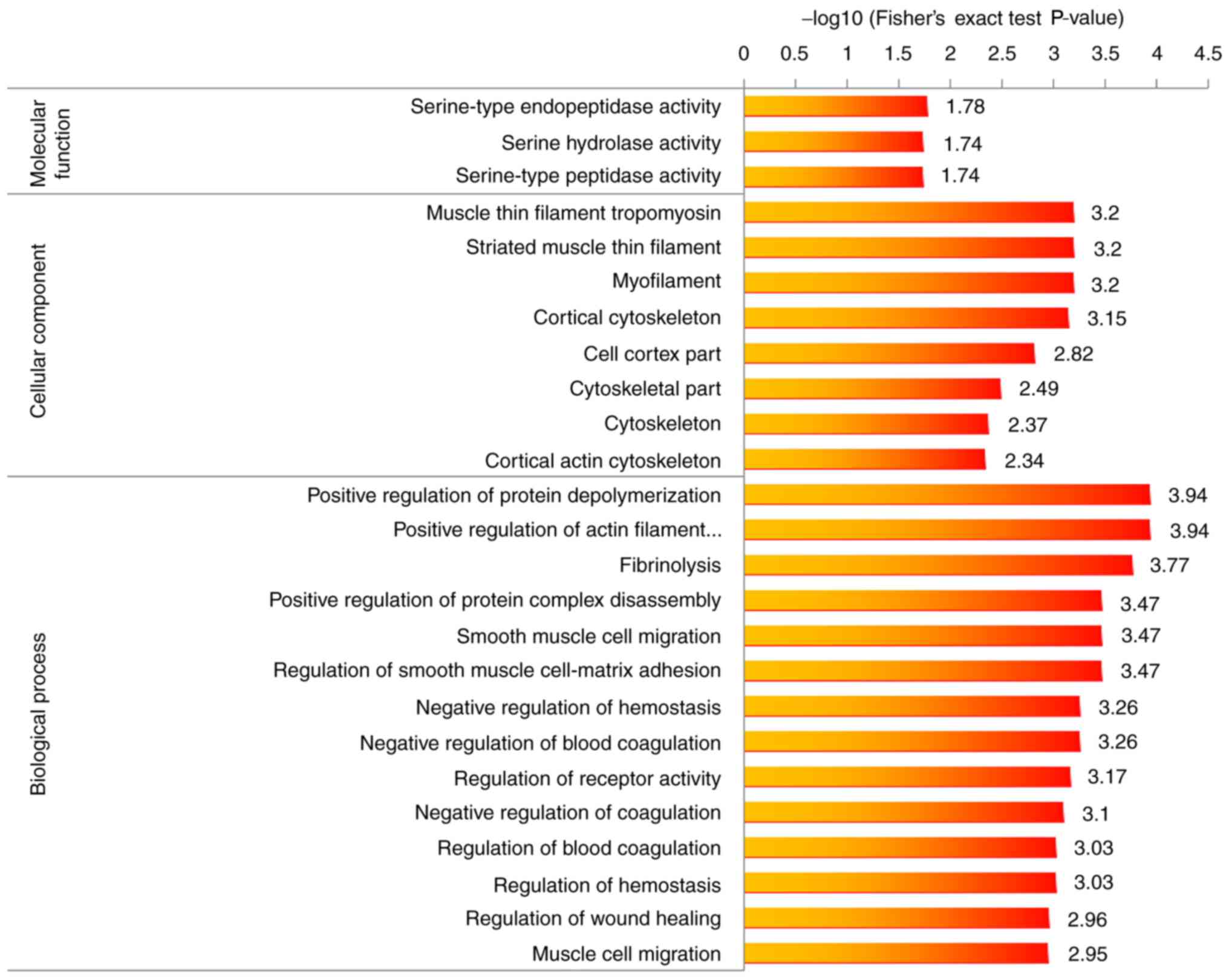
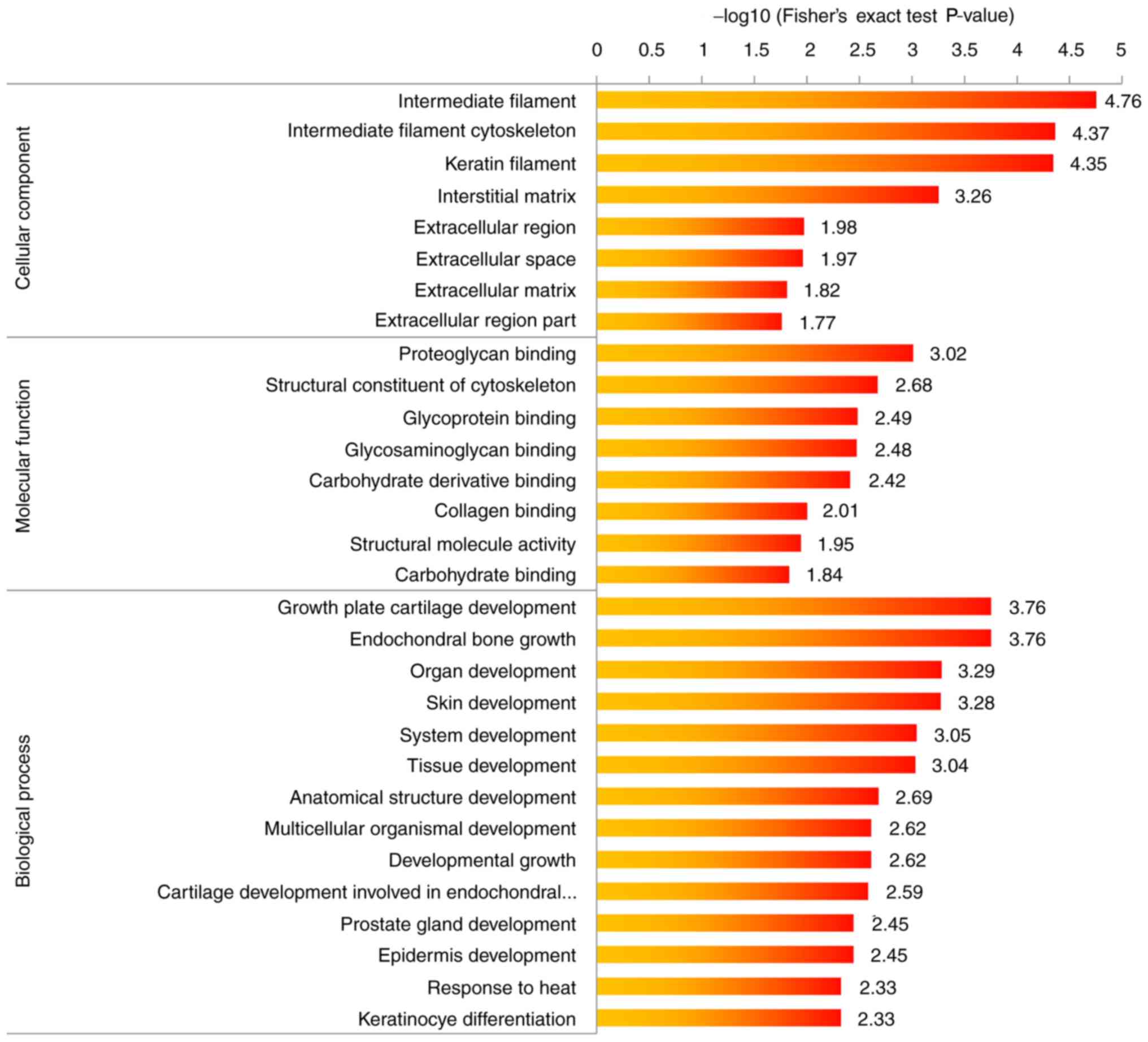
C). Finally, GO enrichment analysis of upregulation and
downregulated proteins was performed (Figs. 5 and 6). Proteins involved in the positive
regulation of protein and actin filament depolymerization were
enriched among the dysregulated proteins.
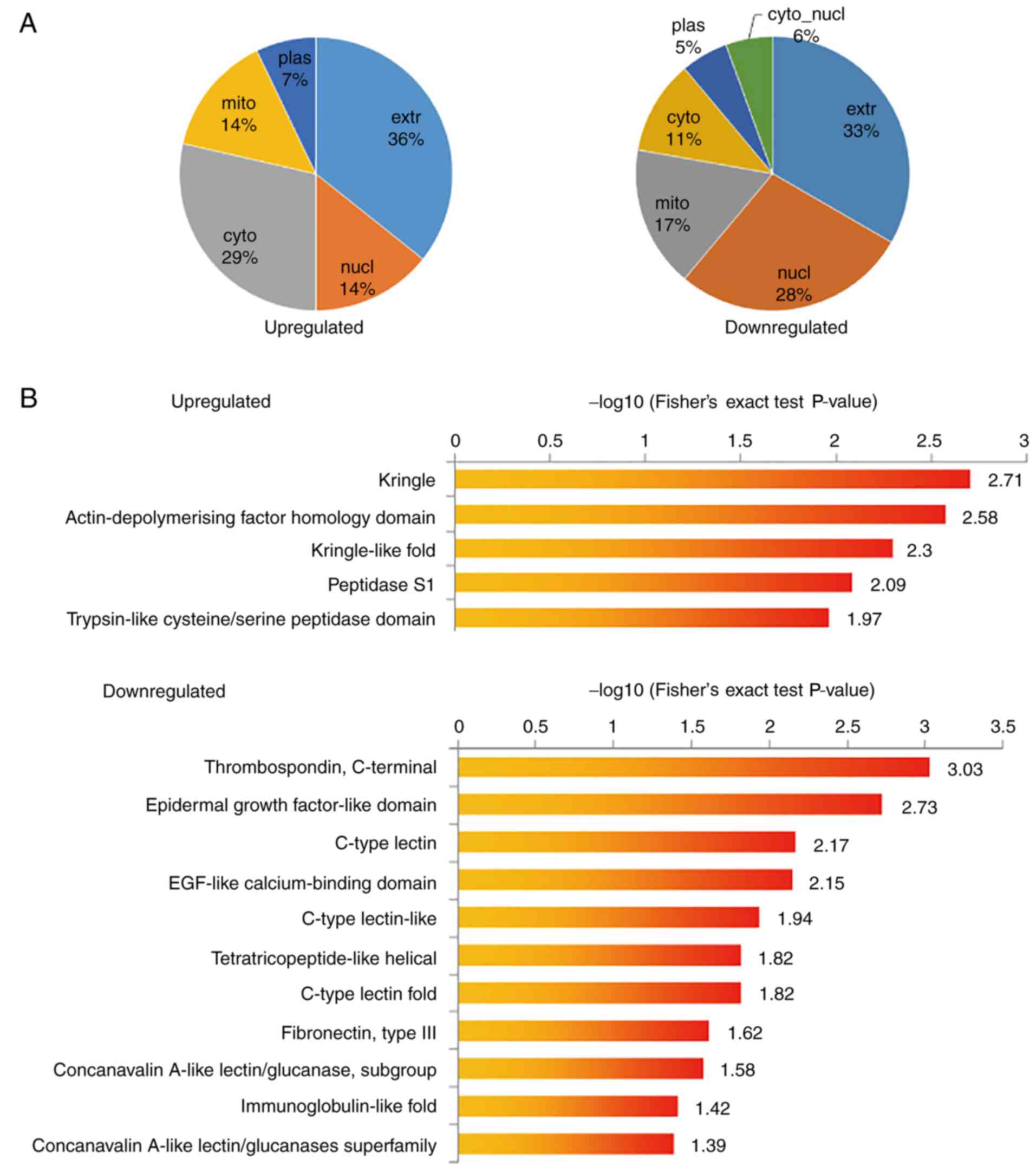 | Figure 4.Subcellular location, protein domain
enrichment and KEGG pathway enrichment analysis of upregulation and
downregulated proteins. (A) Subcellular location of upregulation
and down-regulated proteins (metformin-treated cells vs. control
group). (B) Protein domain enrichment analysis of upregulation and
downregulated proteins. (C) KEGG pathway enrichment analysis of
upregulation and downregulated proteins. KEGG, Kyoto Encyclopedia
of Genes and Genomes; extr, extracellular; plas, plasma; cyto,
cytosol; mito, mitochondria; nucl, nucleus; hsa, Homo
sapiens; ECM, extracellular matrix; PI3K, phosphoinositide-3
kinase; EGF, epidermal growth factor. |
Calml3 exerts a tumor-suppressive role
in gastric cancer cells
The suppressive effect of metformin on the
stimulatory role of TAFs on gastric cancer cells was likely to be
exerted through the upregulation and downregulation of certain
proteins released into the culture medium by TAFs. The further
investigations were focused on the upregulated proteins, which may
have potential antitumor activity. Among the dysregulated proteins
identified in TAFs subjected to metformin treatment, Calml3 was
most significantly upregulated, with a 2.88-fold increase in
metformin-treated TAFs compared with untreated TAFs. To explore
whether Calml3 secreted from gastric TAFs mediated the
tumor-suppressive role of metformin, gastric cancer cells were
incubated with culture medium containing different concentrations
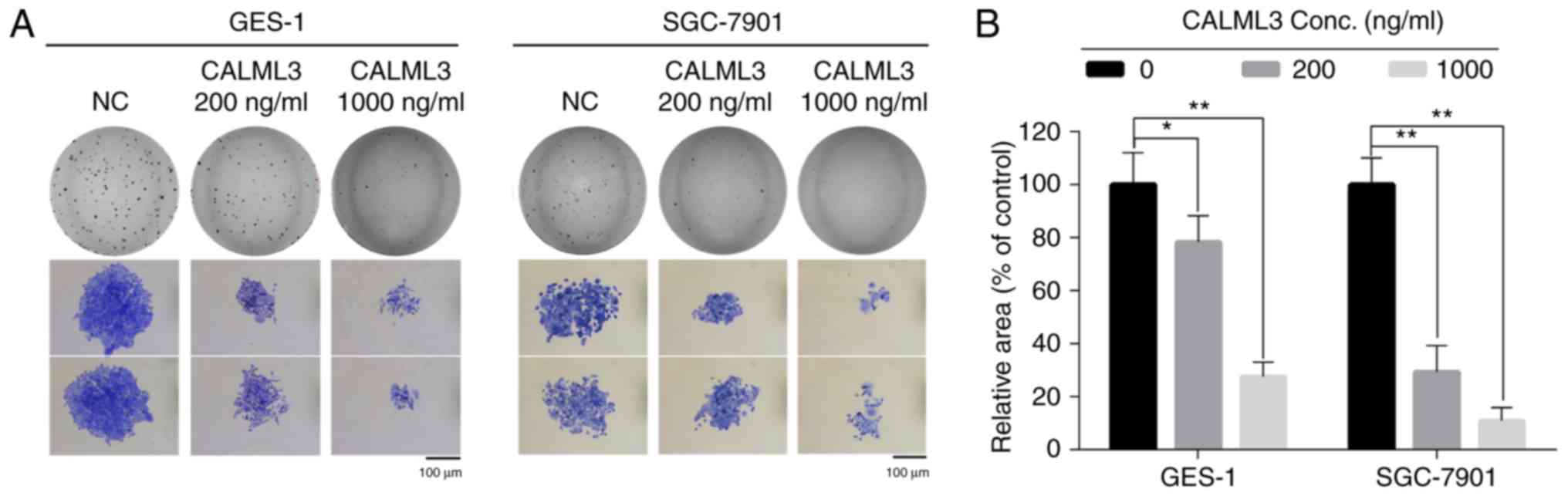
of recombinant Calml3 protein. A clonogenic assay revealed that
Calml3 not only decreased the surviving fraction of gastric cancer
cells, but also decreased the size of cell clones in two gastric
cancer cell lines (Fig. 7A and B).
These results demonstrate that Calml3 secreted from TAFs exerts a
tumor-suppressive role in gastric cancer cells. Calml3 may account
for the antitumor effect of metformin on the tumor
microenvironment.
Discussion
Gastric cancer is one of the leading causes of
cancer-associated mortality worldwide (1–3). It is
clear that tumor-associated fibroblasts have an important role in
the initiation and progression of gastric cancer. In previous
co-culture experiments, TAFs increased the proliferation and
progression of cancer cells and promoted angiogenesis (30,31).
Injection of immortalized prostate epithelial cells together with
TAFs, but not with normal fibroblasts, led to tumor formation in
mice (30). The tumor growth
promoting function of TAFs may be due to the secretion of cytokines
that have a positive regulatory role in cancer (32). Orimo et al (31) reported that stromal fibroblasts
present in invasive human breast carcinomas promote tumor growth
and angiogenesis through elevated stromal cell-derived factor-1α
secretion. Vermeulen et al (33) reported that TAFs secrete hepatocyte
growth factor and activate β-catenin-dependent transcription, and
subsequently the clonogenicity of cancer stem cells. Exosomes
derived from fibroblasts have also emerged as positive mediators of
cancer progression (34–36). Thus, targeting TAFs or inhibiting
the tumor-promoting role of TAFs has emerged as a novel strategy in
cancer treatment (8,9).
Metformin is an oral anti-diabetes drug that has
been widely studied due to its antitumor activity (19–22).
Most previous studies have focused on its direct antitumor effect
on cancer cells. However, it has remained elusive whether metformin
affects the tumor microenvironment or TAFs. In the present study,
gastric cancer cells were co-cultured in a Transwell system with
TAFs that had been pre-treated with metformin. The results
indicated that the proliferation and clonogenicity were reduced in
gastric cancer cells co-cultured with metformin-treated TAFs, which
indicates that metformin inhibited the tumor-promoting role of TAFs
that were isolated from gastric cancer cells. The culture medium
from TAFs was concentrated and analyzed by LC-MS/MS to quantify the
proteomic differences in metformin-treated and untreated TAFs.
Proteomic analysis revealed that metformin affected the secretion
of 32 proteins (14 upregulated and 18 downregulated) in the culture
medium of gastric TAFs. Although the upregulated as well as the
downregulated proteins are likely to be responsible for the
antitumor effect of metformin, the present study we only focused on
upregulated proteins, which may have potential tumor suppressor
activity. Among these, Calml3 was 2.88-fold upregulated in the
culture medium of gastric TAFs after treatment with 0.2 mM
metformin. Further studies on the proteomic profiles of TAF
secretion with other concentrations of metformin are needed.
Calml3 is a 148-amino acid calcium sensor protein
(37). Rogers et al
(38) reported that Calml3 is
downregulated in invasive ductal carcinoma and lobular carcinoma
compared with that in normal breast epithelium. Furthermore, Calml3
is expressed in the normal oral mucosa, and is downregulated during
its malignant transformation (39),
suggesting a potential tumor-suppressive role. A recent study by
Yang et al (27) using
Calml3 gain- and loss-of-function experiments suggested that Calml3
inhibits the genesis and metastasis of HCC. However, their study
focused on Calml3 expressed in HCC cells, and they did not assess
the potential antitumor effects of Calml3 secreted from TAFs. In
the present study, recombinant Calml3 protein was added into the
culture medium of gastric cancer cells to imitate the culture
medium of TAFs treated with metformin. It was revealed that Calml3
exerted a tumor-suppressive effect on these gastric cancer cells by
inhibiting their clonogenicity and proliferation. However, the
physiological protein levels of Calml3 and whether the
concentration of Calml3 used in the clonogenic assay of the present
study (200 ng/ml or 1,000 ng/ml, which were determined in
preliminary experiments) is physiologically achievable remains
elusive. In addition, the mechanism underlying the
tumor-suppressive role of Calml2 warrants further
investigation.
In conclusion, the present study indicated that
metformin alters the proteins secreted from TAFs to reduce their
proliferative effect on gastric cancer cells. In particular, the
levels of Calml3 were increased, which exerted a tumor-suppressive
effect on gastric cancer cells. The present study provides novel
evidence for the antitumor effects of metformin. The present study
may also provide a novel antitumor strategy using Calml3, although
further investigation is required in the future.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant nos. 81602101, 81502038,
81773227 and 31770911), the Key Scientific Development Program of
China (grant no. 2016YFC0904702), the Wuxi Medical Innovation Team
(grant no. CXTD005) and the Social Development Program of Jiangsu
Province (grant nos. BE2017634 and BE2017652).
Availability of data and materials
The data and materials are available from the
corresponding authors on request.
Authors' contributions
SZ and CY conceived and designed the study. GC, CY,
ZT, JZ and QW performed the molecular biology experiments. GC, FA,
JZ and SL drafted the manuscript and prepared the figures. FA, JC,
SL and QW isolated the TAFs. JC, ZT and QZ performed the
statistical analysis. QZ and SZ modified the manuscript. All
authors read and approved the final version of the manuscript.
Ethics approval and consent to
participate
All patients provided written informed consent for
their tissues to be used for scientific research. Ethical approval
of the study was obtained from the First People's Hospital of
Xuzhou (Suzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Van Cutsem E, Sagaert X, Topal B,
Haustermans K and Prenen H: Gastric cancer. Lancet. 388:2654–2664.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Steliarova-Foucher E,
Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D and
Bray F: Cancer incidence and mortality patterns in Europe:
Estimates for 40 countries in 2012. Eur J Cancer. 49:1374–1403.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bornschein J, Selgrad M, Warnecke M,
Kuester D, Wex T and Malfertheiner P: H. pylori infection is a key
risk factor for proximal gastric cancer. Dig Dis Sci. 55:3124–3131.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang F, Meng W, Wang B and Qiao L:
Helicobacter pylori-induced gastric inflammation and gastric
cancer. Cancer Lett. 345:196–202. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jiang L, Yang KH, Guan QL, Zhao P, Chen Y
and Tian JH: Survival and recurrence free benefits with different
lymphadenectomy for resectable gastric cancer: A meta-analysis. J
Surg Oncol. 107:807–814. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Orditura M, Galizia G, Sforza V,
Gambardella V, Fabozzi A, Laterza MM, Andreozzi F, Ventriglia J,
Savastano B, Mabilia A, et al: Treatment of gastric cancer. World J
Gastroenterol. 20:1635–1649. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tao L, Huang G, Song H, Chen Y and Chen L:
Cancer associated fibroblasts: An essential role in the tumor
microenvironment. Oncol Lett. 14:2611–2620. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shiga K, Hara M, Nagasaki T, Sato T,
Takahashi H and Takeyama H: Cancer-associated fibroblasts: Their
characteristics and their roles in tumor growth. Cancers.
7:2443–2458. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chung HW and Lim JB: Role of the tumor
microenvironment in the pathogenesis of gastric carcinoma. World J
Gastroenterol. 20:1667–1680. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kalluri R: The biology and function of
fibroblasts in cancer. Nat Rev Cancer. 16:582–598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tomasek JJ, Gabbiani G, Hinz B, Chaponnier
C and Brown RA: Myofibroblasts and mechano-regulation of connective
tissue remodelling. Nat Rev Mol Cell Biol. 3:349–363. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Marsh T, Pietras K and McAllister SS:
Fibroblasts as architects of cancer pathogenesis. Biochim Biophys
Acta. 1832:1070–1078. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ostman A and Augsten M: Cancer-associated
fibroblasts and tumor growth-bystanders turning into key players.
Curr Opin Genet Dev. 19:67–73. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kalluri R and Zeisberg M: Fibroblasts in
cancer. Nat Rev Cancer. 6:392–401. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Öhlund D, Elyada E and Tuveson D:
Fibroblast heterogeneity in the cancer wound. J Exp Med.
211:1503–1523. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan Y, Wang LF and Wang RF: Role of
cancer-associated fibroblasts in invasion and metastasis of gastric
cancer. World J Gastroenterol. 21:9717–9726. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pang T, Wang X, Gao J, Chen W, Shen XJ,
Nie MM, Luo T, Yin K, Fang G, Wang KX, et al: Fiber-modified
hexon-chimeric oncolytic adenovirus targeting cancer associated
fibroblasts inhibits tumor growth in gastric carcinoma. Oncotarget.
8:76468–76478. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gotlieb WH, Saumet J, Beauchamp MC, Gu J,
Lau S, Pollak MN and Bruchim I: In vitro metformin anti-neoplastic
activity in epithelial ovarian cancer. Gynecol Oncol. 110:246–250.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Akinyeke T, Matsumura S, Wang X, Wu Y,
Schalfer ED, Saxena A, Yan W, Logan SK and Li X: Metformin targets
c-MYC oncogene to prevent prostate cancer. Carcinogenesis.
34:2823–2832. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zakikhani M, Dowling R, Fantus IG,
Sonenberg N and Pollak M: Metformin is an AMP kinase-dependent
growth inhibitor for breast cancer cells. Cancer Res.
66:10269–10273. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu C, Jiao Y, Xue J, Zhang Q, Yang H, Xing
L, Chen G, Wu J, Zhang S, Zhu W and Cao J: Metformin sensitizes
non-small cell lung cancer cells to an epigallocatechin-3-gallate
(EGCG) treatment by suppressing the Nrf2/HO-1 signaling pathway.
Int J Biol Sci. 13:1560–1569. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Safe S, Naira V and Karki K:
Metformin-induced anticancer activities: Recent insights.
Metformin-induced anticancer activities: Recent insights. Biol
Chem. 399:321–335. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Malki A and Youssef A: Antidiabetic drug
metformin induces apoptosis in human MCF breast cancer via
targeting ERK signaling. Oncol Res. 19:275–285. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bhalla K, Hwang BJ, Dewi RE, Twaddel W,
Goloubeva OG, Wong KK, Saxena NK, Biswal S and Girnun GD: Metformin
prevents liver tumorigenesis by inhibiting pathways driving hepatic
lipogenesis. Cancer Prev Res (Philla). 5:544–552. 2012. View Article : Google Scholar
|
|
26
|
Chen G, Feng W, Zhang S, Bian K, Yang Y,
Fang C, Chen M, Yang J and Zou X: Metformin inhibits gastric cancer
via the inhibition of HIF1α/PKM2 signaling. Am J Cancer Res.
5:1423–1434. 2015.PubMed/NCBI
|
|
27
|
Yang B, Li M, Tang W, Liu W, Zhang S, Chen
L and Xia J: Dynamic network biomarker indicates pulmonary
metastasis at the tipping point of hepatocellular carcinoma. Nat
Commun. 9:6782018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang X, Xu X, Zhu J, Zhang S, Wu Y, Wu Y,
Zhao K, Xing C, Cao J, Zhu H, et al: miR-31 affects colorectal
cancer cells by inhibiting autophagy in cancer-associated
fibroblasts. Oncotarget. 7:79617–79628. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu J, Dong X, Wang L, Ji H and Liu A:
Antitumor effects of seleno-β-lactoglobulin (Se-β-Lg) against human
gastric cancer MGC-803 cells. Eur J Pharmacol. 833:109–115. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Olumi AF, Grossfeld GD, Hayward SW,
Carroll PR, Tlsty TD and Cunha GR: Carcinoma-associated fibroblasts
direct tumor progression of initiated human prostatic epithelium.
Cancer Res. 59:5002–5011. 1999.PubMed/NCBI
|
|
31
|
Orimo A, Gupta PB, Sgroi DC,
Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL
and Weinberg RA: Stromal fibroblasts present in invasive human
breast carcinomas promote tumor growth and angiogenesis through
elevated SDF-1/CXCL12 secretion. Cell. 121:335–348. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Klil-Drori AJ, Azoulay L and Pollak MN:
Cancer, obesity, diabetes, and antidiabetic drugs: Is the fog
clearing? Nat Rev Clin Oncol. 14:85–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vermeulen L, De Sousa E, Melo F, van der
Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M,
Merz C, Rodermond H, et al: Wnt activity defines colon cancer stem
cells and is regulated by the microenvironment. Nat Cell Biol.
12:468–476. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Borges FT, Melo SA, Özdemir BC, Kato N,
Revuelta I, Miller CA, Gattone VH II, LeBleu VS and Kalluri R:
TGF-β1-containing exosomes from injured epithelial cells activate
fibroblasts to initiate tissue regenerative responses and fibrosis.
J Am Soc Nephrol. 24:385–392. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kahlert C and Kalluri R: Exosomes in tumor
microenvironment influence cancer progression and metastasis. J Mol
Med. 91:431–437. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Luga V and Wrana JL: Tumor-stroma
interaction: Revealing fibroblast-secreted exosomes as potent
regulators of Wnt-planar cell polarity signaling in cancer
metastasis. Cancer Res. 73:6843–6847. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yaswen P, Smoll A, Peehl DM, Trask DK,
Sager R and Stampfer MR: Down-regulation of a calmodulin-related
gene during transformation of human mammary epithelial cells. Proc
Natl Acad Sci USA. 87:7360–7364. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rogers MS, Foley MA, Crotty TB, Hartmann
LC, Ingle JN, Roche PC and Strehler EE: Loss of immunoreactivity
for human calmodulin-like protein is an early event in breast
cancer development. Neoplasia. 1:220–225. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Brooks MD, Bennett RD, Strehler EE, Sebo
TJ, Eckert SE and Carr AB: Human calmodulin-like protein (CLP)
expression in oral squamous mucosa and in malignant transformation.
J Prosthodont. 18:11–16. 2009. View Article : Google Scholar : PubMed/NCBI
|