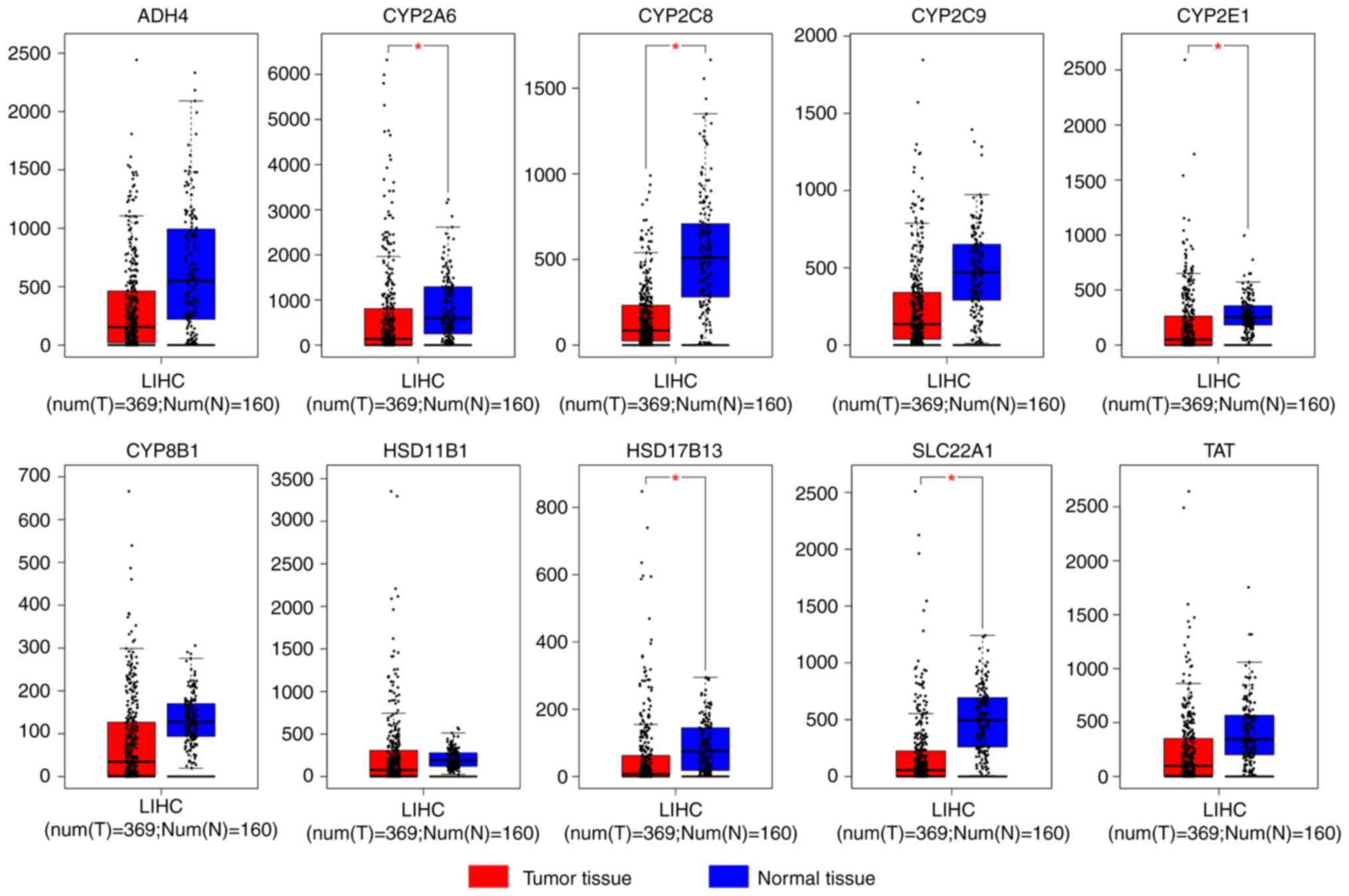
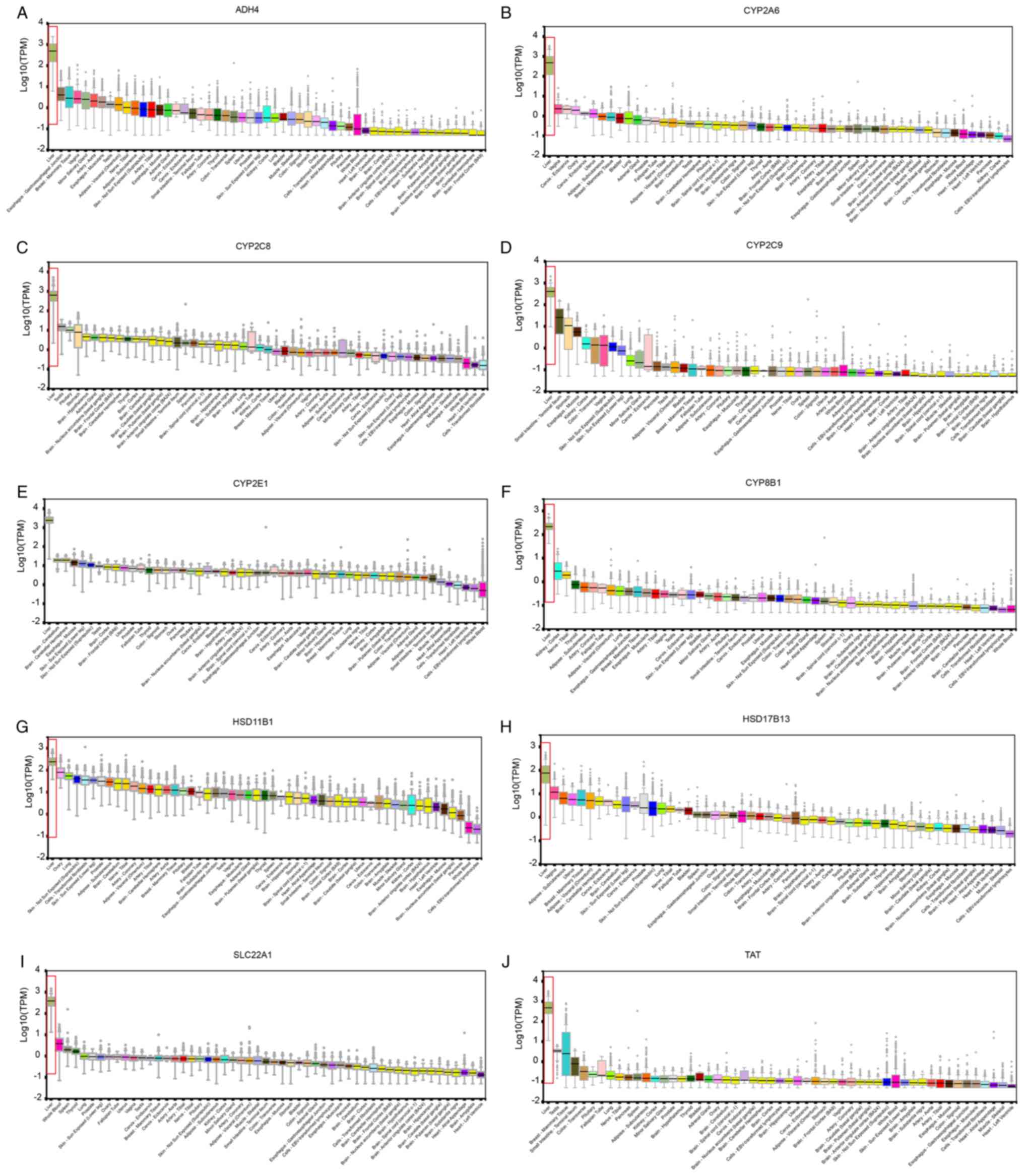
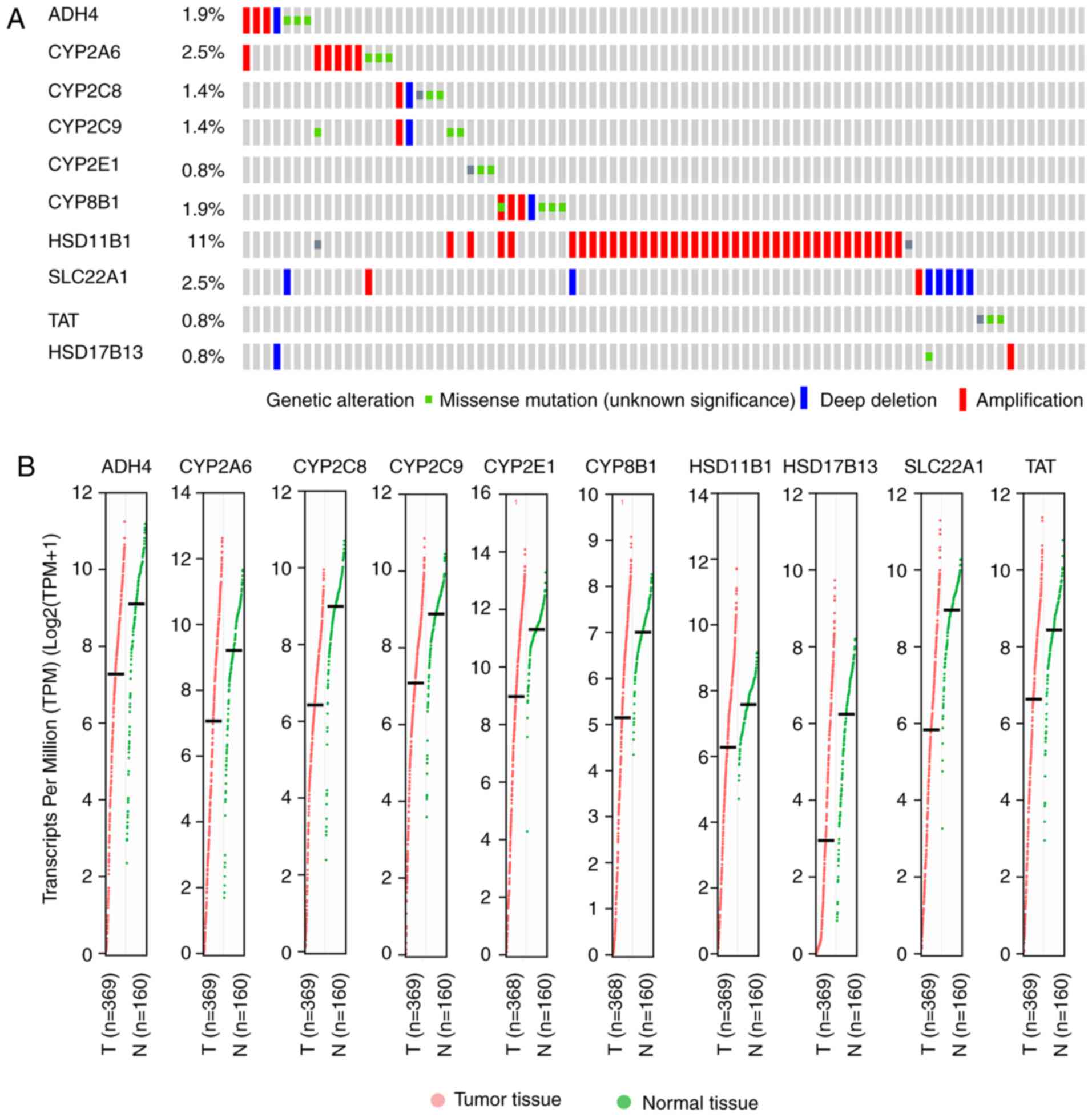
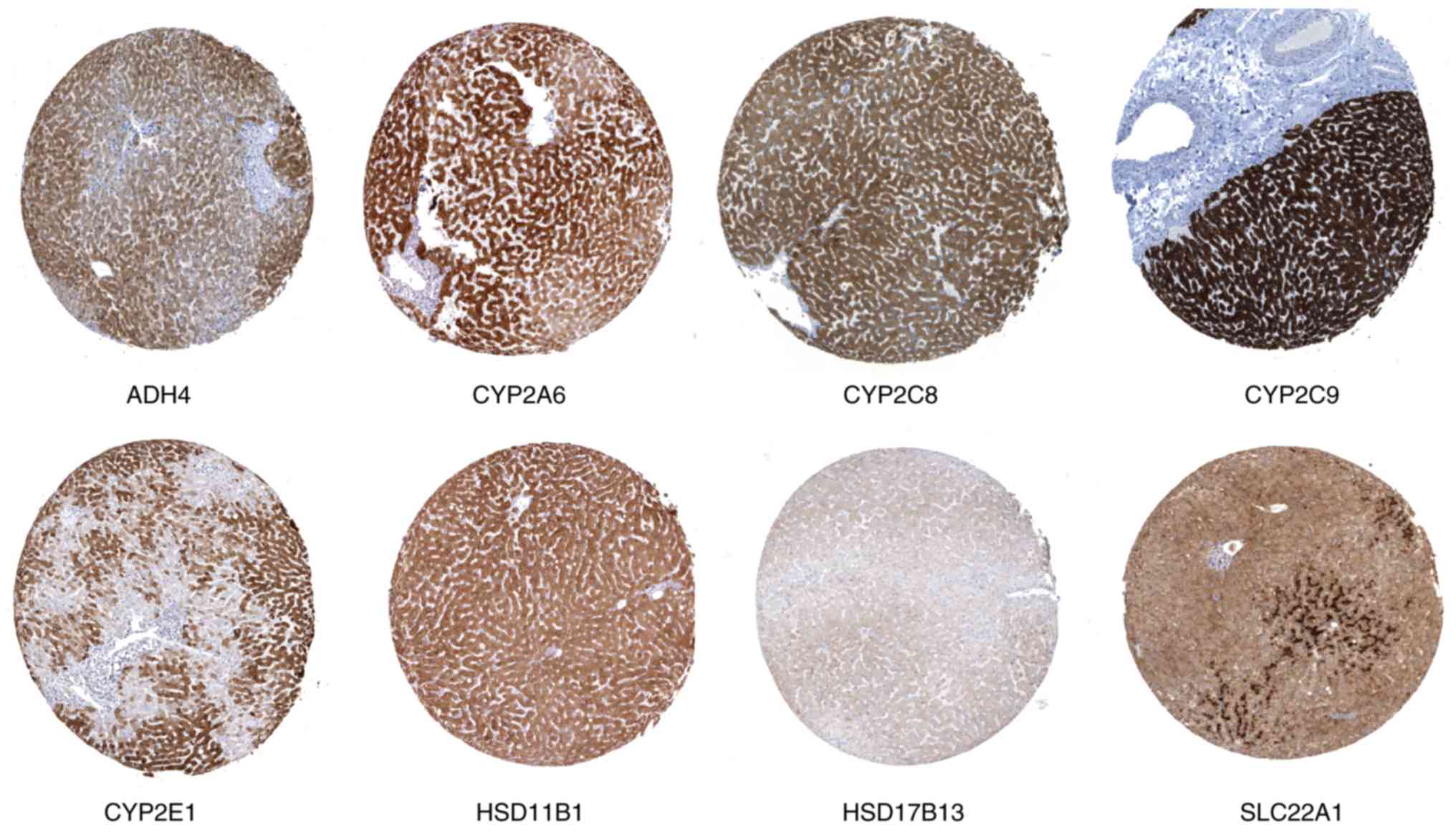
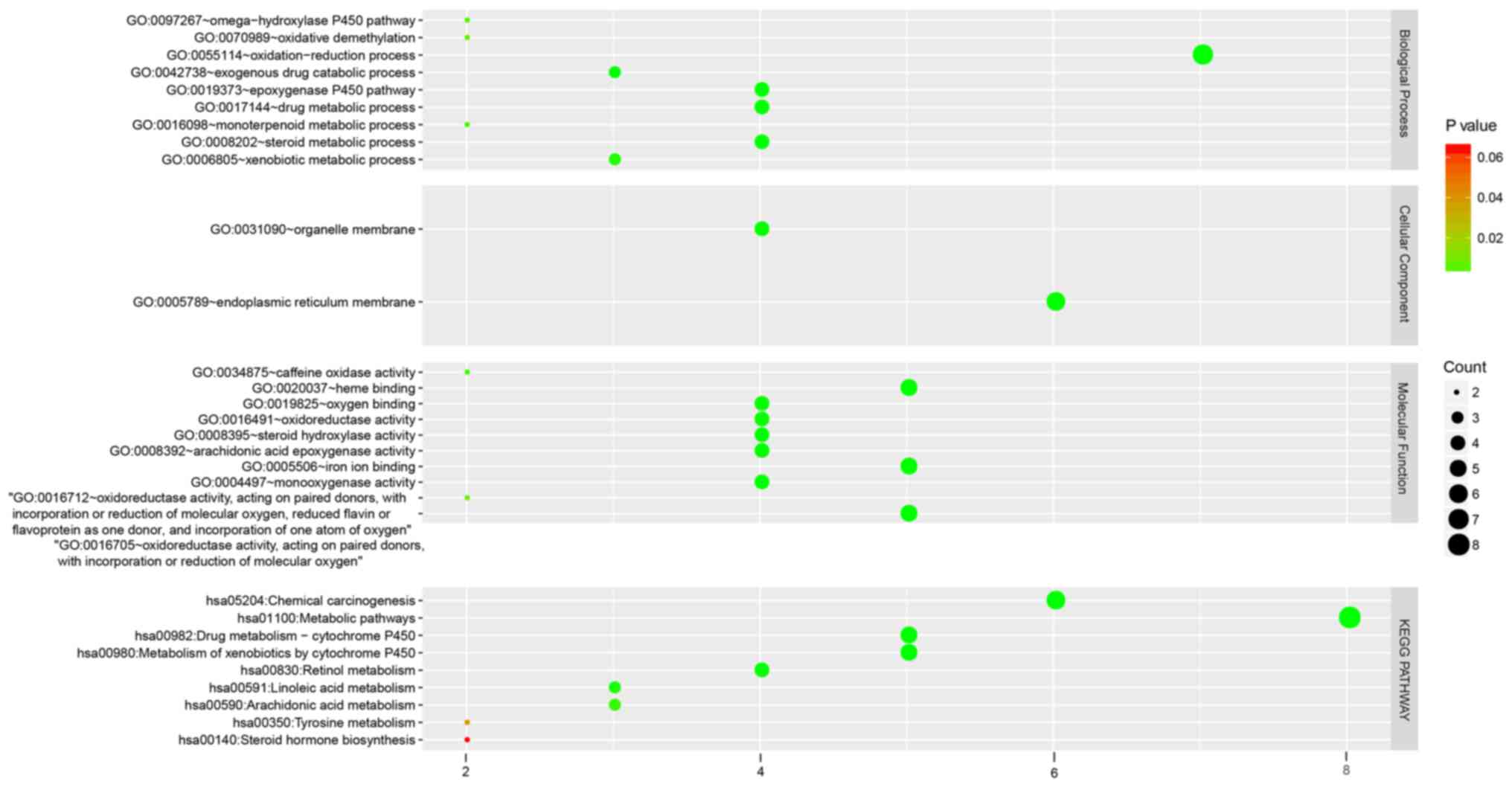
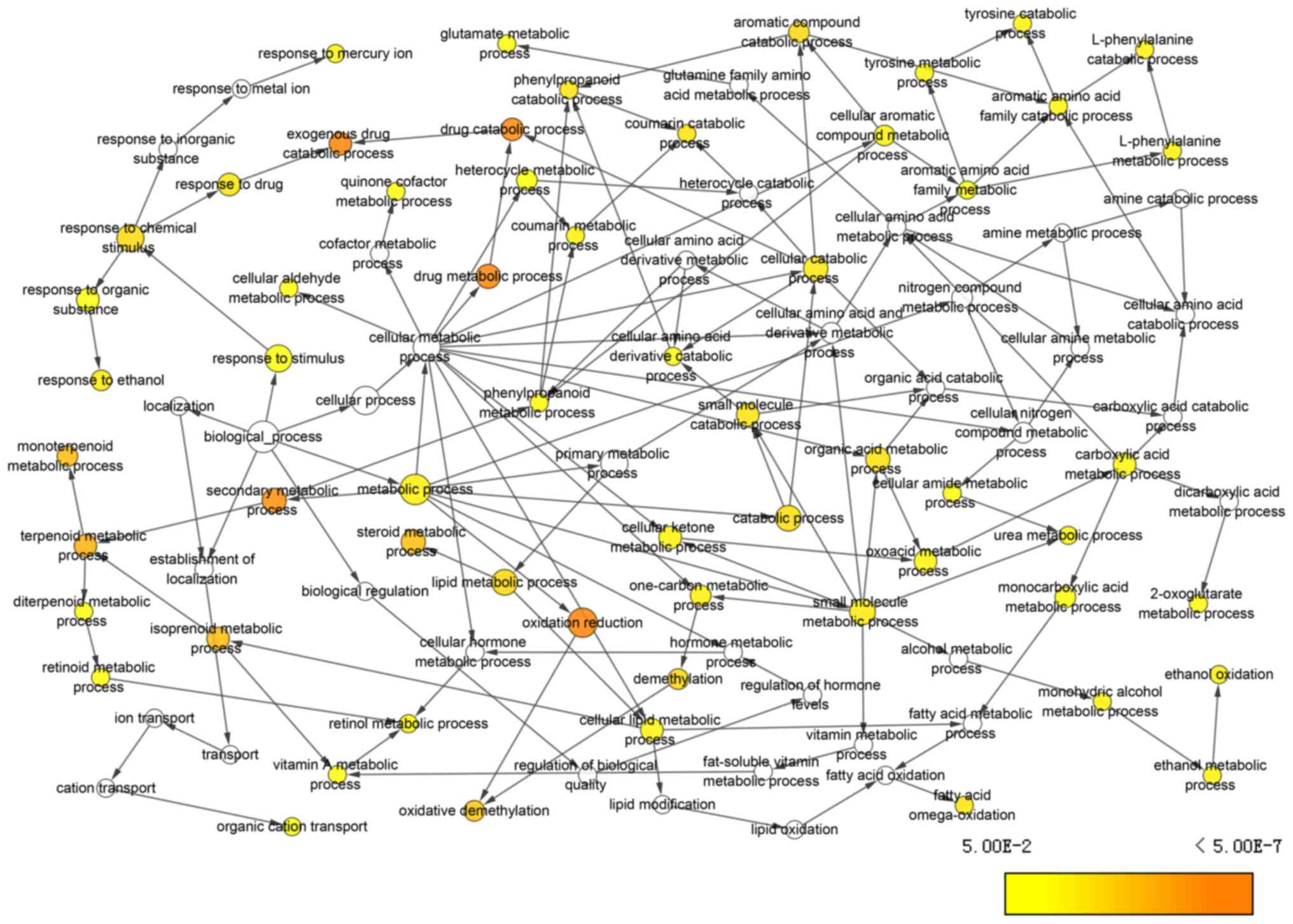
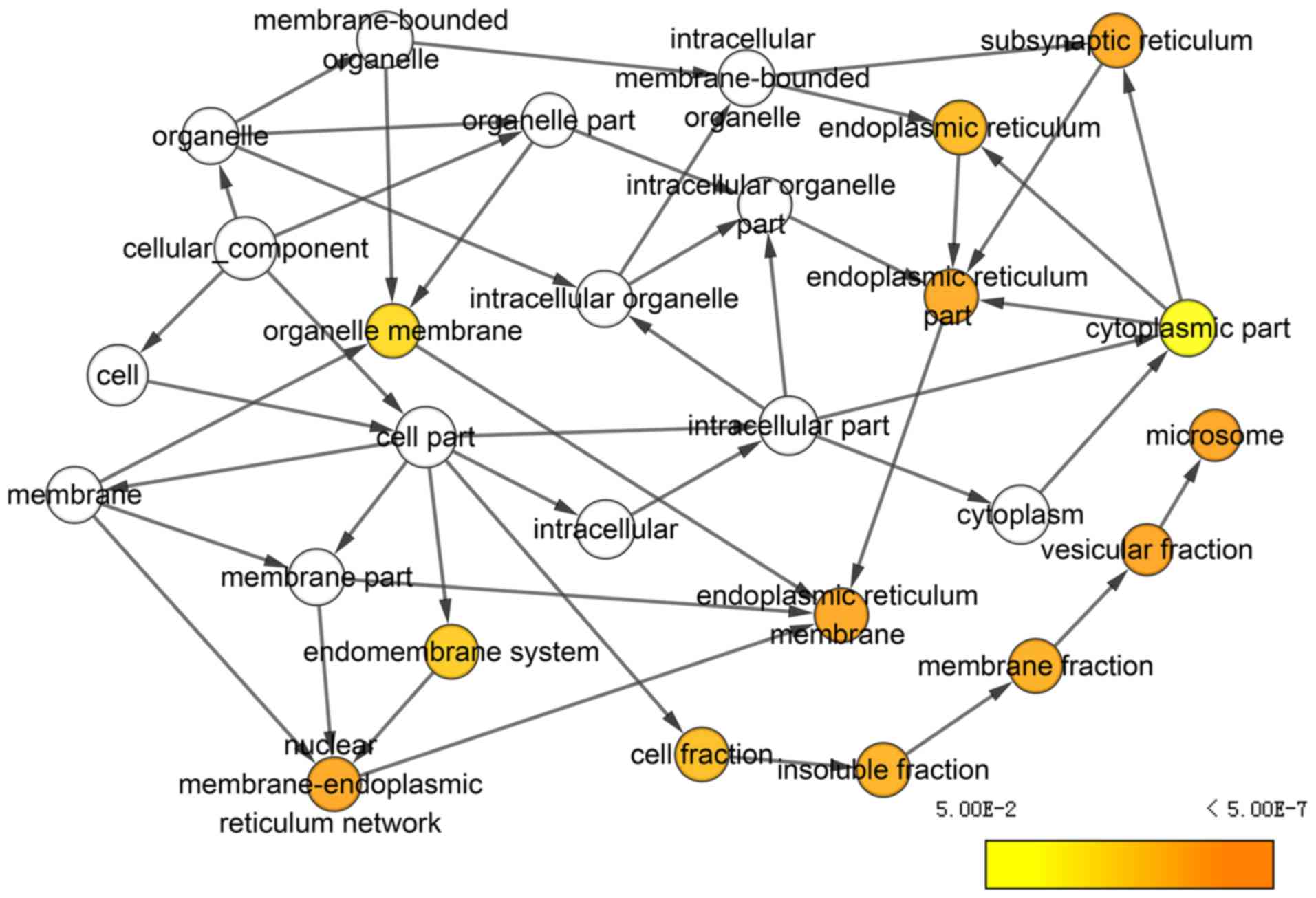
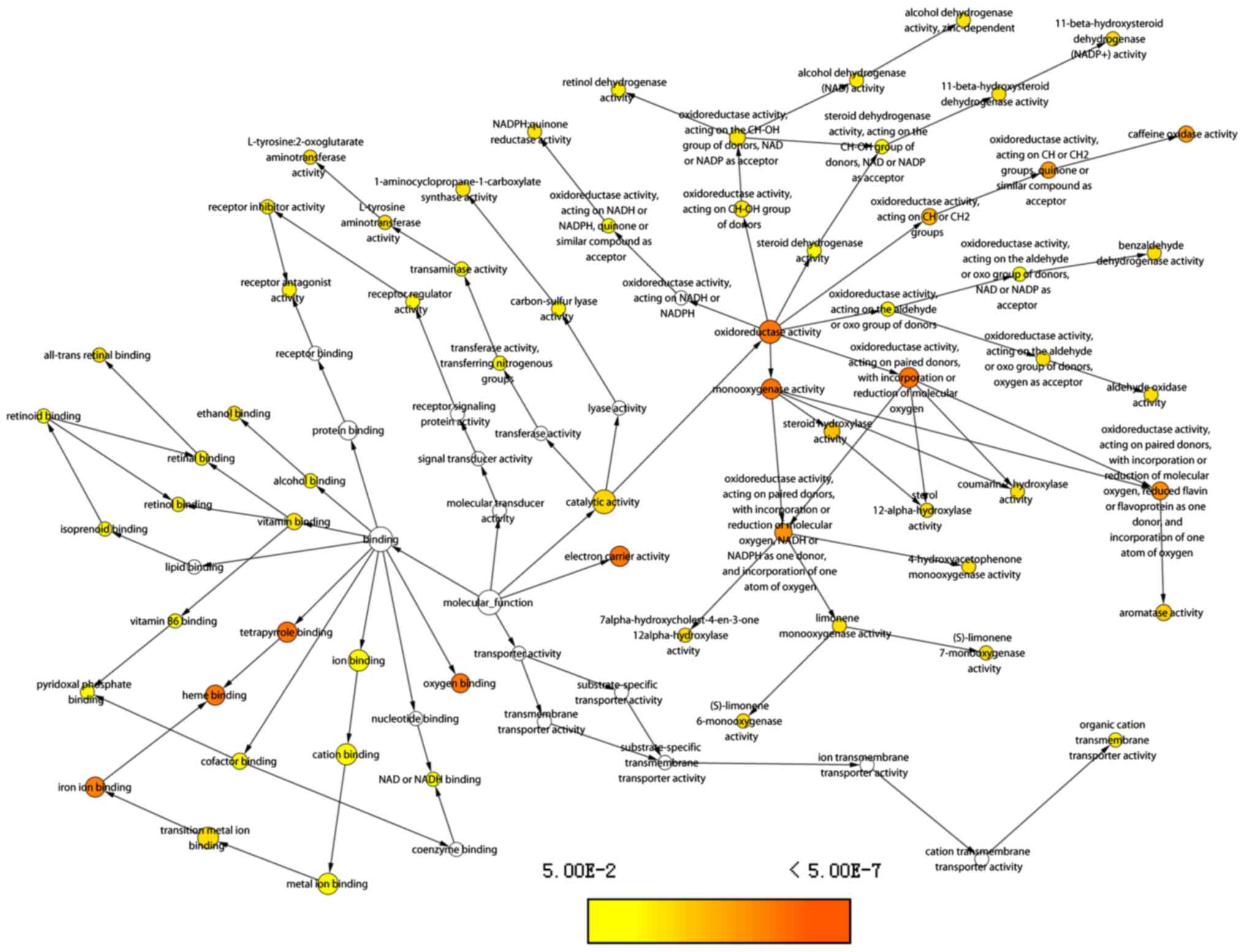
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kolonel LN and Wilkens LR: Migrant
studies. Cancer Epidemiology and Prevention. Schottenfeld D and
Fraumeni JF Jr: 3rd. Oxford University Press, Inc.; New York, NY:
pp. 189–201. 2006, View Article : Google Scholar
|
|
3
|
Kgatle MM, Setshedi M and Hairwadzi HN:
Hepatoepigenetic alterations in viral and nonviral-induced
hepatocellular carcinoma. Biomed Res Int 2016. 39564852016.
|
|
4
|
Marotta F, Vangieri B, Cecere A and
Gattoni A: The pathogenesis of hepatocellular carcinoma is
multifactorial event. Novel immunological treatment in prospect.
Clin Ter. 155:187–199. 2004.PubMed/NCBI
|
|
5
|
Coleman WB: Mechanisms of human
hepatocarcinogenesis. Curr Mol Med. 3:573–388. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dominguez-Malagón H and Gaytan-Graham S:
Hepatocellular carcinoma: An update. Ultrastruct Pathol.
25:497–516. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen G, Wang D, Zhao X, Cao J, Zhao Y,
Wang F, Bai J, Luo D and Li L: miR-155-5p modulates malignant
behaviors of hepatocellular carcinoma by directly targeting CTHRC1
and indirectly regulating GSK-3β-involved Wnt/β-catenin signaling.
Cancer Cell Int. 17:1182017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Balogh J, Victor D III, Asham EH,
Burroughs SG, Boktour M, Saharia A, Li X, Ghobrial RM and Monsour
HP Jr: Hepatocellular carcinoma: A review. J Hepatocell Carcinoma.
3:41–53. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tien AJ, Chien CY, Chen YH, Lin LC and
Chien CT: fruiting bodies of antrodia cinnamomea and its active
triterpenoid, antcin K, ameliorates N-nitrosodiethylamine-induced
hepatic inflammation, fibrosis and carcinogenesis in rats. Am J
Chin Med. 45:173–198. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Quintavalle C, Hindupur SK, Quagliata L,
Pallante P, Nigro C, Condorelli G, Andersen JB, Tagscherer KE, Roth
W, Beguinot F, et al: Phosphoprotein enriched in diabetes
(PED/PEA15) promotes migration in hepatocellular carcinoma and
confers resistance to sorafenib. Cell Death Dis. 8:e31382017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu G and Dang C: CMTM5 is downregulated
and suppresses tumour growth in hepatocellular carcinoma through
regulating PI3K-AKT signalling. Cancer Cell Int. 17:1132017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cao MQ, You AB, Zhu XD, Zhang W, Zhang YY,
Zhang SZ, Zhang KW, Cai H, Shi WK, Li XL, et al: miR-182-5p
promotes hepatocellular carcinoma progression by repressing FOXO3a.
J Hematol Oncol. 11:122018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Maass T, Sfakianakis I, Staib F, Krupp M,
Galle PR and Teufel A: Microarray-based gene expression analysis of
hepatocellular carcinoma. Curr Genomics. 11:261–268. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fu Q, Yang F, Zhao J, Yang X, Xiang T,
Huai G, Zhang J, Wei L, Deng S and Yang H: Bioinformatical
identification of key pathways and genes in human hepatocellular
carcinoma after CSN5 depletion. Cell Signal. 49:79–86. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Davis S and Meltzer PS: GEOquery: A bridge
between the gene expression omnibus (GEO) and BioConductor.
Bioinformatics. 23:1846–1847. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lim HY, Sohn I, Deng S, Lee J, Jung SH,
Mao M, Xu J, Wang K, Shi S, Joh JW, et al: Prediction of
disease-free survival in hepatocellular carcinoma by gene
expression profiling. Ann Surg Oncol. 20:3747–3753. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45(W1):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44572009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Maere S, Heymans K and Kuiper M: BiNGO: A
Cytoscape plugin to assess overrepresentation of gene ontology
categories in biological networks. Bioinformatics. 21:3448–3449.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Montojo J, Zuberi K, Rodriguez H, Kazi F,
Wright G, Donaldson SL, Morris Q and Bader GD: GeneMANIA Cytoscape
plugin: Fast gene function predictions on the desktop.
Bioinformatics. 26:2927–2928. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lu X, Sun W, Tang Y, Zhu L, Li Y, Ou C,
Yang C, Su J, Luo C, Hu Y and Cao J: Identification of key genes in
hepatocellular carcinoma and validation of the candidate gene,
cdc25a, using gene set enrichment analysis, meta-analysis and
cross-species comparison. Mol Med Rep. 13:1172–1178. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
MacDonald JW and Ghosh D: COPA-cancer
outlier profile analysis. Bioinformatics. 22:2950–2951. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Luo X, Zuo L, Kranzler HR, Wang S, Anton
RF and Gelernter J: Recessive genetic mode of an ADH4 variant in
substance dependence in African-Americans: A model of utility of
the HWD test. Behav Brain Funct. 4:422008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Edenberg HJ, Xuei X, Chen HJ, Tian H,
Wetherill LF, Dick DM, Almasy L, Bierut L, Bucholz KK, Goate A, et
al: Association of alcohol dehydrogenase genes with alcohol
dependence: a comprehensive analysis. Hum Mol Genet. 15:1539–1549.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Edenberg HJ, Jerome RE and Li M:
Polymorphism of the human alcohol dehydrogenase 4 (ADH4) promoter
affects gene expression. Pharmacogenetics. 9:25–30. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Guindalini C, Scivoletto S, Ferreira RGM,
Breen G, Zilberman M, Peluso MA and Zatz M: Association of genetic
variants in alcohol dehydrogenase 4 with alcohol dependence in
Brazilian patients. Am J Psychiatry. 162:1005–1007. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Luo X, Kranzler HR, Zuo L, Wang S and
Gelernter J: Personality traits of agreeableness and extraversion
are associated with ADH4 variation. Biol Psychiatry. 61:599–608.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Goode EL, White KL, Vierkant RA, Phelan
CM, Cunningham JM, Schildkraut JM, Berchuck A, Larson MC, Fridley
BL, Olson JE, et al: Xenobiotic-metabolizing gene polymorphisms and
ovarian cancer risk. Mol Carcinog. 50:397–402. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Oze I, Matsuo K, Suzuki T, Kawase T,
Watanabe M, Hiraki A, Ito H, Hosono S, Ozawa T, Hatooka S, et al:
Impact of multiple alcohol dehydrogenase gene polymorphisms on risk
of upper aerodigestive tract cancers in a Japanese population.
Cancer Epidemiol Biomarkers Prev. 18:3097–3102. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wei RR, Zhang MY, Rao HL, Pu HY, Zhang HZ
and Wang HY: Identification of ADH4 as a novel and potential
prognostic marker in hepatocellular carcinoma. Med Oncol.
29:2737–2743. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang X, Yu T, Liao X, Yang C, Han C, Zhu
G, Huang K, Yu L, Qin W, Su H, et al: The prognostic value of CYP2C
subfamily genes in hepatocellular carcinoma. Cancer Med. 7:966–980.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gelboin HV and Krausz K: Monoclonal
antibodies and multifunctional cytochrome P450: Drug metabolism as
paradigm. J Clin Pharmacol. 46:353–372. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Goldstein JA and de Morais SM:
Biochemistry and molecular biology of the human CYP2C subfamily.
Pharmacogenetics. 4:285–299. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hamman MA, Thompson GA and Hall SD:
Regioselective and stereoselective metabolism of ibuprofen by human
cytochrome P450 2C. Biochem Pharmacol. 54:33–41. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jaakkola T, Laitila J, Neuvonen PJ and
Backman JT: Pioglitazone is metabolised by CYP2C8 and CYP3A4 in
vitro: Potential for interactions with CYP2C8 inhibitors. Basic
Clin Pharmacol Toxicol. 99:44–51. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Unger T: Inhibiting angiotensin receptors
in the brain: Possible therapeutic implications. Curr Med Res Opin.
19:449–451. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kiang TK, Ping CH, Anari MR, Tong V,
Abbott FS and Chang TK: Contribution of CYP2C9, CYP2A6, and CYP2B6
to valproic acid metabolism in hepatic microsomes from individuals
with the CYP2C9*1/*1 genotype. Toxicol Sci. 94:261–271. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tzveova R, Naydenova G, Yaneva T, Dimitrov
G, Vandeva S, Matrozova Y, Pendicheva-Duhlenska D, Popov I,
Beltheva O, Naydenov C, et al: Gender-specific effect of CYP2C8*3
on the risk of essential hypertension in bulgarian patients.
Biochem Genet. 53:319–333. 2005. View Article : Google Scholar
|
|
45
|
Bosó V, Herrero MJ, Santaballa A, Palomar
L, Megias JE, de la Cueva H, Rojas L, Marqués MR, Poveda JL,
Montalar J and Aliño SF: SNPs and taxane toxicity in breast cancer
patients. Pharmacogenomics. 15:1845–1858. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu W, Wang B, Ding HU, Wang DW and Zeng
H: A potential therapeutic effect of CYP2C8 overexpression on
anti-TNF-α activity. Int J Mol Med. 34:725–732. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wei X, Zhang D, Dou X, Niu N, Huang W, Bai
J and Zhang G: Elevated 14,15-epoxyeicosatrienoic acid by
increasing of cytochrome P450 2C8, 2C9 and 2J2 and decreasing of
soluble epoxide hydrolase associated with aggressiveness of human
breast cancer. BMC Cancer. 14:8412014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yu D, Green B, Marrone A, Guo Y, Kadlubar
S, Lin D, Fuscoe J, Pogribny I and Ning B: Suppression of CYP2C9 by
microRNA hsa-miR-128-3p in human liver cells and association with
hepatocellular carcinoma. Sci Rep. 5:85342015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang J, Greene S, Eriksson LC, Reihnér E,
Reihner E, Einarsson C, Eggertsen G and Gåfvels M: Human sterol
12a-hydroxylase (CYP8B1) is mainly expressed in hepatocytes in a
homogenous pattern. Histochem Cell Biol. 123:441–446. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Björkhem I and Eggertsen G: Genes involved
in initial steps of bile acid synthesis. Curr Opin Lipidol.
12:97–103. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Xu Y, Li F, Zalzala M, Xu J, Gonzalez FJ,
Adorini L, Lee YK, Yin L and Zhang Y: Farnesoid X receptor
activation increases reverse cholesterol transport by modulating
bile acid composition and cholesterol absorption in mice.
Hepatology. 64:1072–1085. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Qin J, Han TQ, Yuan WT, Zhang J, Fei J,
Jiang ZY, Niu ZM, Zhang KY, Hua Q, Cai XX, et al: Single nucleotide
polymorphism rs3732860 in the 3’-untranslated region of CYP8B1 gene
is associated with gallstone disease in Han Chinese. J
Gastroenterol Hepatol. 28:717–722. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Qin J, Jiang ZY, Niu ZM, Zhang KY, Hua Q,
Jiang ZH, Wang Y, Huang W, Han TQ and Zhang SD: Association of
single nucleotide polymorphism in human CYP8B1 gene with gallstone
disease. Zhonghua Yi Xue Za Zhi. 91:2092–2095. 2011.(In Chinese).
PubMed/NCBI
|
|
54
|
White DL, Saunders VA, Dang P, Engler J,
Venables A, Zrim S, Zannettino A, Lynch K, Manley PW and Hughes T:
Most CML patients who have a suboptimal response to imatinib have
low OCT-1 activity: Higher doses of imatinib may overcome the
negative impact of low OCT-1 activity. Blood. 110:4064–4072. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
de Lima LT, Vivona D, Bueno CT, Hirata RD,
Hirata MH, Luchessi AD, de Castro FA, de Lourdes F, Chauffaille M,
Zanichelli MA, et al: Reduced ABCG2 and increased SLC22A1 mRNA
expression are associated with imatinib response in chronic myeloid
leukemia. Med Oncol. 31:8512014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Singh O, Chan JY, Lin K, Heng CC and
Chowbay B: SLC22A1- ABCB1 haplotype profiles predict imatinib
pharmacokinetics in Asian patients with chronic myeloid leukemia.
PLoS One. 7:e517712012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Rulcova A, Krausova L, Smutny T, Vrzal R,
Dvorak Z, Jover R and Pavek P: Glucocorticoid receptor regulates
organic cation transporter 1 (OCT1, SLC22A1) expression via HNF4α
upregulation in primary human hepatocytes. Pharmacol Rep.
65:1322–1335. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Herraez E, Lozano E, Macias RI, Vaquero J,
Bujanda L, Banales JM, Marin JJ and Briz O: Expression of SLC22A1
variants may affect the response of hepatocellular carcinoma and
cholangiocarcinoma to sorafenib. Hepatology. 58:1065–1073. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Lautem A, Heise M, Gräsel A,
Hoppe-Lotichius M, Weiler N, Foltys D, Knapstein J, Schattenberg
JM, Schad A, Zimmermann A, et al: Downregulation of organic cation
transporter 1 (SLC22A1) is associated with tumor progression and
reduced patient survival in human cholangiocellular carcinoma. Int
J Oncol. 42:1297–1304. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Mehere P, Han Q, Lemkul JA, Vavricka CJ,
Robinson H, Bevan DR and Li J: Tyrosine aminotransferase:
Biochemical and structural properties and molecular dynamics
simulations. Protein Cell. 1:1023–1032. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Maydan G, Andresen BS, Madsen PP, Zeigler
M, Raas- Rothschild A, Zlotogorski A, Gutman A and Korman SH: TAT
gene mutation analysis in three Palestinian kindreds with
oculocutaneous tyrosinaemia type II; characterization of a silent
exonic transversion that causes complete missplicing by exon 11
skipping. J Inherit Metab Dis. 29:620–626. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Bouyacoub Y, Zribi H, Azzouz H, Nasrallah
F, Abdelaziz RB, Kacem M, Rekaya B, Messaoud O, Romdhane L,
Charfeddine C, et al: Novel and recurrent mutations in the TAT gene
in Tunisian families affected with Richner-Hanhart syndrome. Gene.
529:45–49. 2003. View Article : Google Scholar
|
|
63
|
Fu L, Dong SS, Xie YW, Tai LS, Chen L,
Kong KL, Man K, Xie D, Li Y, Cheng Y, et al: Down-regulation of
tyrosine aminotransferase at a frequently deleted region 16q22
contributes to the pathogenesis of hepatocellular carcinoma.
Hepatology. 51:1624–1634. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Su W, Wang Y, Jia X, Wu W, Li L, Tian X,
Li S, Wang C, Xu H, Cao J, et al: Comparative proteomic study
reveals 17β-HSD13 as a pathogenic protein in nonalcoholic fatty
liver disease. Proc Natl Acad Sci USA. 111:11437–11442. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Fujimoto Y, Itabe H, Sakai J, Makita M,
Noda J, Mori M, Higashi Y, Kojima S and Takano T: Identification of
major proteins in the lipid droplet-enriched fraction isolated from
the human hepatocyte cell line HuH7. Biochim Biophys Acta 1644.
47–59. 2004.
|
|
66
|
Chen J, Zhuo JY, Yang F, Liu ZK, Zhou L,
Xie HY, Xu X and Zheng SS: 17-beta-hydroxysteroid dehydrogenase 13
inhibits the progression and recurrence of hepatocellular
carcinoma. Hepatobiliary Pancreat Dis Int. 17:220–226. 2018.
View Article : Google Scholar : PubMed/NCBI
|