Introduction
Histamine is a biogenic amine that is released
throughout the entire body of an organism via the autocrine and/or
paracrine mechanisms (1–3). The multiple actions of histamine are
mediated by its receptors, including H1, H2, H3 and H4, which
belong to the family of G protein-coupled receptors (4,5).
Histamine receptor H3 (HRH3) is an integral membrane protein that
regulates neurotransmitter release (6). In addition, HRH3 increases the
voltage-dependent calcium currents in smooth muscles, and
innervates the heart and blood vessels in the cardiovascular system
(1). In multiple sclerosis
patients, it was observed that HRH3 was highly expressed in
oligodendroglial cells obtained from demyelination lesions,
indicating the existence of a genetic association between
susceptibility to multiple sclerosis and an exonic single
nucleotide polymorphism in HRH3 (7). Decrease in HRH3 function was linked to
epileptic activity in the temporal neocortex and hippocampus of
patients with pharmacoresistant mesial temporal lobe epilepsy
(8). It has also been reported that
histamine and histamine receptors (HRs) are critical molecules
during carcinogenesis and inflammation (9). Clobenpropit, an HRH3 antagonist, was
observed to significantly lower the multiplicity of colonic
adenocarcinoma, suggesting that HRH3 may be a possible oncoprotein
and potential target in tumor therapies (10). Hepatocellular carcinoma (HCC) is one
of the most common malignant tumors, and its incidence rate ranks
the second among malignant tumors and >200,000 people succumb to
HCC every year in China (11).
However, the functional roles of HRH3 in tumor growth and
metastasis remain largely unknown, particularly in HCC.
The full-length HRH3 protein is composed of 445
amino acids (1). Thus far, at least
20 isoforms of the human HRH3 have been identified based on the
various splicing mRNA forms; however, their regional expression and
function currently remain unclear (1). The stimulation of HRH3 negatively
regulates the synthesis of histamine via inhibiting adenylate
cyclase, which then catalyzes the formation of the second messenger
cyclic adenosine monophosphate, followed by the activation of
histidine decarboxylase (1). Thus,
the activation of HRH3 leads to a decrease in histamine synthesis
in neurons. However, the activation of the HRH3 signaling pathway
has been poorly elaborated in cancer cells, particularly in HCC
cells.
In the present study, the expression and functional
roles of HRH3 in HCC were systematically investigated. The study
results improve the understanding on the pathological roles of HRH3
and provide experimental evidence for the application of HRH3 as a
potential therapeutic target in HCC.
Materials and methods
Reagents and antibodies
The HRH3 antagonist clobenpropit and agonist imetit
were purchased from Abcam (Cambridge, UK). The primary antibodies
used in western blot analysis and their working concentrations were
as follows: HRH3 (dilution 1:1,000; cat. no. ab236952; Abcam) and
β-actin (dilution 1:3,000; cat. no. 20536-1-AP; ProteinTech Group,
Inc., Chicago, IL, USA). In immunohistochemical (IHC) analysis, the
HRH3 antibody was used at a dilution of 1:200. In
immunofluorescence analysis, the HRH3 (dilution 1:200) and F-actin
(dilution 1:300; cat. no. ab205; Abcam) antibodies were used.
Cell culture and tissue
collection
The HLF, JHH-2, Huh-1 and HLE cell lines were from
Japanese Collection of Research Biosources Cell Bank (JCRB), and
were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10%
fetal bovine serum (FBS; HyClone; GE Healthcare Life Sciences,
Logan, UT, USA). SNU739, SNU368, SNU354 and SNU878 cell lines were
from the Korean Cell Line Bank (KCLB), and were routinely cultured
in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (HyClone; GE Healthcare Life
Sciences).
Between January 2009 and January 2012, a total of 96
Han Chinese patients with primary HCC were recruited from Xijing
Hospital of the Fourth Military Medical University (Xi'an, China).
There was no previous history of other cancers for all patients.
All patients received surgery within 2 months after diagnosis and
no patient received anticancer treatment before surgery. All tissue
samples were stored in liquid nitrogen. The present study was
approved by the Ethics Committee of the Fourth Military Medical
University (Xi'an, China) and written informed consent was obtained
from all the patients involved in the study.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissue samples or
cultured cells using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). RNA concentration was measured by
spectrophotometer (One Drop Nanjing, China) and was adjusted to 50
ng/µl with ddH2O. RT was performed using the PrimeScript
RT reagent kit (Takara Bio, Inc., Ostu, Japan) according to the
manufacturer's protocol. Next, qPCR was performed using the SYBR 2X
qPCR Master Mix (Takara Bio, Inc.) in order to determine HRH3
expression, with GAPDH serving as the internal control. The cycling
parameters were as follows: 95°C for 15 sec, 55°C for 15 sec and
72°C for 15 sec for 40 cycles. All primer sequences used in qPCR
are listed in Table I. The relative
expression of target genes was determined using the
2−ΔΔCq method (12).
 | Table I.Sequences of primers used in
polymerase chain reaction. |
Table I.
Sequences of primers used in
polymerase chain reaction.
| Gene | Primer sequence (5′
to 3′) |
|---|
| HRH3 | Forward:
GGTACGAAACCTCCTTCTGGC |
|
| Reverse:
CTCTGGTGGGCCACTCACTTC |
| GAPDH | Forward:
GGAGCGAGATCCCTCCAAAAT |
|
| Reverse:
GGCTGTTGTCATACTTCTCATGG |
Western blot analysis
Cells or tissues were lysed in cold RIPA buffer and
protein concentrations were determined using a BCA protein assay
kit (Beyotime Institute of Biotechnology, Haimen, China). A total
of 50 µG protein was separated by 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis, and then proteins were
transferred to polyvinylidene difluoride (PVDF) membranes, which
was blocked by soaking in 5% non-fat milk for 1 h at room
temperature. The membranes were incubated with specific primary
antibodies against HRH3 and β-actin overnight at 4°C, and then with
horseradish peroxidase-conjugated secondary antibody (dilution
1:10,000; cat. no. SA00001-2; ProteinTech Group, Inc.) at room
temperature for 2 h. Finally, the protein bands were visualized
using an enhanced chemiluminescence detection system (Clinx Science
Instruments Co., Ltd., Shanghai, China). ImageJ software (NIH;
National Institutes of Health, Bethesda, MD, USA) was used to
quantify the protein band intensity.
IHC analysis
HCC tissues were fixed in 4% paraformaldehyde and
embedded in paraffin. IHC analysis was then performed on 4-µm
paraffin-embedded sections using an IHC kit (Invitrogen; Thermo
Fisher Scientific, Inc.). The expression level of target proteins
was blindly evaluated by two pathologists according to the
proportion and intensity of positive cells within five microscopic
visual fields/slide, as previously described (13). The final score between 0–3 was
determined as low expression and a score >3 was determined as
high expression.
Immunofluorescence analysis
HCC cells were washed with PBS and fixed with 4%
paraformaldehyde for 15 min. Cells were then permeabilized with
0.2% Triton X-100 in PBS followed by blocking in blocking buffer
(2% BSA in PBS) for 30 min. Cells were incubated overnight at 4°C
with HRH3 antibody at 1:200 dilution or F-actin antibody at 1:300
dilution followed by immunofluorescence-goat anti-rabbit IgG
H&L (FITC) (dilution 1:100; cat. no. ab6717; Abcam). The
nuclear counterstain is DAPI (dilution 1:5,000; cat. no. C1002;
Beyotime Institute of Biotechnology).
Cell viability assay
SNU-739 and SNU-368 cells were seeded into 96-well
plates at a density of 1,000 cells/well and incubated for 100 µM
imetit, or 100 µM clobenpropit, or dimethyl sulfoxide (DMSO) for 24
h, and the cell viability was measured by an MTS assay (Beyotime
Institute of Biotechnology) according to the manufacturer's
protocol. The microplates were read in a spectrophotometer at 490
nm.
Colony formation assay
SNU-739 and SNU-368 cells (1×103/well)
were seeded into 6-well plates and cultured for two weeks.
Subsequent to staining with 1% crystal violet, the number of
colonies formed was counted.
Wound healing assay
SNU-739 and SNU-368 cell migration was measured by a
wound healing assay. Briefly, ~105 cells were seeded
into a 6-well plate and incubated until they reached 80%
confluence. Next, a scratch wound was made in each well using a
pipette tip, and this time-point was marked as 0 h. Images of the
cells were captured at 0 and 48 h in order to assess the migration
ability.
Transwell migration assay
For Transwell migration assays, Transwell insert
(Becton Dickinson Biosciences, San Jose, USA) were placed into the
wells of 24-well culture plates, SNU-739 and SNU-368 cells
(~104) cultured in medium with 1% FBS was added to the
upper chamber. In the lower chamber of the Transwell insert, medium
with 10% FBS was added. Following incubation for 48 h, cells that
had migrated to the lower surface of the filter were stained with
1% crystal violet, and then the non-migrated cells remaining on the
upper membrane were removed with a cotton wool. Migration was
determined by counting the number of cells in five random
microscopic fields/well.
In vivo assays for tumor growth
A total of 18 BALB/c male nude mice (5 weeks old;
body weight, 18–22 g) were purchased the from Animal Center of the
Fourth Military Medical University (Xi'an, China), and were housed
under a 12-h light/dark cycles at 20–22 with 60% humidity and had
free access to food and water. The mice were randomly divided into
three groups (n=6/group). Mice were subjected to a subcutaneous
injection of 1×107 SNU-368 cells into their back in
order to contract xenografts. After one week, imetit (40
µg/kg/mouse in 100 µl DMSO) or clobenpropit (20 µg/kg/mouse in 100
µl DMSO) or DMSO (100 µl/mouse) was administered by tail vein
injection every three days for one month. The tumor volume
(mm3) was measured every three days, as previously
described (14). After one month of
treatment, the mice were sacrificed by injection of an overdose of
pentobarbital sodium, the tumor nodules were harvested, and images
of tumor tissues were captured. All animal experimental procedures
were approved by the Ethics Committee of the Fourth Military
Medical University.
Statistical analysis
Experiments were repeated three times, where
appropriate. Data are represented as the mean ± standard error of
the mean. SPSS software (version 17.0; SPSS, Inc., Chicago, IL,
USA) was used to conduct statistical analyses, and P<0.05 was
considered to denote a statistically significant difference. Paired
or unpaired t-tests were used for comparisons between two groups,
while one-way analysis of variance (ANOVA) was used for comparisons
among three or more groups. Overall survival (OS) and
recurrence-free survival (RFS) in relation to HFH3 expression were
evaluated by the Kaplan-Meier survival curve and the log-rank
test.
Results
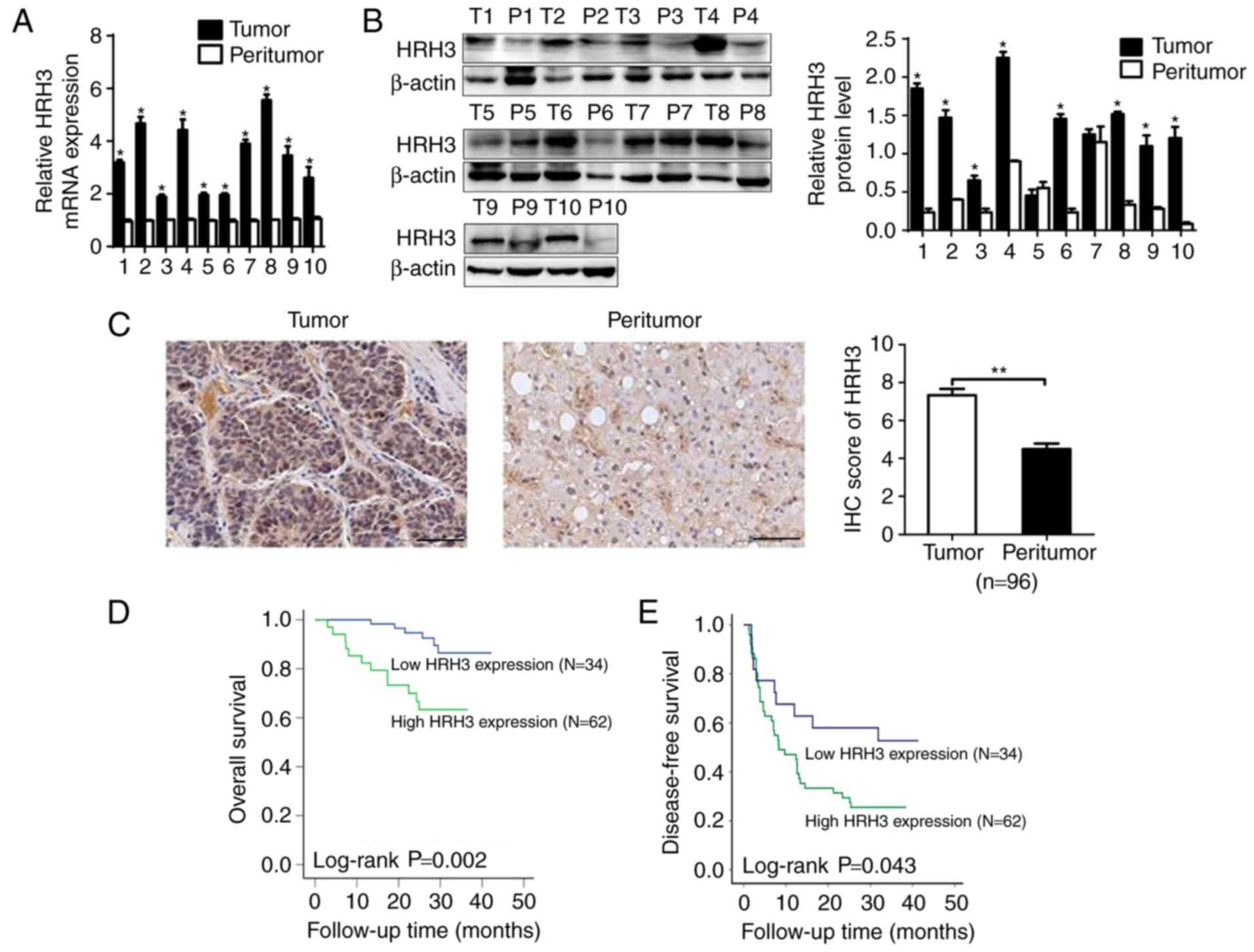
HRH3 is upregulated in HCC tissues and
contributes to tumor progression and worse prognosis
The mRNA and protein expression levels of HRH3 were
first evaluated in 10 paired tumor and peritumor tissues from HCC
patients. RT-qPCR and western blot analysis data demonstrated that
HRH3 was significantly upregulated in HCC tissues when compared
with peritumor tissues (Fig. 1A and
B). The expression of HRH3 was then further evaluated by IHC
analysis in 96 paired tumor and peritumor tissues from HCC
patients. As shown in Fig. 1C, HRH3
was significantly upregulated in HCC tissues when compared with the
paired peritumor tissues.
The prognostic significance of HRH3 was also
assessed in the 96 HCC patients based on the IHC data from the
tumor tissues. Kaplan-Meier survival analysis revealed that HCC
patients with high HRH3 expression had significantly shorter OS and
RFS as compared with those in patients exhibiting low HRH3
expression (Fig. 1D and E). Taken
together, these data indicated that HRH3 contributes to the
progression and poor prognosis of HCC.
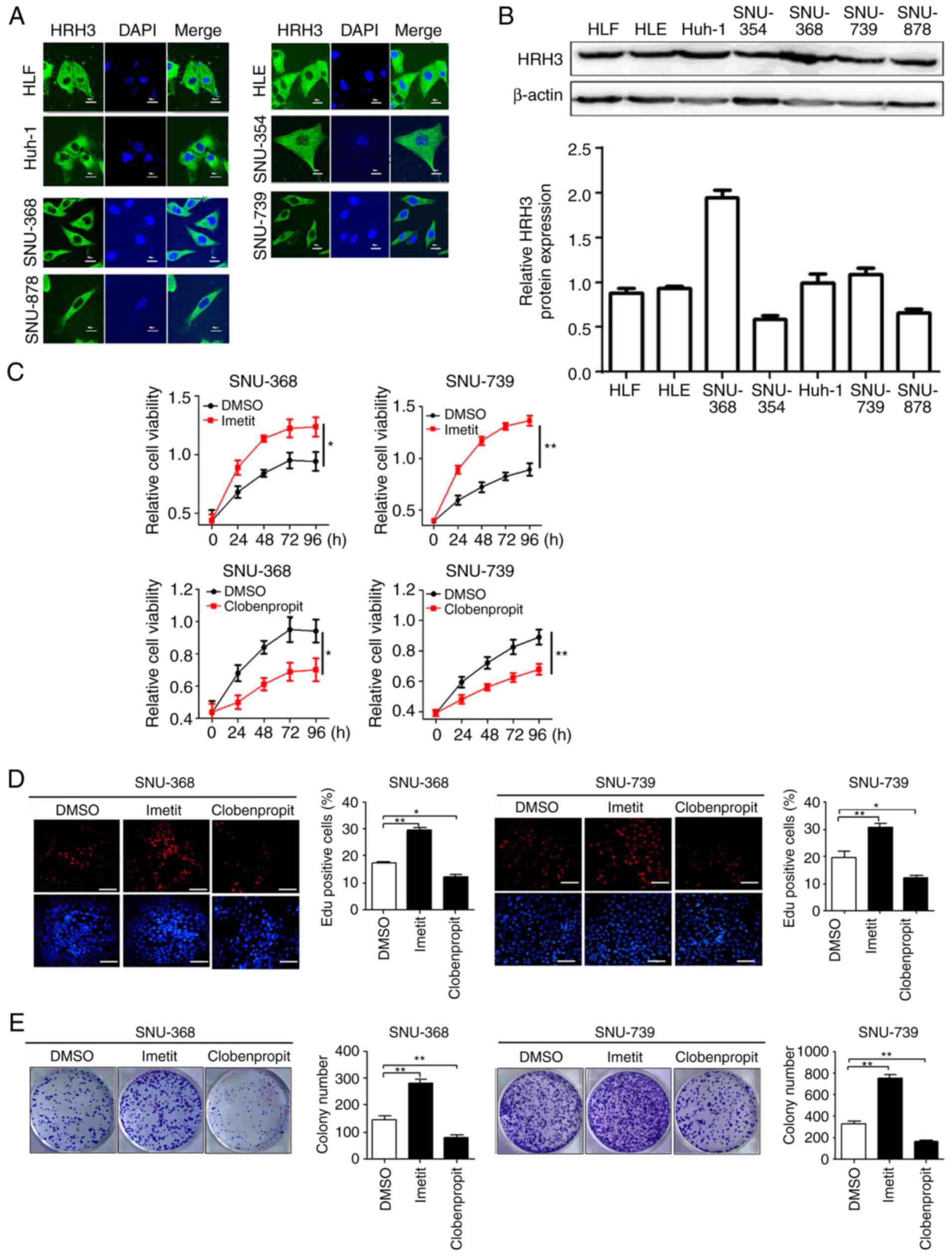
HRH3 promotes HCC cell growth in
vitro
To explore the potential function of HRH3 in cell
growth, a series of in vitro biological experiments were
performed using an agonist (imetit) or antagonist (clobenpropit) of
HRH3. Immunofluorescence and western blot data demonstrated that
the expression of HRH3 in SNU-368 and SNU-739 cells was relatively
high among the seven HCC cell lines examined (Fig. 2A and B). Next, the MTS assay
revealed that HCC cells treated with HRH3 agonist (imetit)
exhibited a significantly faster growth compared with the control
(DMSO-treated) cells, whereas HCC cells treated with HRH3
antagonist (clobenpropit) exhibited significantly slower growth
than the control cells (Fig. 2C).
Consistent with this, the number of colonies and
5-ethynyl-2′-deoxyuridine (EdU)-positive cells were significantly
increased in HCC cells after treatment with imetit, whereas these
were significantly decreased in HCC cells following treatment with
clobenpropit, when compared with the respective control group
(Fig. 2D and E).
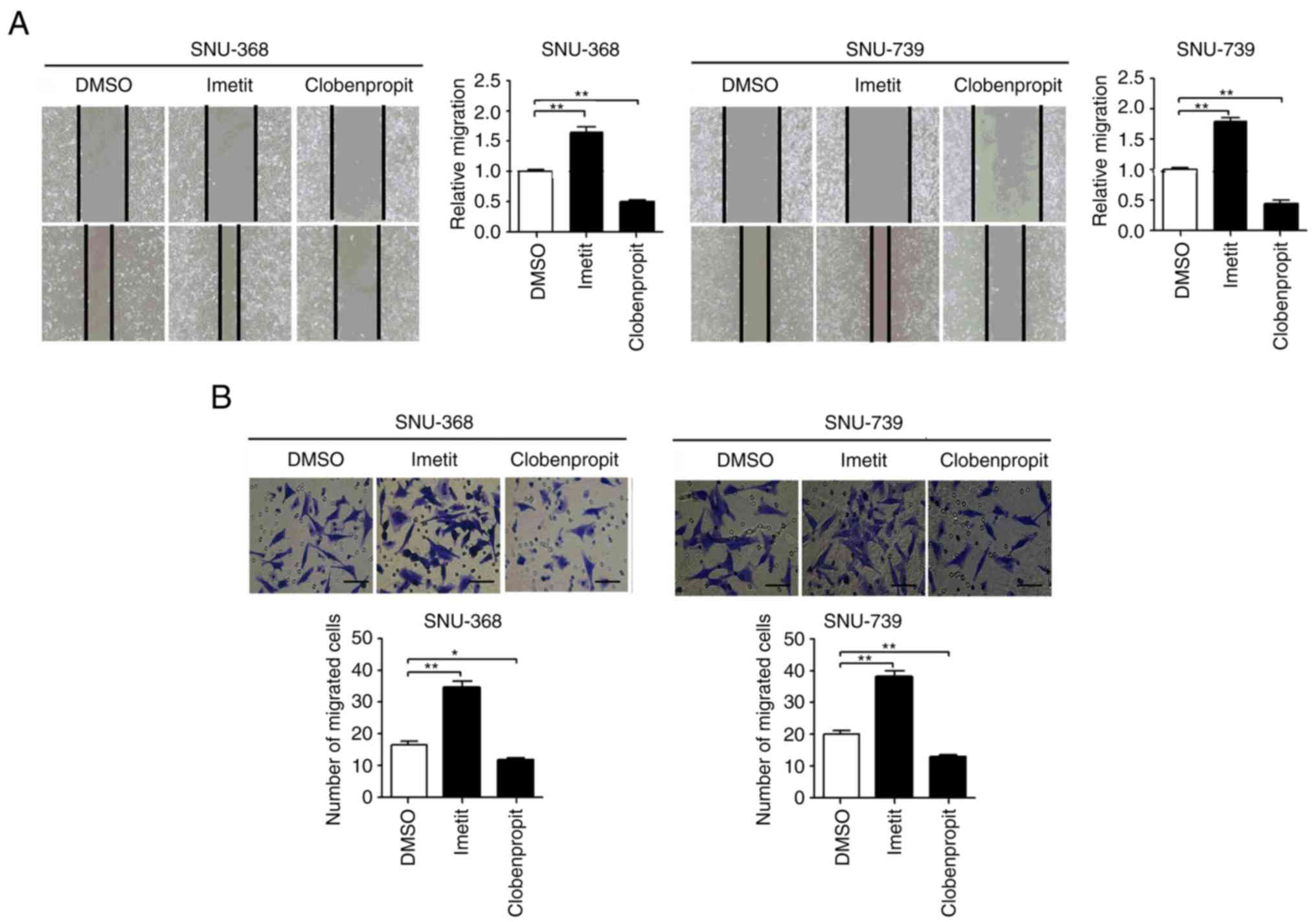
HRH3 promotes the migration of HCC
cells in vitro
The effects of HRH3 on the migration of HCC cells
were also investigated using scratch wound healing and Transwell
assays. The scratch wound healing assay revealed that the migration
ability of HCC cells was markedly increased when HRH3 was activated
by imetit. By contrast, the migration capability of HCC cells was
significantly decreased following treatment with clobenpropit
(Fig. 3A). Similarly, the Transwell
migration assay also indicated that activation of HRH3 by imetit
significantly enhanced the migration ability of HCC cells, while
inhibition of HRH3 by clobenpropit had the opposite effect
(Fig. 3B). These results indicated
that HRH3 increased the migration abilities of HCC cells.
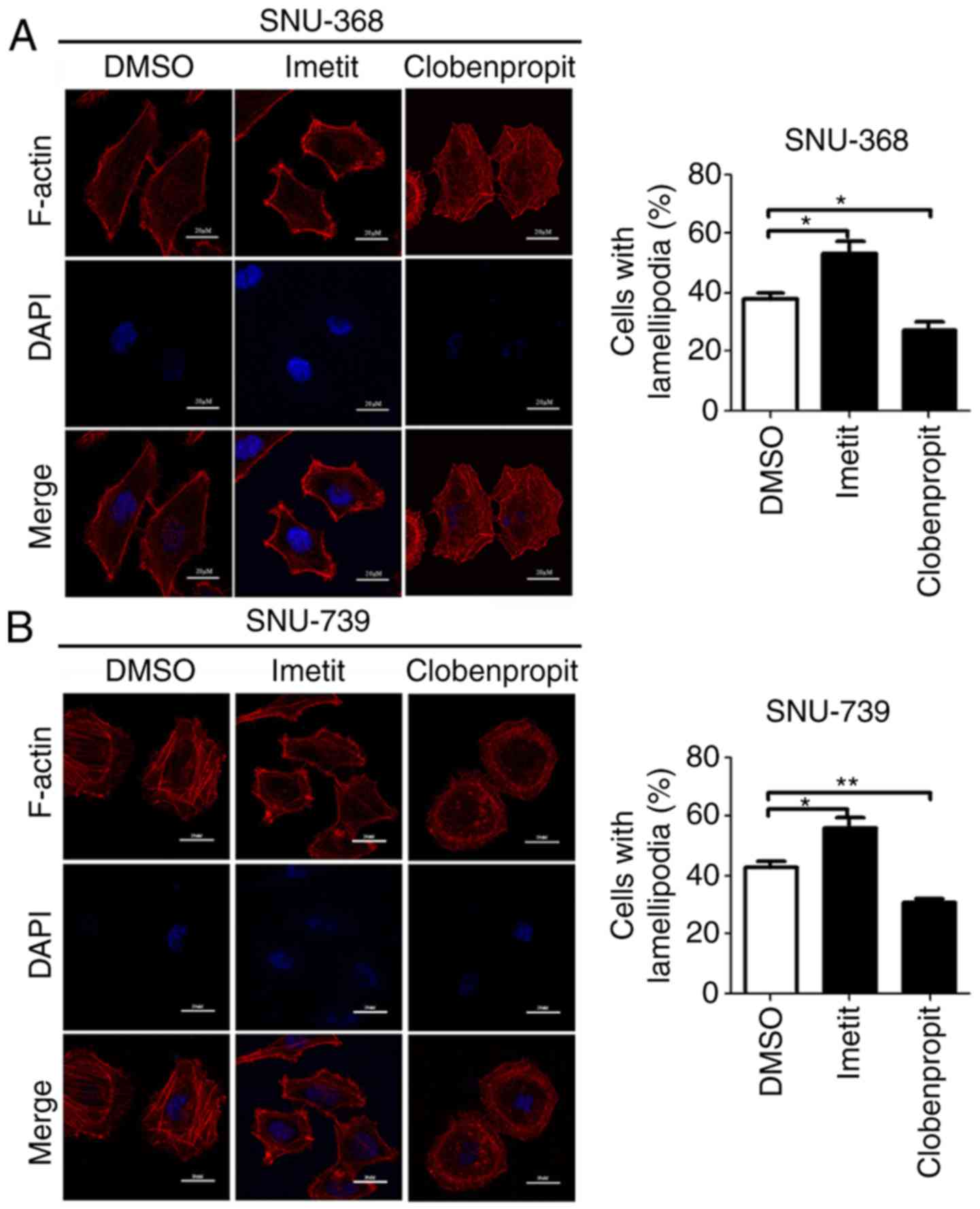
HRH3 promotes the migration of HCC by
inducing lamellipodium formation
Lamellipodia are flattened extensions of the cell
that serve a key role in cell migration. To explore the mechanisms
by which HRH3 promotes HCC cell migration, lamellipodium formation
was evaluated in SNU-368 and SNU-739 cells following treatment with
imetit or clobenpropit. F-actin staining results demonstrated that
stimulation of HRH3 through binding of the agonist imetit markedly
increased lamellipodium formation in HCC cells. By contrast,
inhibition of HRH3 through binding of the antagonist clobenpropit
significantly decreased lamellipodium formation in HCC (Fig. 4). These results suggested that HRH3
promoted the migration of HCC cells mainly through regulating actin
rearrangement.
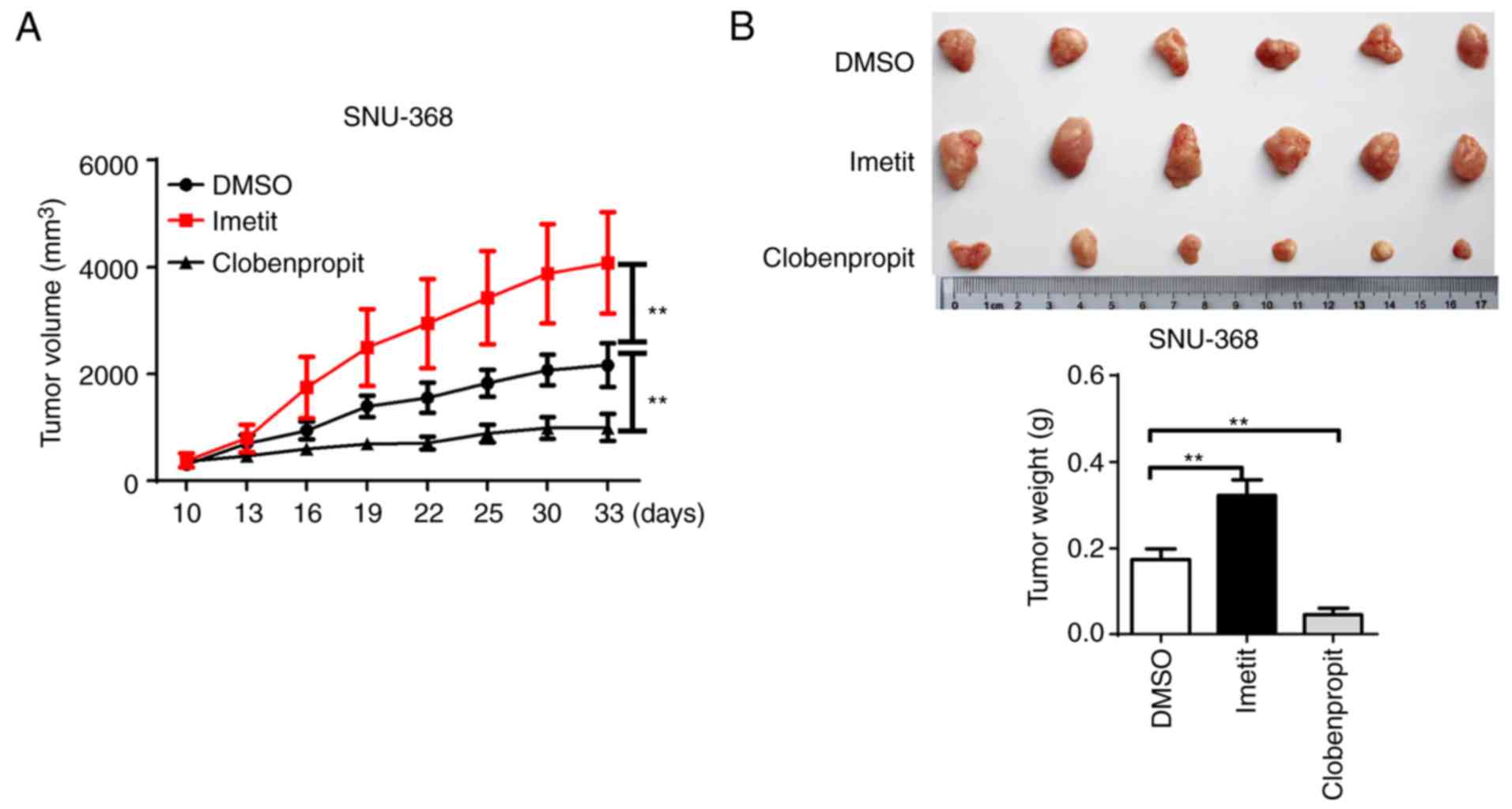
HRH3 promotes HCC cell growth in
vivo
To further investigate the tumorigenic effect of
HRH3 in vivo, a xenograft model was constructed in nude mice
using the SNU-368 cell line. As shown in Fig. 5A and B, a significant increase in
the growth capacity (tumor volume and weight) was observed in
xenograft tumors treated with imetit when compared with the control
xenograft tumors. By contrast, clobenpropit treatment significantly
inhibited the growth of xenograft tumors.
Discussion
Recently, high expression levels of HRs have been
reported in different cancer types, including gastrointestinal
cancer, melanoma, pancreatic and breast cancer (6,9). The
results in the present study demonstrated that HRH3 was
significantly upregulated in HCC tissues and contributed to tumor
progression, suggesting that increased HRH3 level may serve as a
potential prognostic marker for HCC patients. Consistent with these
results, a previous study in breast cancer reported that the
expression of HRH3 was significantly higher in carcinoma tissues
compared with that in benign lesions and non-tumor breast tissues
(15). Similarly, HRH3 expression
was significantly higher in adrenocortical cancer samples in
comparison with that in normal adrenal cortex and benign
adrenocortical adenomas (16).
Another study reported that HRH3 was upregulated in glioblastoma
and glioma cell lines when compared with normal brain tissue and
astrocytes, while overexpression of HRH3 was associated with glioma
progression (17). However, He
et al (18) reported that
gene polymorphisms of HRH3 are irrelevant in breast cancer. Taken
together, all these results indicate that HRH3 is upregulated in
malignant carcinomas, suggesting that it may serve as a novel
therapeutic target in a number of malignancies.
Previous functional studies have reported that HRH3
induces dual effects on tumor proliferation, and promotes cell
proliferation and tumor growth in several types of cancer. For
instance, Lin et al (17)
have indicated that inhibition of HRH3 suppresses glioblastoma cell
growth. Furthermore, it has been demonstrated that histamine
increased MDA-MB-231 cell proliferation via HRH3, suggesting that
HRH3 is involved in the regulation of breast cancer growth
(19). In addition, the
proliferation of breast cancer cells was markedly suppressed upon
treatment with an HRH3 antagonist, such as OUP-186 and clobenpropit
(19). A study by Cricco et
al (20) indicated that HRH3
was involved in pancreatic carcinoma cell growth, and that the
proliferation was increased through activating HRH3. By contrast,
the stimulation of HRH3 through binding of an agonist inhibited the
growth of cholangiocarcinoma in vitro and in vivo
(21), while it also inhibited the
biliary growth of BDL rats (22).
In addition, the results of Davenas et al (23) demonstrated that HRH3 antagonists
increased McA-RH7777 hepatoma cell proliferation, suggesting that
HRH3 may negatively regulate McA-RH7777 cell growth. Consistent
with previous studies (17,19,20),
the results reported in the present study indicated that
stimulation of HRH3 by imetit significantly increased the
proliferation ability of HCC cells, whereas inhibition of HRH3 by
clobenpropit markedly suppressed HCC cell proliferation, which
indicated that overexpression of HRH3 promoted HCC progression. The
dual roles of HRH3 in cancer cell growth may be cell-type specific,
however, this warrants more comprehensive investigation.
Previous experiments have proven that HRH3 serves an
important role in promoting tumor invasion and metastasis. For
example, the decrease in cholangiocarcinoma invasion was proven to
be mediated by the activation of HRH3 (21). In addition, the study of Lin et
al (17) demonstrated that
inhibition of HRH3 suppressed glioblastoma tumor invasion.
Furthermore, the histamine-HRH3 axis signaling prompted MDA-MB-231
cell migration, suggesting that HRH3 may be involved in the
regulation of breast cancer progression (19). Consistent with these previous
studies, the present study demonstrated that activation of HRH3
promoted HCC cell migration, while the inhibition of HRH3
suppressed the metastasis of HCC cells, which further supports the
functional roles of HRH3 in promoting the metastasis of malignant
carcinomas.
Thus far, the mechanisms by which HRH3 promotes
malignant carcinoma growth and metastasis remain undefined. Francis
et al (21) demonstrated
that HRH3 inhibited cholangiocarcinoma growth by PKCα-dependent
ERK1/2 dephosphorylation. In addition, HRH3 has been proven to
increase proliferation through regulation of the cell cycle in
normal and tumor epithelial tissues (24–26).
The results of Lin et al (17) also indicated that inhibition of HRH3
led to suppressed invasion and epithelial-mesenchymal transition of
glioblastoma by inactivating the PI3K/Akt and MEK/ERK pathways. The
lamellipodium is a dynamic surface extension of the cell, which
serves a pivotal role in cell migration (27). The present study demonstrated that
activation of HRH3 promoted HCC cell migration with increased
formation of lamellipodia, whereas inhibition of HRH3 suppressed
the migration of HCC cells, with decreased formation of
lamellipodia. Furthermore, phosphorylation of paxillin has been
demonstrated to be involved in the progression and metastasis of
different malignancies (28,29). A
previous study from our group has also reported that
phosphorylation of paxillin was involved in the formation of
lamellipodia in HCC cells (30).
Considering that mounting evidences have indicated that PKC and PKA
are commonly triggered by the activation of HRs, it can be
speculated that HRH3 induces the formation of lamellipodia mainly
by activating PKC or PKA, although this requires further
investigation.
In conclusion, the present study revealed that HRH3
was upregulated in HCC tissues and was closely correlated with poor
prognosis in HCC patients. Furthermore, upregulation of HRH3
facilitated HCC cell growth and metastasis. In our future study,
small interference RNA-based experiments will be applied to provide
further support for the findings of the present study. Taken
together, the results of the present study suggest that HRH3 may be
used as a novel therapeutic target in HCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81472298 and
81502085).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
DY performed most experiments and wrote the
manuscript. YW and QZ participated in the cell proliferation and
metastasis analyses. JZhao participated in the in vivo
experiments and immunofluorescence assay. JH performed data
analysis. JL was involved in the conception of the study. JZhu
designed the overall study, analyzed the results and also wrote the
manuscript. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
All human and animal experiments conducted in the
present study were approved by the Ethics Committee of the Fourth
Military Medical University (Xi'an, China). Written informed
consent was obtained from all patients involved in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Singh M and Jadhav HR: Histamine H3
receptor function and ligands: Recent developments. Mini Rev Med
Chem. 13:47–57. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Coruzzi G, Adami M and Pozzoli C: Role of
histamine H4 receptors in the gastrointestinal tract. Front Biosci.
4:226–239. 2012. View
Article : Google Scholar
|
|
3
|
Hegyesi H, Darvas Z, László V, Pós Z,
Pállinger E, Hirschberg K, Kovács P and Falus A: Retinoic acid
enhances histamine content and H1 receptor expression in human
neuroblastoma cell line Paju. Anticancer Res. 24:1657–1663.
2004.PubMed/NCBI
|
|
4
|
Kitakaze M: Clinical evidence of the role
of histamine in heart failure. J Am Coll Cardiol. 67:1553–1555.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
de Esch IJ, Thurmond RL, Jongejan A and
Leurs R: The histamine H 4 receptor as a new therapeutic target for
inflammation. Trends Pharmacol Sci. 26:462–469. 2005.PubMed/NCBI
|
|
6
|
Medina VA and Rivera ES: Histamine
receptors and cancer pharmacology. Brit J Pharmacol. 161:755–767.
2010. View Article : Google Scholar
|
|
7
|
Chen Y, Zhen W, Guo T, Zhao Y, Liu A,
Rubio JP, Krull D, Richardson JC, Lu H and Wang R: Histamine
receptor 3 negatively regulates oligodendrocyte differentiation and
remyelination. PLoS One. 12:e1893802017. View Article : Google Scholar
|
|
8
|
Bhowmik M, Khanam R and Vohora D:
Histamine H3 receptor antagonists in relation to epilepsy and
neurodegeneration: A systemic consideration of recent progress and
perspectives. Brit J Pharmacol. 167:1398–1414. 2012. View Article : Google Scholar
|
|
9
|
Yang XD, Ai W, Asfaha S, Bhagat G,
Friedman RA, Jin G, Park H, Shykind B, Diacovo TG, Falus A and Wang
TC: Histamine deficiency promotes inflammation-associated
carcinogenesis through reduced myeloid maturation and accumulation
of CD11b+Ly6G+ immature myeloid cells. Nat
Med. 17:87–95. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tanaka T, Kochi T, Shirakami Y, Mori T,
Kurata A, Watanabe N, Moriwaki H and Shimizu M: Cimetidine and
clobenpropit attenuate inflammation-associated colorectal
carcinogenesis in male ICR mice. Cancers. 8(pii): E252016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wong MC, Jiang JY, Goggins WB, Liang M,
Fang Y, Fung FD, Leung C, Wang HH, Wong GL, Wong VW, et al:
International incidence and mortality trends of liver cancer: A
global profile. Sci Rep. 7:458462017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang Q, Lei Z, Cao H, Li J, Lyu Y, Guo X,
Zhang J, Ji L, Ren T, An J, et al: Increased mitochondrial fission
promotes autophagy and hepatocellular carcinoma cell survival
through the ROS-modulated coordinated regulation of the NFKB and
TP53 pathways. Autophagy. 12:999–1014. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu J, Jin M, Wang J, Zhang H, Wu Y, Li D,
Ji X, Yang H, Yin C, Ren T, et al: TNFα induces Ca2+
influx to accelerate extrinsic apoptosis in hepatocellular
carcinoma cells. J Exp Clin Canc Res. 37:432018. View Article : Google Scholar
|
|
15
|
Medina V, Croci M, Crescenti E, Mohamad N,
Sanchez-Jiménez F, Massari N, Nuñez M, Cricco G, Martin G, Bergoc
R, et al: The role of histamine in human mammary carcinogenesis: H3
and H4 receptors as potential therapeutic targets for breast cancer
treatment. Cancer Biol Ther. 7:28–35. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Szabó PM, Wiener Z, Tömböl Z, Kovács A,
Pócza P, Horányi J, Kulka J, Riesz P, Tóth M, Patócs A, et al:
Differences in the expression of histamine-related genes and
proteins in normal human adrenal cortex and adrenocortical tumors.
Virchows Arch. 455:133–142. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin JJ, Zhao TZ, Cai WK, Yang YX, Sun C,
Zhang Z, Xu YQ, Chang T and Li ZY: Inhibition of histamine receptor
3 suppresses glioblastoma tumor growth, invasion, and
epithelial-to-mesenchymal transition. Oncotarget. 6:17107–17120.
2015.PubMed/NCBI
|
|
18
|
He GH, Lin JJ, Cai WK, Xu WM, Yu ZP, Yin
SJ, Zhao CH and Xu GL: Associations of polymorphisms in histidine
decarboxylase, histamine N-methyltransferase and histamine receptor
H3 genes with breast cancer. PLoS One. 9:e977282014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tanaka S, Sakaguchi M, Yoneyama H, Usami Y
and Harusawa S: Histamine H3 receptor antagonist OUP-186
attenuates the proliferation of cultured human breast cancer cell
lines. Biochem Biophys Res Commun. 480:479–485. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cricco GP, Mohamad NA, Sambuco LA, Genre
F, Croci M, Gutiérrez AS, Medina VA, Bergoc RM, Rivera ES and
Martín GA: Histamine regulates pancreatic carcinoma cell growth
through H3 and H4 receptors. Inflamm Res. 57
(Suppl 1):S23–S24. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Francis H, Onori P, Gaudio E, Franchitto
A, DeMorrow S, Venter J, Kopriva S, Carpino G, Mancinelli R, White
M, et al: H3 histamine receptor-mediated activation of protein
kinase Calpha inhibits the growth of cholangiocarcinoma in vitro
and in vivo. Mol Cancer Res. 7:1704–1713. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Francis H, Franchitto A, Ueno Y, Glaser S,
DeMorrow S, Venter J, Gaudio E, Alvaro D, Fava G, Marzioni M, et
al: H3 histamine receptor agonist inhibits biliary growth of BDL
rats by downregulation of the cAMP-dependent PKA/ERK1/2/ELK-1
pathway. Lab Invest. 87:473–487. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Davenas E, Rouleau A, Morisset S and
Arrang JM: Autoregulation of McA-RH7777 hepatoma cell proliferation
by histamine H3 receptors. J Pharmacol Exp Ther.
326:406–413. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kennedy L, Hodges K, Meng F, Alpini G and
Francis H: Histamine and histamine receptor regulation of
gastrointestinal cancers. Transl Gastrointest Cancer. 1:215–227.
2012.PubMed/NCBI
|
|
25
|
Grandi D, Schunack W and Morini G:
Epithelial cell proliferation is promoted by the histamine
H3 receptor agonist (R)-α-methylhistamine throughout the
rat gastrointestinal tract. Eur J Pharmacol. 538:141–147. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Medina V, Cricco G, Nuñez M, Martín G,
Mohamad N, Correa-Fiz F, Sanchez-Jimenez F, Bergoc R and Rivera ES:
Histamine-mediated signaling processes in human malignant mammary
cells. Cancer Biol Ther. 5:1462–1471. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Machesky LM: Lamellipodia and filopodia in
metastasis and invasion. Febs Lett. 582:2102–2111. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Brown MC, Perrotta JA and Turner CE:
Serine and threonine phosphorylation of the paxillin LIM domains
regulates paxillin focal adhesion localization and cell adhesion to
fibronectin. Mol Biol Cell. 9:1803–1816. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang C, Jacobson K and Schaller MD: A
role for JNK-paxillin signaling in cell migration. Cell Cycle.
3:4–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ren T, Zhang H, Wang J, Zhu J, Jin M, Wu
Y, Guo X, Ji L, Huang Q, Zhang H, et al: MCU-dependent
mitochondrial Ca2+ inhibits NAD+/SIRT3/SOD2
pathway to promote ROS production and metastasis of HCC cells.
Oncogene. 36:5897–5909. 2017. View Article : Google Scholar : PubMed/NCBI
|